Abstract
Background
LncRNA PRNCR1 has been reported to be involved in LPS-induced inflammation, which contributes to osteoarthritis (OA). We predicted that miR-377-3p could bind to PRNCR1.MiR-377-3p can suppress OA development. We therefore analyzed the potential interaction between them in OA.
Methods
Expression of miR-377-3p and PRNCR1 in both OA (n = 40) and control (n = 40) samples were analyzed by RT-qPCR. MiR-377-3p or PRNCR1 were overexpressed in synoviocytes to explore their potential interaction. The subcellular location of PRNCR1 was analyzed by nuclear fractionation assay. The direct interaction between miR-377-3p and PRNCR1 was analyzed by RNA-pull down assay. The proliferation and apoptosis of synoviocytes were analyzed by BrdU and apoptosis assay, respectively.
Results
PRNCR1 was overexpressed in OA, while miR-377-3p was downexpressed in OA. PRNCR1 was detected in the cytoplasm and directly interacted with miR-377-3p. Interestingly, overexpression of PRNCR1 and miR-377-3p showed no regulatory role in each other’s expression. LPS treatment increased PRNCR1 expression and decreased miR-377-3p expression. PRNCR1 overexpression decreased LPS-induced synoviocyte proliferation and increased LPS-induced synoviocyte apoptosis. MiR-377-3p played opposite roles in cell proliferation and apoptosis. Moreover, PRNCR1 suppressed the role of miR-377-3p.
Conclusions
Therefore, PRNCR1 is was detected in cytoplasm and regulates synoviocyte proliferation and apoptosis in OA by sponging miR-377-3p.
Similar content being viewed by others
Introduction
Osteoarthritis (OA), the most common type of chronic joint conditions, is the degeneration of joint tissues, including synovium, articular cartilage, and subchondral bone [1, 2]. Almost all populations are experiencing a high incidence of OA. It is estimated that more than 10% of people over 60 years are suffering from OA [3]. With the obesity epidemic and the growth of the aging population, the incidence of OA is estimated to continuously increase for the long term [4]. Arthritis is usually treated with joint replacement surgery and anti-inflammatory drugs, although anti-inflammatory drugs can have some side effects [5]. The development of OA requires a series of molecular and cellular processes [6, 7]. A considerable molecular factors have shown critical roles in OA [8]. Increased understating of the molecular alterations involved in OA will facilitate the development of novel targeted therapies by affecting gene expression to improve OA [9]. At present, the major challenge of the development of OA-targeted therapy is the lack of safe and effective targets [10]. Alterations in the expression of lncRNAs, which lack the information of protein-coding but affect protein accumulation to regulate cellular processes, are frequently observed in OA patients [11]. Therefore, certain lncRNAs with critical roles in OA may be targeted to treat OA. PRNCR1 has been reported to promote LPS-induced inflammation [12], which contributes to osteoarthritis (OA) [13]. Besides that lncRNA PRNCR1 was reported to contribute to osteolysis via regulating CXCR4 expression [14] and osteogenic differentiation in osteolysis [15]. We hypothesized that PRNCR1 could interact with miR-377-3p, which suppresses OA development via alleviating chondrocyte apoptosis and cartilage degradation [16], and analyzed the potential interaction between PRNCR1 and miR-377-3p and their roles in OA.
Materials and methods
Articular cartilage tissues
This study included 40 OA articular cartilage tissues and 40 normal articular cartilage tissues from 40 OA patients and patients with femoral neck fracture (OA or rheumatic arthritis was not diagnosed), respectively. All patients were enrolled at the Affiliated Hospital of Southwest Medical University from January 2019 and January 2021. All OA patients were diagnosed with radiography following the criteria of the American College of Rheumatology. All participants signed informed consent. The clinical data of both groups are listed in Additional file 1: Table S1. The study was approved by the Ethical Committee of the School of Pharmacy, Southwest Medical University (No. SP536, 201901).
Synoviocytes
In vitro experiments were conducted using OA synoviocytes (type B primary cells, Cat. 408OA-05A, Sigma-Aldrich), which were derived from an OA patient. OA synoviocytes were cultured following the instructions from Sigma-Aldrich. LPS treatment was performed by incubating synoviocytes with 0, 3, 9, 12, and 15 μg/ml LPS (Sigma-Aldrich) for 48 h [17].
Electroporation transfection
PRNCR1 expression vector was constructed using pcDNA3.1 vector as the backbone by GeneCopoeia (Guangzhou, China). NC mimic and miR-377-3p mimic were synthesized by Beyotime Biotechnology (Shanghai, China). OA synoviocytes were transfected with vectors used in this study using Lipofectamine 2000 (Life Technologies) [18].
RNA sample preparations
RNAs were isolated using Direct-zol (ZYMO RESEARCH) and treated with DNase I to remove genomic DNAs. RNA integrity was analyzed using Agilent 2100 Bioanalyzer (Agilent Technologies, USA). RNA preparations were repeated if the quality was low [19].
RT-qPCRs
cDNAs were prepared from 2 μg total RNAs through reverse transcriptions. PRNCR1 and miR-377-3p expression was quantified using qPCRs with 18S rRNA as the endogenous control. The method used for Ct value analysis was 2−ΔΔCt [20].
RNA interaction analysis
The binding of miR-377-3p to PRNCR1 was predicted using the online program IntaRNA 2.0 (default parameters). To confirm the interaction, a pull-down assay was carried out with biotin-ligated miR-377-3p (Bio- miR-377-3p) or negative control (NC) miRNA (Bio-NC). Bio-miR-377-3p and Bio-NC were transfected into OA synoviocytes, and cells were lysed at 48 h post-transfection. RNA pull-down was performed with streptavidin agarose magnetic beads (Life Technologie), and PRNCR1 expression in both samples was quantified after isolations of total RNA [21].
Nuclear fractionation assay
OA synoviocytes were subjected to the preparations of both nuclear (N) and cytoplasm (C) samples. Fractions of both N and C samples were separated by centrifugation at 2500 g at 4 °C for 15 min. After that, total RNA was extracted from both samples, and RT-PCRs were performed to analyze PRNCR1 expression in both fractions. EB-stained gels were observed with MyECL image (Bio-Rad) and photographed [22].
BrdU assay
BrdU incorporation (cell proliferation) assay was performed according to the manufacture’s protocol. Cells were seeded onto a 96-well plate with 3000 cells per well. After the addition of BrdU (10 µM), cells were cultured for another 24 h. After fixation and anti-BrdU-antibody incubation, peroxidase substrate incubation was performed for 1 h, and optical density (OD) at 450 nm was measured [23].
Cell apoptosis assay
OA synoviocytes were seeded onto a 96-well plate with 5000 cells per well at 48 h post-transfections. After that, cells were treated with 15 μg/ml LPS was for 24 h. After ice-cold PBS washing, cells were resuspended in annexin binding buffer (0.5 ml) and stained with Annexin V FITC and PI (Beyotime) for 15 min in the dark. Cell apoptosis was then analyzed using flow cytometry [24].
Statistical analysis
Differences between two independent groups and among multiple independent groups were analyzed by unpaired t test and ANOVA Tukey’s test, respectively. p < 0.05 was considered statistically significant.
Results
PRNCR1 and miR-377-3p accumulation in OA and their correlations
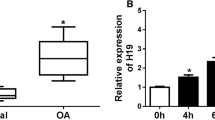
PRNCR1 and miR-377-3p levels in both 40 OA and normal articular cartilage tissues were analyzed using RT-qPCR. PRNCR1 was overexpressed in OA (Fig. 1A, p < 0.01), while miR-377-3p was downexpressed in OA (Fig. 1B, p < 0.01). Correlation analysis using Pearson’s correlation coefficient showed that PRNCR1 and miR-377-3p were not closely correlated with each other across OA (Fig. 1C) and normal (Fig. 1D) samples.
PRNCR1 and miR-377-3p expression in OA and their correlations. PRNCR1 (A) and miR-377-3p (B) expression in both 40 OA and normal articular cartilage tissues were analyzed by RT-qPCR. The correlations between PRNCR1 and miR-377-3p across OA (C) and normal (D) samples were analyzed using Pearson’s correlation coefficient. **p < 0.01
Subcellular location of PRNCR1 in OA synoviocytes and its interaction with miR-377-3p
Although PRNCR1 was detected in both nuclear (N) and cytoplasm (C) samples. In contrast, GAPDH was detected only in cytoplasm because it is a cytoplasmic marker (Fig. 2A). IntaRNA 2.0 prediction illustrated that PRNCR1 and miR-377-3p could form multiple base pairs (Fig. 2B). To further confirm the interaction between PRNCR1 and miR-377-3p, RNA pulldown was performed using biotin-ligated miR-377-3p (Bio-miR-377-3p) or negative control (NC) miRNA (Bio-NC). PRNCR1 level was significantly higher in Bio-miR-377-3p group than in Bio-NC group (Fig. 2C, p < 0.01).
Subcellular location of PRNCR1 in OA synoviocytes and its interaction with miR-377-3p. The subcellular location of PRNCR1 in the nuclear (N) and cytoplasm (C) fractions from OA synoviocytes was analyzed using nuclear fractionation assay (A). The binding of miR-377-3p to PRNCR1 was predicted using IntaRNA 2.0 (B) and confirmed using RNA pull-down assays using biotin-ligated miR-377-3p (Bio-miR-377-3p) or negative control (NC) miRNA (Bio-NC) (C). **p < 0.01
The regulatory role of PRNCR1 and miR-377-3p in each other’s expression
PRNCR1 or miR-377-3p was overexpressed in OA synoviocytes (Fig. 3A, B, both p < 0.05). RT-qPCR analysis illustrated that PRNCR1 showed no role in regulating miR-377-3p expression (Fig. 3C). Moreover, miR-377-3p overexpression also failed to significantly affect PRNCR1 expression (Fig. 3D).
Regulatory role of PRNCR1 and miR-377-3p in each other’s expression. PRNCR1 was overexpressed in OA synoviocytes, and PRNCR1 overexpression was confirmed by RT-qPCR every 24 h until 72 h (A). MiR-377-3p was overexpressed in OA synoviocytes, and miR-377-3p overexpression was confirmed by RT-qPCR every 24 h until 72 h (B). The role of PRNCR1 in regulating miR-377-3p expression (C) and the role of miR-377-3p in regulating PRNCR1 expression (D) were analyzed using RT-qPCR. *p < 0.05
The role of PRNCR1 and miR-377-3p in the proliferation and apoptosis of OA synoviocytes
Synoviocytes were treated with 0, 3, 9, 12, and 15 μg/ml LPS (Sigma-Aldrich) for 48 h, and the levels of PRNCR1 and miR-377-3p were measured using RT-qPCR after RNA isolation. The results showed that LPS increased PRNCR1 expression and decreased miR-377-3p expression (Fig. 4A, B, both p < 0.05). Cell proliferation and apoptosis analysis showed that PRNCR1 overexpression decreased synoviocyte proliferation (Fig. 4C, p < 0.05), increased synoviocyte apoptosis induced by LPS (Fig. 4D, p < 0.05), and promoted IL-1β expression (Fig. 4E, p < 0.05). By contrast, MiR-377-3p overexpression exerted opposite effects on cell proliferation and apoptosis (Fig. 4C–E, p < 0.05). Moreover, PRNCR1 suppressed the role of miR-377-3p in synoviocyte proliferation and apoptosis.
Role of PRNCR1 and miR-377-3p in the proliferation and apoptosis of OA synoviocytes. OA synoviocytes were treated with 0, 3, 9, 12, and 15 μg/ml LPS (Sigma-Aldrich) for 48 h, and PRNCR1 expression (A) and miR-377-3p expression (B) was analyzed using RT-qPCR after RNA isolation. The role of PRNCR1 and miR-377-3p in regulating the proliferation (C) and apoptosis of synoviocytes induced by LPS (D) were analyzed using cell proliferation and apoptosis analyses. IL-1β expression level was analyzed using qRT-PCR (E). *p < 0.05
Discussion
The study analyzed the involvement of PRNCR1 and miR-377-3p in OA and their interactions in OA synoviocytes and, for the first time, showed the differential expression of PRNCR1 and miR-377-3p in OA and revealed the role of PRNCR1 and miR-377-3p in the proliferation and apoptosis of OA synoviocytes.
Previous studies of PRNCR1 have mainly focused on its role in cancers [25, 26]. A recent study revealed the role of PRNCR1 in increasing pulmonary vascular endothelial cell injury induced by LPS via interacting with miR-330-5p/TLR4 axis [12]. It has been well-established that LPS-induced inflammation is a critical contributor to the development of OA. Therefore, we speculated on the involvement of PRNCR1 in OA. The present study showed PRNCR1 was overexpressed in OA. During the development of OA, synoviocytes produce IL-1β, a major pathogenic cytokine in OA that can promote inflammatory responses [27]. The study showed that LPS treatment enhanced PRNCR1 expression, and PRNCR1 overexpression suppressed OA synoviocyte proliferation and increased OA synoviocyte apoptosis and IL-1β expression level in OA induced by LPS. Therefore, PRNCR1 overexpression may promote OA progression by promoting inflammatory responses. Our data and the previous study [12] suggested that PRNCR1 may play opposite roles in LPS-induced cell apoptosis in different cell types.
Interestingly, Tu et al. showed that miR-377-3p could suppress chondrocyte apoptosis induced by IL-1β in OA [16]. This study showed that miR-377-3p increased OA synoviocyte proliferation and decreased OA synoviocyte apoptosis induced by LPS, suggesting its pro-inflammatory roles in OA. Therefore, depending on different cell types, miRNA-377-3p may play different roles in different cells involved in OA.
It has been well-established that mature miRNAs are located in the cytoplasm [28]. The study revealed that PRNCR1 was detected in cytoplasm and could directly interact with mature miR-377-3p. However, PRNCR1 and miR-377-3p showed no significant effects on the expression of each other. Interestingly, PRNCR1 suppressed the role of miR-337-3p in cell proliferation, apoptosis, and IL-1β expression. It has been well-established that the role of miRNA sponges, or competing RNAs is to suppress the role of miRNAs but may not affect their expression [29, 30]. Therefore, PRNCR1 may sponge miR-377-3p to suppress its role in OA. Our results should be further confirmed in OA animal models in the future. The function of miRNAs in OA has been extensively analyzed in previous studies and some miRNAs have been characterized as potential targets to treat OA [31,32,33]. Therefore, analyzing the interaction between lncRNAs and miRNAs may provide a novel way to study the role of miRNAs in cancer biology, therefor providing novel insights to the regulation of the role miRNAs.
Conclusion
PRNCR1 was overexpressed in OA, and miR-377-3p was underexpressed in OA. PRNCR1 may sponge miR-377-3p to suppress its role in promoting OA synoviocyte proliferation and inhibiting OA synoviocyte apoptosis. These results suggested that PRNCR1 may be a potential target for the diagnosis and treatment of OA patients.
Availability of data and materials
The data that support the findings of this study are not publicly available due to their containing information that could compromise the privacy of research participants, but are available on reasonable requests from the corresponding author.
Abbreviations
- OA:
-
Osteoarthritis
- LncRNA PRNCR1:
-
Prostate cancer non-coding RNA 1
- LPS:
-
Lipopolysaccharide
- OD value:
-
Optical density value
References
Glyn-Jones S, Palmer AJ, Agricola R, Price AJ, Vincent TL, Weinans H, et al. Osteoarthritis. Lancet. 2015;386(9991):376–87.
Vina ER, Kwoh CK. Epidemiology of osteoarthritis: literature update. Curr Opin Rheumatol. 2018;30(2):160–7.
Prieto-Alhambra D, Judge A, Javaid MK, Cooper C, Diez-Perez A, Arden NK. Incidence and risk factors for clinically diagnosed knee, hip and hand osteoarthritis: influences of age, gender and osteoarthritis affecting other joints. Ann Rheum Dis. 2014;73(9):1659–64.
Rahmati M, Mobasheri A, Mozafari M. Inflammatory mediators in osteoarthritis: a critical review of the state-of-the-art, current prospects, and future challenges. Bone. 2016;85:81–90.
Bay-Jensen AC, Slagboom E, Chen-An P, Alexandersen P, Qvist P, Christiansen C, et al. Role of hormones in cartilage and joint metabolism: understanding an unhealthy metabolic phenotype in osteoarthritis. Menopause. 2013;20(5):578–86.
Kim JR, Yoo JJ, Kim HA. Therapeutics in osteoarthritis based on an understanding of its molecular pathogenesis. Int J Mol Sci. 2018;19(3):674.
Liu-Bryan R, Terkeltaub R. Emerging regulators of the inflammatory process in osteoarthritis. Nat Rev Rheumatol. 2015;11(1):35–44.
Xia B, Di C, Zhang J, Hu S, Jin H, Tong P. Osteoarthritis pathogenesis: a review of molecular mechanisms. Calcif Tissue Int. 2014;95(6):495–505.
Mobasheri A. The future of osteoarthritis therapeutics: targeted pharmacological therapy. Curr Rheumatol Rep. 2013;15(10):364.
Goldring MB, Berenbaum F. Emerging targets in osteoarthritis therapy. Curr Opin Pharmacol. 2015;22:51–63.
Wu Y, Lu X, Shen B, Zeng Y. The therapeutic potential and role of miRNA, lncRNA, and circRNA in osteoarthritis. Curr Gene Ther. 2019;19(4):255–63.
Yu Y, Sun H, Zhu L, Ji L, Liu H. Downregulating lncRNA PRNCR1 ameliorates LPS-induced pulmonary vascular endothelial cell injury by modulating miR-330-5p/TLR4 axis. J Biochem Mol Toxicol. 2021;35(2):e22644.
Mendez ME, Sebastian A, Murugesh DK, Hum NR, McCool JL, Hsia AW, et al. LPS-induced inflammation prior to injury exacerbates the development of post-traumatic osteoarthritis in mice. J Bone Miner Res. 2020;35(11):2229–41.
Gong ZM, Tang ZY, Sun XL. LncRNA PRNCR1 regulates CXCR4 expression to affect osteogenic differentiation and contribute to osteolysis after hip replacement. Gene. 2018;673:251–61.
Gong ZM, Tang ZY, Sun XL. LncRNA PRNCR1 regulates osteogenic differentiation in osteolysis after hip replacement by targeting miR-211-5p. Biosci Rep. 2018;05(42):25–30.
Tu Y, Ma T, Wen T, Yang T, Xue L, Cai M, et al. MicroRNA-377-3p alleviates IL-1β-caused chondrocyte apoptosis and cartilage degradation in osteoarthritis in part by downregulating ITGA6. Biochem Biophys Res Commun. 2020;523(1):46–53.
Qi K, Lin R, Xue C, Liu T, Wang Y, Zhang Y, et al. Long non-coding RNA (LncRNA) CAIF is downregulated in osteoarthritis and inhibits LPS-induced interleukin 6 (IL-6) upregulation by downregulation of MiR-1246. Med Sci Monit Int Med J Exp Clin Res. 2019;25:8019–24.
Zhao W, Geng D, Li S, Chen Z, Sun M. LncRNA HOTAIR influences cell growth, migration, invasion, and apoptosis via the miR-20a-5p/HMGA2 axis in breast cancer. Cancer Med. 2018;7(3):842–55.
Wang L, Luan T, Zhou S, Lin J, Yang Y, Liu W, et al. LncRNA HCP5 promotes triple negative breast cancer progression as a ceRNA to regulate BIRC3 by sponging miR-219a-5p. Cancer Med. 2019;8(9):4389–403.
Wu H, Liu B, Chen Z, Li G, Zhang Z. MSC-induced lncRNA HCP5 drove fatty acid oxidation through miR-3619-5p/AMPK/PGC1alpha/CEBPB axis to promote stemness and chemo-resistance of gastric cancer. Cell Death Dis. 2020;11(4):233.
Zhuang ST, Cai YJ, Liu HP, Qin Y, Wen JF. LncRNA NEAT1/miR-185-5p/IGF2 axis regulates the invasion and migration of colon cancer. Mol Genet Genom Med. 2020;8(4):e1125.
Liu HT, Fang L, Cheng YX, Sun Q. LncRNA PVT1 regulates prostate cancer cell growth by inducing the methylation of miR-146a. Cancer Med. 2016;5(12):3512–9.
Li R, Liu J, Qi J. Knockdown of long non-coding RNA CCAT1 suppresses proliferation and EMT of human cervical cancer cell lines by down-regulating Runx2. Exp Mol Pathol. 2020;113:104380.
Zhang L, Kang W, Lu X, Ma S, Dong L, Zou B. LncRNA CASC11 promoted gastric cancer cell proliferation, migration and invasion in vitro by regulating cell cycle pathway. Cell Cycle. 2018;17(15):1886–900.
Chu H, Chen Y, Yuan Q, Hua Q, Zhang X, Wang M, et al. The HOTAIR, PRNCR1 and POLR2E polymorphisms are associated with cancer risk: a meta-analysis. Oncotarget. 2017;8(26):43271–83.
Yu WL, Yao JJ, Xie ZZ, Huang YJ, Xiao S. LncRNA PRNCR1 rs1456315 and CCAT2 rs6983267 polymorphisms on 8q24 associated with lung cancer. Int J Gen Med. 2021;14:255–66.
Chou CH, Jain V, Gibson J, Attarian DE, Haraden CA, Yohn CB, et al. Synovial cell cross-talk with cartilage plays a major role in the pathogenesis of osteoarthritis. Sci Rep. 2020;10(1):10868.
Murchison EP, Hannon GJ. miRNAs on the move: miRNA biogenesis and the RNAi machinery. Curr Opin Cell Biol. 2004;16(3):223–9.
Wang B, Hua P, Zhang L, et al. LncRNA-IUR up-regulates PTEN by sponging miR-21 to regulate cancer cell proliferation and apoptosis in esophageal squamous cell carcinoma. Esophagus. 2020;17(3):298–304.
Cui K, Zhu G. LncRNA CTBP1-AS2 regulates miR-216a/PTEN to suppress ovarian cancer cell proliferation. J Ovarian Res. 2020;13(1):84.
Giordano L, Porta GD, Peretti GM, et al. Therapeutic potential of microRNA in tendon injuries. Br Med Bull. 2020;133(1):79–94.
Oliviero A, Della Porta G, Peretti GM, et al. MicroRNA in osteoarthritis: physiopathology, diagnosis and therapeutic challenge. Br Med Bull. 2019;130(1):137–47.
Gargano G, Oliviero A, Oliva F, et al. Small interfering RNAs in tendon homeostasis. Br Med Bull. 2021;138(1):58–67.
Acknowledgements
We thank Luzhou People's Government - Southwest Medical University Cooperation project (No. 2020LZXNYDF02) for approving.
Funding
Luzhou People's Government - Southwest Medical University Cooperation project (No. 2020LZXNYDF02).
Author information
Authors and Affiliations
Contributions
G.W. and Y.L. involved in study concepts, literature research, clinical studies, data analysis, experimental studies, and manuscript writing and review; C.L. involved in study design, literature research, experimental studies, and manuscript editing; X.Z. involved in definition of intellectual content, clinical studies, data acquisition, and statistical analysis; L.T. involved in data acquisition, manuscript preparation, and data analysis. All authors have read and approved the submission of the manuscript.
Corresponding author
Ethics declarations
Ethical approval and consent to participate
All patients signed the written informed consent. All procedures were approved by the Ethics Committee of the Affiliated Hospital of Southwest Medical University and completed in keeping with the standards set out in the Announcement of Helsinki and the Laboratory Guidelines of Research in China.
Consent for publication
Not applicable.
Competing interests
All other authors declare that we do not have any commercial or associative interest that represents a conflict of interest in connection with the work submitted.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1.
Clinical data of OA patients and normal participants.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Wang, G., Li, C., Zhang, X. et al. Long non-coding PRNCR1 regulates the proliferation and apoptosis of synoviocytes in osteoarthritis by sponging miR-377-3p. J Orthop Surg Res 17, 238 (2022). https://doi.org/10.1186/s13018-022-03035-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13018-022-03035-2