Abstract
Background
Contralateral breast cancer (CBC) is the most common second primary cancer diagnosed in breast cancer survivors, yet the understanding of the genetic susceptibility of CBC, particularly with respect to common variants, remains incomplete. This study aimed to investigate the genetic basis of CBC to better understand this malignancy.
Findings
We performed a genome-wide association analysis in the Women’s Environmental Cancer and Radiation Epidemiology (WECARE) Study of women with first breast cancer diagnosed at age < 55 years including 1161 with CBC who served as cases and 1668 with unilateral breast cancer (UBC) who served as controls. We observed two loci (rs59657211, 9q32, SLC31A2/FAM225A and rs3815096, 6p22.1, TRIM31) with suggestive genome-wide significant associations (P < 1 × 10–6). We also found an increased risk of CBC associated with a breast cancer-specific polygenic risk score (PRS) comprised of 239 known breast cancer susceptibility single nucleotide polymorphisms (SNPs) (rate ratio per 1-SD change: 1.25; 95% confidence interval 1.14–1.36, P < 0.0001). The protective effect of chemotherapy on CBC risk was statistically significant only among patients with an elevated PRS (Pheterogeneity = 0.04). The AUC that included the PRS and known breast cancer risk factors was significantly elevated.
Conclusions
The present GWAS identified two previously unreported loci with suggestive genome-wide significance. We also confirm that an elevated risk of CBC is associated with a comprehensive breast cancer susceptibility PRS that is independent of known breast cancer risk factors. These findings advance our understanding of genetic risk factors involved in CBC etiology.
Similar content being viewed by others
Introduction
Women with breast cancer have a twofold to sixfold increased risk of developing a new primary cancer in the contralateral breast (CBC) compared with the risk of developing a first primary breast cancer among the general population [1]. Genetic factors play a critical role in CBC development, including germline pathogenic variants in BRCA1/2, TP53, CHEK2, and PALB2 [2,3,4]. Previous studies have investigated individual common variants in high- or moderate-penetrance breast cancer susceptibility genes [5] or drug metabolizing genes [6] and reported associations of breast cancer susceptibility variants identified from genome-wide association studies (GWAS) [7] with CBC risk. Studies have further demonstrated a positive cumulative effect of genetic variants, i.e., the polygenic risk score (PRS), on CBC risk, using a limited number of SNPs [8], in high-risk populations [9] or with limited adjustment for covariates [10]. However, a comprehensive GWAS assessing the associations between common variants and CBC risk has not been reported.
To advance the understanding of the genetic susceptibility of CBC for a large and growing population of breast cancer survivors, we carried out a GWAS in the Women’s Environmental Cancer and Radiation Epidemiology (WECARE) Study and evaluated the association between the updated breast cancer PRS [11] and CBC risk.
Methods
Study participants
The WECARE Study is a multi-center, population-based case–control study of CBC conducted in two phases: the WECARE I Study (2001–2004) and WECARE II Study (2009–2012) [12, 13]. Due to the word limit, we described the study design and participants in details in Additional file 1. The final analytic data set included 2829 participants (1161 cases and 1668 controls) for the main analysis and 2483 (1017 cases and 1466 controls) for the PRS analysis involving non-Hispanic White women only.
CBC GWAS analysis
The genome-wide association analysis was performed in the combined data of the WECARE I and II Studies. Details about genotyping, quality control and imputation could be found in Additional file 1. Conditional logistic regression models with adjustment for the top five principal components (PCs) and age at first breast cancer diagnosis were performed to test additive effects of genetic variants. Genome-wide statistical significance was determined by the threshold of P < 5 × 10–8 with P < 1 × 10–6 considered as suggestive significance. The functional annotation was performed using Functional Mapping and Annotation (FUMA) [14]. We applied the Sum of Single Effects (SuSiE) method to identify credible sets in each identified locus [15]. Stratified analyses were further performed, and heterogeneity was assessed using the likelihood ratio test for nested models.
PRS analysis
We constructed a weighted PRS, consisting of the 313 known breast cancer risk susceptibility SNPs [11]. Genotyping data were available at 239 of the 313 loci and proxies were determined for 18 of the remaining 74 loci. Detailed information could be found in Additional file 1. The association of the PRS with CBC risk was assessed using the continuous PRS, per standard deviation (SD) of the PRS, and PRS categorized by median, and quartile cut points based on UBC controls. Multivariable adjusted rate ratios (RR) and corresponding 95% confidence interval (CI) were estimated by fitting conditional logistic regression models adjusted for age at first breast cancer diagnosis, the top five genetic PCs, and known or suspected CBC risk factors. Area under the curve (AUC) of receiver operating characteristic (ROC) curves for various nested models were compared using the DeLong test [16].
All analyses were performed using R v4.1.3 or SAS v9.4 (The SAS Institute, Cary, NC).
Results
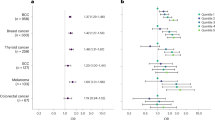
The quantile–quantile (Q–Q) plot is shown in Additional file 2: Fig. S1. The inflation factor of the genome-wide scan was 1.034, indicating that the population structure was not an issue for the current analysis. Two loci associated with an elevated but not statistically significant CBC risk, 9q32 (rs59657211, P = 2.96 × 10–7, SLC31A2/FAM225A) and 6p22.1 (rs3815096, P = 9.58 × 10–7, TRIM31), were identified (Fig. 1a and Additional file 2: Fig. S2). One credible set, consisting of rs10817445, rs12337704, rs59657211, and rs9632905, was identified for 9q32 using SuSiE. However, SuSiE failed to identify any credible set for 6p22.1. There was no heterogeneity in associations of CBC with rs59657211 and rs3815096 by age at first breast cancer diagnosis, first-degree family history of breast cancer, ER and PR status, and chemotherapy or radiotherapy for the first breast cancer (Fig. 1b).
a Manhattan plot of GWAS for contralateral breast cancer risk in the WECARE STUDY; b stratified analyses for the two loci with suggestive genome-wide significant associations (P < 1 × 10–6) in the WECARE Study. CI, confidence interval; ER, estrogen receptor; No., number; PR, progesterone receptor; RR, rate ratio
Among non-Hispanic White women, the weighted PRS without proxies (239 SNPs) was associated with an increased CBC risk of 46% (RR = 1.46, 95% CI 1.25–1.71 per weighted risk allele; RR = 1.25, 95% CI 1.14–1.36 per SD estimate). PRS were evaluated both as below and above the median and by quartiles; the above the median PRS category and the highest PRS quartile were both statistically significantly associated with increased CBC risk (Table 1). ROC curves were generated and AUCs were estimated to compare the discrimination ability of two models in the combined WECARE data (Table 2): CBC risk factors alone and PRS plus risk factors. The AUC of PRS plus risk factors model was 62.4 (95% CI 60.2–64.7), which was significantly higher than the model of the risk factors alone (AUC: 60.73, 95% CI 58.5–63.0, P = 0.01). We repeated the analysis in the WECARE I Study where information regarding BRCA1/2 mutations was known. The AUC of model with PRS, risk factors, and BRCA1/2 mutations was 67.7 (95% CI 64.7–70.7), significantly higher than the model with risk factors alone (AUC: 63.0, 95% CI 60.0–66.1, P < 0.0001) and the model with PRS plus risk factors (AUC: 65.1, 95% CI 62.0–68.1, P = 0.01). The association of PRS with CBC risk was modified by chemotherapy (Pheterogeneity = 0.04) such that the association between the PRS and CBC risk was stronger among women who did not receive chemotherapy for their first primary breast cancer compared to women who had chemotherapy (data not shown in Tables). When focused on the effects of chemotherapy, our data showed a reduced CBC risk among patients with higher PRS (RR = 0.61, 95% CI 0.46–0.81), but no association among patients with lower PRS (RR = 0.90, 95% CI 0.67–1.22) (Table 3). There was no heterogeneity in the association of the PRS with CBC risk by age at first diagnosis, family history of breast cancer, radiation treatment, ER status of first breast cancer, PR status of first breast cancer, hormone replacement therapy at first breast cancer, age at menopause, or parity (Table 3). Similar findings were observed when using the PRS with proxies (257 SNPs) (Table 1).
Discussion
The present study is the largest population-based GWAS analysis of CBC risk to date and identified two loci with suggestive genome-wide significance. rs59657211 at the FAM225A locus has been reported to be involved in the tumorigenesis and metastasis of several types of cancers, including nasopharyngeal, colorectal, and esophageal squamous cell cancer [17, 18]. rs3815096, an intronic variant of TRIM31, is located within the major histocompatibility complex (MHC) region. It has also been reported to be nominally associated with risk of first primary breast cancer (OR 1.02, 95% CI 1.00–1.03, P = 0.007) in a prior GWAS [19], consistent with our results. TRIM31, a member of the TRIM family and acting as an E3 ubiquitin ligase, may play a promoting or suppressing role in malignant processes of multiple cancers [20, 21]. In breast cancer, TRIM31 was found to suppress the cancer progression through the stabilization and activation of p53 [22]. Further investigation into these loci is needed to determine the underpinning mechanisms involved in CBC development.
We further confirmed an elevated risk of CBC is associated with the established breast cancer susceptibility PRS after the adjustment for other risk factors. Our findings corroborate prior studies that found a PRS consisting of the 313 breast cancer susceptibility SNPs associated with CBC risk [9, 10]. Moreover, the AUC that included the PRS and known breast cancer risk factors with or without BRCA1/2 mutations was significantly higher than that of the risk factors alone, suggesting the PRS may add additional predictive values in identifying breast cancer patients with an elevated risk of CBC. Our study also reported novel findings: i.e., chemotherapy was found to be protective among patients with higher PRS but was not among those with lower PRS. This suggests that breast cancer survivors with an unfavorable genetic background may benefit more from chemotherapy; when chemotherapy was not a viable option or the patients declined to receive the treatment, a more intense surveillance strategy might serve better for these patients for an early detection of CBC and treatment.
Our study has several strengths. Most notably, we included the largest number of CBCs reported in a GWAS study with available detailed risk factors, treatment, and clinical information. One primary limitation pertains to the generalizability across racial and ethnic groups as the WECARE Study included predominantly women of European ancestry and we lacked the statistical power to examine subgroups of interest.
In summary, our findings further the understanding of the genetic risk involved in CBC etiology, conferred by common SNPs. In turn, these results will be useful for the development of prevention strategies for CBC as well as for the long-term management of patients with breast cancer.
Availability of data and materials
Access to the WECARE data could be requested by submission of an inquiry to Dr. Jonine L. Bernstein (BernsteJ@mskcc.org) and WECARE Study Collaborative Group.
Abbreviations
- AUC:
-
Area under the curve
- CBC:
-
Contralateral breast cancer
- CI:
-
Confidence interval
- ER:
-
Estrogen receptor
- FUMA:
-
Functional mapping and annotation
- GWAS:
-
Genome-wide association studies
- HWE:
-
Hardy-Weinberg equilibrium
- LD:
-
Linkage disequilibrium
- MAF:
-
Minor allele frequency
- MHC:
-
Major histocompatibility complex
- OR:
-
Odds ratio
- PMRA:
-
Precision medicine diversity array
- PRS:
-
Polygenic risk score
- PR:
-
Progesterone receptor
- QC:
-
Quality control
- Q-Q:
-
Quantile-quantile
- ROC:
-
Receiver operating characteristic
- RR:
-
Rate ratios
- SNPs:
-
Single nucleotide polymorphisms
- SuSiE:
-
Sum of single effects
- TOPMed:
-
Trans-omics for precision medicine
- UBC:
-
Unilateral breast cancer
- WECARE:
-
Women’s Environmental Cancer and Radiation Epidemiology
References
Chen Y, Thompson W, Semenciw R, Mao Y. Epidemiology of contralateral breast cancer. Cancer Epidemiol Biomark Prev. 1999;8(10):855–61.
Malone KE, Begg CB, Haile RW, Borg A, Concannon P, Tellhed L, Xue S, Teraoka S, Bernstein L, Capanu M, et al. Population-based study of the risk of second primary contralateral breast cancer associated with carrying a mutation in BRCA1 or BRCA2. J Clin Oncol. 2010;28(14):2404–10.
Morra A, Mavaddat N, Muranen TA, Ahearn TU, Allen J, Andrulis IL, Auvinen P, Becher H, Behrens S, Blomqvist C, et al. The impact of coding germline variants on contralateral breast cancer risk and survival. Am J Hum Genet. 2023;110:475–86.
Kuchenbaecker KB, Hopper JL, Barnes DR, Phillips KA, Mooij TM, Roos-Blom MJ, Jervis S, van Leeuwen FE, Milne RL, Andrieu N, et al. Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA. 2017;317(23):2402–16.
Yadav S, Boddicker NJ, Na J, Polley EC, Hu C, Hart SN, Gnanaolivu RD, Larson N, Holtegaard S, Huang H, et al. Contralateral breast cancer risk among carriers of germline pathogenic variants in ATM, BRCA1, BRCA2, CHEK2, and PALB2. J Clin Oncol. 2023;41:1703.
Brooks JD, Comen EA, Reiner AS, Orlow I, Leong SF, Liang X, Mellemkjaer L, Knight JA, Lynch CF, John EM, et al. CYP2D6 phenotype, tamoxifen, and risk of contralateral breast cancer in the WECARE Study. Breast Cancer Res. 2018;20(1):149.
Teraoka SN, Bernstein JL, Reiner AS, Haile RW, Bernstein L, Lynch CF, Malone KE, Stovall M, Capanu M, Liang X, et al. Single nucleotide polymorphisms associated with risk for contralateral breast cancer in the Women’s Environment, Cancer, and Radiation Epidemiology (WECARE) Study. Breast Cancer Res. 2011;13(6):R114.
Robson ME, Reiner AS, Brooks JD, Concannon PJ, John EM, Mellemkjaer L, Bernstein L, Malone KE, Knight JA, Lynch CF, et al. Association of common genetic variants with contralateral breast cancer risk in the WECARE study. J Natl Cancer Inst. 2017;109:10.
Lakeman IMM, van den Broek AJ, Vos JAM, Barnes DR, Adlard J, Andrulis IL, Arason A, Arnold N, Arun BK, Balmana J, et al. The predictive ability of the 313 variant-based polygenic risk score for contralateral breast cancer risk prediction in women of European ancestry with a heterozygous BRCA1 or BRCA2 pathogenic variant. Genet Med. 2021;23(9):1726–37.
Kramer I, Hooning MJ, Mavaddat N, Hauptmann M, Keeman R, Steyerberg EW, Giardiello D, Antoniou AC, Pharoah PDP, Canisius S, et al. Breast cancer polygenic risk score and contralateral breast cancer risk. Am J Hum Genet. 2020;107(5):837–48.
Mavaddat N, Michailidou K, Dennis J, Lush M, Fachal L, Lee A, Tyrer JP, Chen TH, Wang Q, Bolla MK, et al. Polygenic risk scores for prediction of breast cancer and breast cancer subtypes. Am J Hum Genet. 2019;104(1):21–34.
Bernstein JL, Langholz B, Haile RW, Bernstein L, Thomas DC, Stovall M, Malone KE, Lynch CF, Olsen JH, Anton-Culver H, et al. Study design: evaluating gene-environment interactions in the etiology of breast cancer—the WECARE study. Breast Cancer Res. 2004;6(3):R199-214.
Langballe R, Mellemkjaer L, Malone KE, Lynch CF, John EM, Knight JA, Bernstein L, Brooks J, Andersson M, Reiner AS, et al. Systemic therapy for breast cancer and risk of subsequent contralateral breast cancer in the WECARE Study. Breast Cancer Res. 2016;18(1):65.
Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8(1):1826.
Wang G, Sarkar A, Carbonetto P, Stephens M. A simple new approach to variable selection in regression, with application to genetic fine mapping. J R Stat Soc Ser B Stat Methodol. 2020;82(5):1273–300.
DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44(3):837–45.
Zhang C, Luo Y, Cao J, Wang X, Miao Z, Shao G. Exosomal lncRNA FAM225A accelerates esophageal squamous cell carcinoma progression and angiogenesis via sponging miR-206 to upregulate NETO2 and FOXP1 expression. Cancer Med. 2020;9(22):8600–11.
Zhang X, Shi H, Yao J, Li Y, Gao B, Zhang Y, Wang C, Zhou H, Zhang L. FAM225A facilitates colorectal cancer progression by sponging miR-613 to regulate NOTCH3. Cancer Med. 2020;9(12):4339–49.
Zhang H, Ahearn TU, Lecarpentier J, Barnes D, Beesley J, Qi G, Jiang X, O’Mara TA, Zhao N, Bolla MK, et al. Genome-wide association study identifies 32 novel breast cancer susceptibility loci from overall and subtype-specific analyses. Nat Genet. 2020;52(6):572–81.
Wang H, Yao L, Gong Y, Zhang B. TRIM31 regulates chronic inflammation via NF-kappaB signal pathway to promote invasion and metastasis in colorectal cancer. Am J Transl Res. 2018;10(4):1247–59.
Li H, Zhang Y, Zhang Y, Bai X, Peng Y, He P. TRIM31 is downregulated in non-small cell lung cancer and serves as a potential tumor suppressor. Tumour Biol. 2014;35(6):5747–52.
Guo Y, Li Q, Zhao G, Zhang J, Yuan H, Feng T, Ou D, Gu R, Li S, Li K, et al. Loss of TRIM31 promotes breast cancer progression through regulating K48- and K63-linked ubiquitination of p53. Cell Death Dis. 2021;12(10):945.
Acknowledgements
We thank the research staff and participants of the WECARE Study.
Funding
The WECARE Study was supported by the U.S. National Cancer Institute (U01CA083178, R01CA097397, R01CA129639, R21CA234752) and MSK Cancer Center Support Grant (P30CA008748); Xiaohui Sun is supported by R00CA230205, R21CA234752 and China Scholarship Council (CSC) (202108330197); Xiang Shu is supported, in part, by R00CA230205.
Author information
Authors and Affiliations
Contributions
LM, KEM, CFL, EMJ, JAK, JLB and PC contributed to conception and design; LM, KEM, CFL, EMJ, JAK, XL, MW and JLB involved in data collection and assembly. XS, ASR, APT, JHO, XS, AD, XL, GPW and JLB involved in data analysis and interpretation; XS, ASR, APT, JHO, XS and JLB involved in manuscript drafting; all authors involved in manuscript revising; all authors gave final approval for the manuscript; JLB and JHO acquired fundings.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was approved by the institutional ethics review boards at the University of Iowa (IRB-01), Fred Hutchinson Cancer Research Center, Cancer Prevention Institute of California, Mount Sinai Hospital, and Memorial Sloan Kettering Cancer Center and by the Committee for the Protection of Human Subjects of the State of California. All study participants provided written informed consent.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing financial or non-financial interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1
. Description of methods for the current study.
Additional file 2
. Supplementary Figures.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Sun, X., Reiner, A.S., Tran, A.P. et al. A genome-wide association study of contralateral breast cancer in the Women’s Environmental Cancer and Radiation Epidemiology Study. Breast Cancer Res 26, 16 (2024). https://doi.org/10.1186/s13058-024-01765-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13058-024-01765-1