Abstract
Breast cancer is common worldwide. Phosphoglycerate mutase 5 (PGAM5) belongs to the phosphoglycerate mutase family and plays an important role in many cancers. However, research on its role in breast cancer remains unclear. The present investigation highlights the significant expression of PGAM5 in breast cancer and its essential role in cell proliferation, invasion, apoptosis and the regulation of ferroptosis in breast cancer cells. Overexpression or knockdown of ubiquitin-specific protease 11 (USP11) promotes or inhibits the growth and metastasis of breast cancer cells, respectively, in vitro and in vivo. Mechanistically, USP11 stabilizes PGAM5 via de-ubiquitination, protecting it from proteasome-mediated degradation. In addition, the USP11/PGAM5 complex promotes breast cancer progression by activating iron death-related proteins, indicating that the synergy between USP11 and PGAM5 may serve as a predictor of disease outcome and provide a new treatment strategy for breast cancer.
Similar content being viewed by others
Background
In 2020, breast cancer emerged as the predominant form of cancer worldwide, surpassing other cancer types, according to a report by the World Health Organization (2021) [1]. To alleviate the global burden of cancer, there is an immediate necessity for the development of more cost-effective therapeutic approaches and strategies. PGAM5, a member of the phosphoglycerate mutase family, exhibits a distinctive role among its counterparts by operating as a serine/threonine protein phosphatase instead of a phosphoglycerate mutase [2]. Its involvement in cell apoptosis and necrosis occurs via mitochondrial damage-induced autophagy [3,4,5]. It has been found that the expression of PGAM5 is upregulated in many cancer types, promoting tumor progression [6,7,8]. Despite its widespread expression in various cancers, the function and regulatory mechanisms of PGAM5 in breast cancer remain unclear. Through bioinformatics analysis, we successfully identified high expression of PGAM5 in breast cancer. Subsequent investigations have shed further light on the crucial role of PGAM5 in various cellular processes, including proliferation, invasion, apoptosis, and ferroptosis, in breast cancer cells.
Ferroptosis, a form of iron-dependent cell death, is characterized by lipid peroxidation. Disturbances in the cysteine/glutamate antiporter (system xc-) and antioxidant systems can result in iron overload. Mechanistically, SLC7A11, a member of the solute carrier family 7, supplies cysteine for glutathione (GSH) synthesis. Subsequently, GSH is utilized by GSH-dependent glutathione peroxidase 4 (GPX4) to safeguard against lipid peroxidation by catalyzing lipid peroxide degradation [9]. Dihydroorotate dehydrogenase and GTP cyclohydrolase 1 can inhibit lipid peroxidation in a GPX4/GSH-independent manner [10, 11]. Breast cancer cells escape ferroptosis despite requiring large amounts of iron and reactive oxygen species to maintain active metabolism and proliferation. However, the underlying mechanisms remain unclear.
USP11 has emerged as a key regulator of many cancer-related signaling pathways, offering promising avenues for targeted therapeutic strategies. However, the role of USP11 in various cancer types remains poorly understood. In breast cancer, elevated USP11 levels are associated with increased invasiveness in vitro and enhanced metastatic potential in vivo [12]. Interestingly, a cohort study involving patients with breast cancer has shown a significant association between high USP11 expression and reduced survival [13]. In this study, we found an interaction between PGAM5 and USP11 where de-ubiquitination enhances this interaction, leading to the stabilization of PGAM5. This finding is significant because it clarifies the key role of the USP11/PGAM5 signaling axis in inhibiting breast cancer-induced iron-dependent cell death. Therefore, the potential of this signaling axis as a promising therapeutic target for the treatment of breast cancer has been emphasized.
Materials and methods
Cell culture and transfection
ATCC supplied the breast cancer cell lines (MCF-7 and MDA-MB-231) for this study. Cells were cultured in DMEM supplemented with 10% fetal bovine serum. Plasmids and mimics were introduced into the cells using Lipofectamine 3000 reagent (Invitrogen, USA), and the effectiveness of transfection was assessed by western blot analysis. Additional experiments were conducted 24 h after transfection.
CCK-8 assay
Cell growth was detected using the CCK8 assay. Cells were seeded in a 96-well plate with a volume of 100 µL. Subsequently, 10 µL of the CCK8 solution was added to each well. After 4 h of uninterrupted incubation, the culture plate was transferred to a microplate reader to measure absorbance at 450 nm. Each experimental group included three replicate control wells.
Colony formation assay
Cells were evenly seeded onto six-well plates at a density of 2000 cells per well. Following a 14-d culture period, cells were fixed with 4% paraformaldehyde for 30 min. Subsequently, cells were stained with 0.1% crystal violet for 20 min. After rinsing, photographs were captured, and the number of cell colonies was determined.
Flow cytometric analysis
Apoptosis was detected by flow cytometry. After transfection, the cells were collected after 48 h and subjected to double staining with an apoptosis kit (Invitrogen) using Annexin V-FITC/PI. The cells were then treated at room temperature for 15 min and supplemented with 400 µL of PBS.
Transwell assay
Following the cellular digestion process, the cells were thoroughly washed in serum-free medium, and approximately 10,000 cells were subsequently introduced into the upper chamber of the transwell. An additional 500 µL of DMEM medium was carefully introduced into the lower chamber. The incubation phase lasted for 24 h in an incubator. The cells were fixed with 4% paraformaldehyde for 30 min, followed by staining with 0.1% crystal violet for 20 min.
Ferroptosis analysis experiment
First, the breast cancer cell lines MCF-7 and MDA-MB-231 were transfected with plasmids containing NC, sh-USP11, or ov-PGAM5. Following the transfection, Erastin (10 µg/mL) or RSL3 (10 ng/mL) was introduced into the culture medium on the third day. The cells were then incubated for an additional 3 d before assessing the levels of ferrous ions, malondialdehyde (MDA) content, and fluorescence intensity of C11-BODIPY. The Ferrous Iron Colorimetric Assay Kit (Elabscience), MDA Colorimetric Assay Kit (Elabscience), and C11-BODIPY 581/591 (Maokang Bio) were used to measure the respective ferroptosis-related indicators.
Western blot analysis
The bicinchoninic acid method was used to measure protein concentration following the extraction procedure. Following the gel loading process, the protein sample was introduced into each well and separated by constant-current electrophoresis. Proteins were subsequently transferred onto polyvinylidene fluoride membranes. After blocking for 2 h using skim milk powder, the membrane was subjected to a TBST wash, followed by overnight incubation with the primary antibody. The next day, after rinsing with TBST, a secondary antibody labeled with HRP was introduced and incubated at room temperature for 1 h. Subsequently, an automated chemiluminescence imaging system was used to capture images.
Cellular immunofluorescence
Paraformaldehyde was used to fix the hepatoma cells, followed by permeabilization with 1% Triton X-100 for 15 min. Subsequently, the cells were washed three times with PBS for 5 min each. The cells were then sealed with a blocking solution. The primary antibody was added and incubated overnight. After an overnight incubation, the cells were washed with PBS and incubated with a secondary antibody for 2 h. After washing, DAPI was added, and microscopic examination was performed.
Coimmunoprecipitation (co-IP)
The complete cellular lysate was mixed with 5 µg of the primary antibody or IgG antibody overnight. Subsequently, 5 µL of Protein A + G agarose beads (Absin, Shanghai, China) were added, and the mixture was incubated for an additional 2 h. The protein-antibody complex was rinsed with PBS three times. Finally, the proteins were separated via sodium dodecyl sulfate-polyacrylamide gel electrophoresis and analyzed by western blotting.
Statistical analysis
SPSS 26.0 software was used for statistical analysis, and the data were calculated as mean ± standard deviation. An independent sample t-test was used to compare the two groups of data, and p < 0.05 was considered statistically significant.
Results
Predicting high expression of PGAM5 in breast cancer through bioinformatics analysis
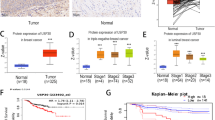
By analyzing the difference between breast cancer tissue and normal tissue using the TCGA database, we found that the composition of the tumor and normal groups showed clear separation (Fig. 1A). We found that PGAM5 was highly expressed in breast cancer through cluster analysis of differential genes and mining of differential genes (Fig. 1B). We further validated the data using the GEPIA database and found that PGAM5 was highly expressed in various cancers, such as colon and lung cancer (Fig. 1C). This includes breast cancer as well (Fig. 1D). In addition, we found that PGAM5 was closely related to breast cancer prognosis (Fig. 1E). We conducted GO analysis of the differentially expressed genes and found a correlation between mitotic nuclear division, collagen-containing extracellular matrix, and glycosaminoglycan binding (Fig. 1F). Through KEGG analysis, we found that the occurrence of breast cancer was closely related to neuroactive ligand-receptor interactions, cytokine receptor interactions, and the cell cycle (Fig. 1G).
Predicting high expression of PPGAM5 in breast cancer through bioinformatics analysis (A) The difference between breast cancer tissue and normal tissue was analyzed through TCGA database, and the composition of tumor group and normal group had obvious separation. (B) cluster analysis was conducted on differentially expressed genes. (C-D) PGAM5 was found to be highly expressed in various cancers through the GEPIA database. (E) PGAM5 is closely related to the prognosis of breast cancer. (F) GO analysis of differentially expressed genes. (G) KEGG analysis of differentially expressed genes
Effects of PGAM5 on proliferation, apoptosis and invasion in breast cancer
In our investigation into the expression of PGAM5 in breast cancer, we performed qPCR to detect its expression in both breast cancer tissues and paired paracancerous tissues. The results demonstrated a notable increase in the expression of PGAM5 in breast cancer cells (Fig. 2A). To validate these results, we performed immunohistochemistry, which revealed the predominant cytoplasmic expression of PGAM5 in breast cancer tissues (Fig. 2B). To gain a deeper understanding of PGAM5’s role in breast cancer, we used interference plasmids to decrease its expression in MCF-7 and MDA-MB-231 cells. The findings showed significant interference effects on PGAM5 using both sh-PGAM5#1 and sh-PGAM5#2 (Fig. 2C). An experiment involving clone formation was performed to evaluate the effects of sh-PGAM5 on cell growth. The results of the clonogenic assay revealed that cell colony size was smaller in the group with PGAM5 interference than in the negative control group (Fig. 2D). Subsequently, flow cytometry was used to assess the effect of PGAM5 knockdown on apoptosis. These findings demonstrated a significant increase in the regulatory ability of apoptosis in breast cancer cells after PGAM5 knockdown (Fig. 2E). To investigate the effect of PGAM5 on the invasive capabilities of breast cancer cells, a transwell assay was conducted. The results of the transwell assay showed that PGAM5 knockdown hindered the invasion of MCF-7 and MDA-MB-231 cells (Fig. 2F).
Effects of PGAM5 on proliferation, apoptosis and invasion in breast cancer (A) Detection of PGAM5 expression in 27 pairs of breast cancer tissues and adjacent tissues by qPCR. (B) Detection of PGAM5 expression in breast cancer tissue by immunohistochemistry(The original magnification is respectively 5x ,10x, 20x,. (C) After transfecting NC, sh-PGAM5# 1 and sh-PGAM5# 2 into MCF-7 and MDA-MB-231cells, the expression of PGAM5 in the cells was detected using Western blotting(D) Clone formation experiment were used to detect the effect of sh-PGAM5#1 and sh-PGAM5#2 on growth, respectively. **, P < 0.01. (E) Analyzing the effect of sh-PGAM5#1 and sh-PGAM5#2on apoptosis in MCF-7 and MDA-MB-231 cells through flow cytometry analysis. (F) The effect of sh-PGAM5#1 and sh-PGAM5#2 on the migration of hepatocellular carcinoma Cell invasion was detected by Transwell(stained with crystal violet, original magnification ×200). **, P < 0.01
Effect of PGAM5 on ferroptosis in breast cancer
The sh-NC, sh-PGAM5#1, and sh-PGAM5#2 plasmids were used to transfect the breast cancer cell lines MCF-7 and MDA-MB-231. To understand the mechanism underlying the action of PGAM5 in breast cancer, we knocked down PGAM5 and examined the expression of ferroptosis-related genes, namely, SLC7A11 and GPX4. The data indicated that the knockdown of PGAM5 markedly suppressed the expression of SLC7A11 and GPX4, suggesting a potential function of PGAM5 in facilitating tumor progression by inhibiting ferroptosis in breast cancer cells (Fig. 3A).After 3 d of transfection, the culture medium was supplemented with Erastin (10 µg/mL) or RSL3 (10 ng/mL) for an additional 3 d. We measured the levels of ferrous ions, MDA content, and the fluorescence intensity of C11-BODIPY in the cells. The results demonstrated a significant increase in ferrous ion (Fig. 3B and C) and MDA content (Fig. 3D and E) in MCF-7 and MDA-MB-231 cells following the knockdown of PGAM5. Additionally, the C11-BODIPY experiment revealed a shift in the maximum fluorescence emission from 591 nm to 510 nm in MCF-7 and MDA-MB-231 cells after PGAM5 knockdown (Fig. 3F and G). In summary, PGAM5 knockdown enhanced the ferroptosis capability of breast cancer cells.
Effect of PGAM5 on ferroptosis in breast cancer. After transfecting NC, sh-PGAM5#1 and sh-PGAM5#2 into MCF-7 and MDA-MB-231 cells. (A) The sh-PGAM5# 1 and sh-PGAM5# 2 markedly suppressed the expression of SLC7A11 and GPX4 by Western blotting.Erastin (10 µ g/ml) or RSL3 (10ng/ml) were introduced into the culture medium on the third day.We measured the levels of ferrous ions (B, C), MDA content (D, E), and fluorescence intensity of C11-BODIPY (F, G) in the cells
There is an interaction relationship between USP11 and PGAM5
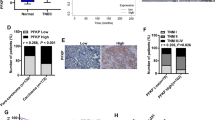
Using qPCR, we detected a substantial level of USP11 expression in breast cancer-affected tissues (Fig. 4A). Furthermore, we observed a positive correlation between USP11 and PGAM5 expression in breast cancer tissues (Fig. 4B). Through exogenous co-IP experiments, we confirmed the existence of protein interactions between USP11 and PGAM5 (Fig. 4C). Endogenous co-IP experiments further confirmed these interactions (Fig. 4D). To investigate the specific regions involved in the interaction between USP1 and PGAM5, we generated a truncated mutant fragment of USP11 to determine the binding site (Fig. 4E). Transfection experiments in MDA-MB-231 cells showed that the absence of amino acids 475–963 and 285–963 of USP11 weakened its ability to bind to PGAM5, whereas the absence of amino acids 1–284 had no effect (Fig. 4F), indicating that the key region for PGAM5 binding is between amino acids 285–785 of USP11. Remarkably, significant inhibition of SLC7A11 and GPX4 expression was observed upon the knockdown of USP11. However, the inhibitory effects of sh-USP11 on SLC7A11 and GPX4 expression were partially reversed by overexpression of PGAM5 (Fig. 4G). In conclusion, USP11 regulates the expression of SLC7A11 and GPX4 via PGAM5.
There is an interaction relationship between USP11 and PGAM5 (A) The expression of USP11 in tissues affected by breast cancer, revealing a substantial level of expression by qPCR.(B) Positive correlation between the expression of USP11 and PGAM5 in breast cancer tissue. (C) The interaction between USP11 and PGAM5 protein was detected by exogenous co-ip assay (D) The interaction between USP11 and PGAM5 protein was detected by endogenous co-ip assay. (E) Constructing a truncated body of USP11 based on its protein structure. (F) Verify the structural domain of USP11 and PGAM5 bonding through COIP experiments. (G) Western blot was used to detect the inhibitory effects of sh-USP11 on PGAM5, GPX4, and SLC7A11, as well as the recovery of sh-USP11 by OV-PGAM
USP11 regulates ferroptosis and tumor growth in cancer through PGAM5
The culture medium of MCF-7 and MDA-MB-231 breast cancer cells was supplemented with NC, sh-USP11, or ov-PGAM5 plasmids for transfection. After a transfection period of 3 d, Erastin (10 µg/mL) or RSL3 (10 ng/mL) was introduced into the medium for an additional 3 d. The levels of ferrous ions (Fig. 5A) and MDA (Fig. 5B) in the cells were evaluated. The findings revealed a significant increase in ferrous ion levels and MDA content in both MCF-7 and MDA-MB-231 cells after USP11 knockdown. The overexpression of PGAM5 partially reinstated the levels of ferrous ions and MDA content. In addition, to assess subcutaneous tumorigenesis in nude mice, we found that knocking down USP11 hindered the subcutaneous tumorigenesis ability of MDA-MB-231 cells, whereas the overexpression of PGAM5 reversed the inhibitory effect of sh-USP11 (Fig. 5C). In conclusion, these findings indicate that USP11 regulates iron-related cell death and tumor growth in breast cancer cells in vitro and in vivo through PGAM5.
USP11 regulates ferroptosis in breast cancer through PGAM5 After transfecting NC, sh-USP11 and ov-PGAM5 into MCF-7 and MDA-MB-231 cells, Erastin (10 µ g/ml) or RSL3 (10ng/ml) were introduced into the culture medium on the third day.We measured the levels of ferrous ions (A), MDA content (B). (C) Subcutaneous tumor formation experiment in nude mice
USP11 prevents degradation of PGAM5 by deubiquitination
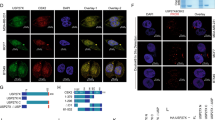
In this study, CHX, a compound that inhibits protein synthesis, was used to treat MCF-7 and MDA-MB-231 cells with USP11 depleted. Our findings demonstrated that the depletion of USP11 (sh-USP11) accelerated the degradation of PGAM5 in both MCF-7 and MDA-MB-231 cells (Fig. 6A). Furthermore, we observed an increase in the ubiquitination of PGAM5 in cells following the knockdown of USP11 (Fig. 6B). These results indicate that USP11 plays a vital role in facilitating the de-ubiquitination and stability of PGAM5. Ubiquitin analysis showed that USP11 overexpression significantly inhibited PGAM5 polyubiquitination (Fig. 6C). In addition, overexpression of wild-type USP11 instead of the catalytic inactivation mutant (C318A) increased PGAM5 protein levels (Fig. 6D).
USP11 Mediated Deubiquitination and Stability of PGAM5 (A) sh-USP11accelerated the degradation of PGAM5 in both MCF-7 and MDA-MB-231 cells. (B) increase in the ubiquitination of PGAM5 in cells subsequent to the knockdown of USP11 in MCF-7 and MDA-MB-231 cells. (C) Overexpression of wild-type USP11 can inhibit the deubiquitination of PGAM5, while mutant USP11 cannot. (D) Overexpression of wild-type USP11 can inhibit protein degradation of PGAM5
Mechanism of USP11 mediated deubiquitination and stability of PGAM5
The two primary forms of polyubiquitination involve ubiquitin chains joined by K48 and K63. Our findings demonstrate that the ubiquitinated K48 chain is selectively cleaved from PGAM5 by USP11 rather than the K63 chain (Fig. 7A). To investigate this further, we incorporated stable point mutations that hindered ubiquitination in K48, thereby eliminating the USP11-facilitated ubiquitination of PGAM5 (Fig. 7B). In summary, our investigation revealed the role of USP11 in modulating the expression of PGAM5 in breast cancer cells by eliminating the ubiquitination chain connected to K48. We co-transfected Myc-PGAM5 and HA ubiquitin plasmids into MDA-MB-231 cells transfected with the sh-USP11 plasmid or empty vector. USP11 knockdown by shRNA increased the polyubiquitination level of the PGAM5 protein in MDA-MB-231 cells (Fig. 7C). To determine lysine binding between PGAM5 and Ub, a point mutation was generated by replacing lysine with arginine in PGAM5. The protein stability of the K191R mutant was significantly increased (Fig. 7D), and compared to the wild-type PGAM5, the ubiquitination of K191R PGAM5 was reduced (Fig. 7E).
USP11 regulates the stability of PGAM5 by removing the ubiquitin chain linked to K48 in breast cancer cells (A) the ubiquitinated K48 chain is selectively cleaved from PGAM5 by USP11, instead of the K63 chain. (B) Introducing stable point mutations that hinder ubiquitination in K48 eliminates the restriction of PGAM5 ubiquitination promoted by USP11. (C) Sh-USP11 increases the ubiquitination level of PGAM5. (D) We replaced lysine with arginine to generate point mutations in candidate sites, and the protein stability of the K191R mutant was significantly increased. (E) And compared with the wild-type PGAM5, the ubiquitination in K191R PGAM5 is reduced
Discussion
In recent years, significant research has focused on molecular targeted therapies for breast cancer. As such, it is of utmost importance to identify key therapeutic targets that are effective in combating this type of cancer [14, 15]. The occurrence and progression of breast cancer are influenced by various oncogenes and tumor suppressor genes [16,17,18]. Investigations have shown that PGAM5 is highly expressed in tissues affected by skin melanoma, and its increased expression is closely related to a negative prognosis of this disease [7]. Additionally, PGAM5 forms trimers with ubiquitin-specific peptidase 10 and S100A9. This trimer formation leads to the de-ubiquitination and stabilization of PGAM5, ultimately resulting in mitochondrial division and reactive oxygen species production. These processes further contribute to the growth and spread of hepatocellular carcinoma [19].
In this study, breast cancer was analyzed using TCGA data to identify differentially expressed genes, and functional enrichment analysis was performed on these genes. Our findings indicate a high level of PGAM5 expression in breast cancer. To gain further insight into the effect of PGAM5 on breast cancer, knockdown experiments were conducted. The expression of SLC7A11 and GPX4 decreased following the knockdown of PGAM5. Ferroptosis is a newly discovered iron-dependent cell death mode that differs from other cell death pathways, such as autophagy, apoptosis, necrosis, and pyroptosis [20]. This form of cell death is characterized by mitochondrial shrinkage, membrane condensation and thickening, decreased activity of the SLC7A11 and GPX4 proteins [21], depletion of GSH, and excessive accumulation of reactive oxygen species, leading to increased lipid peroxidation of the cell membrane. Studies have shown that ferroptosis is also an important regulator of tumor growth [22], with SLC7A11 and GPX4 being the main regulatory factors. SLC7A11 is involved in extracellular cysteine uptake and glutamate release, and it promotes GSH synthesis. GPX4 can inactivate lipid peroxides via GSH, thereby inhibiting cell ferroptosis [23]. Furthermore, a series of experiments were carried out, including colony formation, flow cytometry, transwell, MDA, and ferrous ion level detection. The results revealed that the proliferation and invasion of breast cancer cells were reduced upon knockdown of PGAM5, whereas apoptosis and ferroptosis were increased in breast cancer cells. These results suggest that PGAM5 plays a crucial role in the occurrence and progression of breast cancer by affecting the proliferation, invasion, apoptosis, and ferroptosis of breast cancer cells. These findings highlight PGAM5 as a promising therapeutic target for breast cancer treatment.
To further investigate the underlying mechanism of PGAM5 in breast cancer, we used BioGRID to identify the interacting genes associated with PGAM5. Our findings suggest a potential correlation between PGAM5 and USP11. USP11 is a well-known de-ubiquitinating enzyme that belongs to the ubiquitin-specific processing protease family. It plays a significant role in numerous signal cascades, including TGF-β and the p53 signaling pathways, ultimately controlling the stability of its downstream substrates [24,25,26,27]. USP11 has been closely studied in other types of cancer, indicating its high correlation with tumor occurrence [28,29,30]. Recent investigations have reported elevated USP11 expression in GC, demonstrating its promotion of cancer growth and migration when overexpressed. Additionally, when USP11 is depleted, migration is hindered through RhoA-mediated pathways, and growth and survival may be inhibited through Ras-mediated pathways [31]. It has been experimentally proven that USP11 significantly contributes to breast cancer progression [32]. Using qPCR analysis, we examination the increased expression levels of USP11 in breast cancer tissues, which showed a positive correlation with PGAM5. This finding was solidified through co-IP experiments, which confirmed the protein interaction between PGAM5 and USP11. Moreover, our findings suggested that USP11 plays a pivotal role in the regulation of SLC7A11 and GPX4 expression in breast cancer via PGAM5, thereby affecting ferroptosis. Additionally, our results indicate that USP11 alters the stability of PGAM5 by eliminating the ubiquitin chain connected to K48 in breast cancer cells. To summarize, this study provides groundbreaking evidence that USP11 serves as a mediator of PGAM5 de-ubiquitination within the cytoplasm, underscoring the interconnection between PGAM5 and USP11 in breast cancer. Specifically, we found that USP11 enhances breast cancer progression and ferroptosis through PGAM5.
Conclusions
This study confirmed, for the first time, the elevated expression of PGAM5 in breast cancer and found that PGAM5 plays an important role in promoting the occurrence and development of breast cancer. We also found that USP11 regulates the biological function of breast cancer cells by stabilizing PGAM5 and plays a role in promoting cancer, which is a novel finding. Therefore, this study deepens our understanding of breast cancer and reveals that the USP11/PGAM5 axis may become a new molecular marker for breast cancer, providing new ideas for the clinical diagnosis and treatment of breast cancer and potentially becoming a target for molecular therapies. However, the results of this study have certain limitations. Although we found that PGAM5 is highly expressed in breast cancer through bioinformatics analysis and further validated this through qPCR and immunohistochemistry, our sample size was small. In the future, we will increase the sample size to further explore the relation between PGAM5 and clinical outcomes and investigate its potential applications in clinical practice.
Data availability
No datasets were generated or analysed during the current study.
Abbreviations
- PGAM5:
-
Phosphoglycerate mutase 5
- USP11:
-
Ubiquitin-specific protease 11
- GSH:
-
Glutathione
- GPX4:
-
Glutathione peroxidase 4
- MDA:
-
Malondialdehyde
References
Jiang XT, Liu Q. mRNA vaccination in breast cancer: current progress and future direction. J Cancer Res Clin Oncol. 2023;149(11):9435–50.
Takeda K, Komuro Y, Hayakawa T, Oguchi H, Ishida Y, Murakami S, Noguchi T, Kinoshita H, Sekine Y, Iemura S, et al. Mitochondrial phosphoglycerate mutase 5 uses alternate catalytic activity as a protein serine/threonine phosphatase to activate ASK1. Proc Natl Acad Sci U S A. 2009;106(30):12301–5.
He GW, Gunther C, Kremer AE, Thonn V, Amann K, Poremba C, Neurath MF, Wirtz S, Becker C. PGAM5-mediated programmed necrosis of hepatocytes drives acute liver injury. Gut. 2017;66(4):716–23.
Ramachandran A, Jaeschke H. PGAM5: a new player in immune-mediated liver injury. Gut. 2017;66(4):567–8.
Wang Z, Jiang H, Chen S, Du F, Wang X. The mitochondrial phosphatase PGAM5 functions at the convergence point of multiple necrotic death pathways. Cell. 2012;148(1–2):228–43.
Meng L, Hu P, Xu A. PGAM5 promotes tumorigenesis of gastric cancer cells through PI3K/AKT pathway. Pathol Res Pract. 2023;244:154405.
Peng J, Wang T, Yue C, Luo X, Xiao P. PGAM5: a necroptosis gene associated with poor tumor prognosis that promotes cutaneous melanoma progression. Front Oncol. 2022;12:1004511.
Jiang X, Stockwell BR, Conrad M. Ferroptosis: mechanisms, biology and role in disease. Nat Rev Mol Cell Biol. 2021;22(4):266–82.
Fu G, Li ST, Jiang Z, Mao Q, Xiong N, Li X, et al. PGAM5 deacetylation mediated by SIRT2 facilitates lipid metabolism and liver cancer proliferation. Acta Biochim Biophys Sin (Shanghai). 2023;55(9):1370–9.
Mao C, Liu X, Zhang Y, Lei G, Yan Y, Lee H, Koppula P, Wu S, Zhuang L, Fang B, et al. DHODH-mediated ferroptosis defence is a targetable vulnerability in cancer. Nature. 2021;593(7860):586–90.
Kraft VAN, Bezjian CT, Pfeiffer S, Ringelstetter L, Muller C, Zandkarimi F, Merl-Pham J, Bao X, Anastasov N, Kossl J, et al. GTP cyclohydrolase 1/Tetrahydrobiopterin counteract ferroptosis through lipid remodeling. ACS Cent Sci. 2020;6(1):41–53.
Garcia DA, Baek C, Estrada MV, Tysl T, Bennett EJ, Yang J, Chang JT. USP11 enhances TGFbeta-Induced epithelial-mesenchymal plasticity and human breast Cancer Metastasis. Mol Cancer Res. 2018;16(7):1172–84.
Dwane L, O’Connor AE, Das S, Moran B, Mulrane L, Pinto-Fernandez A, Ward E, Blumel AM, Cavanagh BL, Mooney B, et al. A functional genomic screen identifies the deubiquitinase USP11 as a Novel Transcriptional Regulator of ERalpha in breast Cancer. Cancer Res. 2020;80(22):5076–88.
Salem IM, Mostafa SM, Salama I, El-Sabbagh OI, Hegazy WAH, Ibrahim TS. Design, synthesis and antitumor evaluation of novel pyrazolo[3,4-d]pyrimidines incorporating different amino acid conjugates as potential DHFR inhibitors. J Enzyme Inhib Med Chem. 2023;38(1):203–15.
Liu WJ, Wang LY, Sheng Z, Zhang B, Zou X, Zhang CY. RNA methylation-driven assembly of fluorescence-encoded nanostructures for sensitive detection of m(6)a modification writer METTL3/14 complex in human breast tissues. Biosens Bioelectron. 2023;240:115645.
Zhang S, Guo A, Wang H, Liu J, Dong C, Ren J, Wang G. Oncogenic MORC2 in cancer development and beyond. Genes Dis. 2024;11(2):861–73.
Sadri F, Hosseini SF, Rezaei Z, Fereidouni M. Hippo-YAP/TAZ signaling in breast cancer: reciprocal regulation of microRNAs and implications in precision medicine. Genes Dis. 2024;11(2):760–71.
Papatsirou M, Scorilas A, Sideris DC, Kontos CK. Targeted nanopore sequencing for the identification of novel PRMT1 circRNAs unveils a diverse transcriptional profile of this gene in breast cancer cells. Genes Dis. 2024;11(2):589–92.
Zhong C, Niu Y, Liu W, Yuan Y, Li K, Shi Y, Qiu Z, Li K, Lin Z, Huang Z, et al. S100A9 Derived from Chemoembolization-Induced Hypoxia governs mitochondrial function in Hepatocellular Carcinoma Progression. Adv Sci (Weinh). 2022;9(30):e2202206.
Yan HF, Zou T, Tuo QZ, Xu S, Li H, Belaidi AA, Lei P. Ferroptosis: mechanisms and links with diseases. Signal Transduct Target Ther. 2021;6(1):49.
Yuan Y, Zhai Y, Chen J, Xu X, Wang H. Kaempferol Ameliorates Oxygen-Glucose Deprivation/Reoxygenation-Induced Neuronal Ferroptosis by Activating Nrf2/SLC7A11/GPX4 Axis. Biomolecules 2021, 11(7).
Chen X, Kang R, Kroemer G, Tang D. Broadening horizons: the role of ferroptosis in cancer. Nat Rev Clin Oncol. 2021;18(5):280–96.
Liu M, Kong XY, Yao Y, Wang XA, Yang W, Wu H, Li S, Ding JW, Yang J. The critical role and molecular mechanisms of ferroptosis in antioxidant systems: a narrative review. Ann Transl Med. 2022;10(6):368.
Jacko AM, Nan L, Li S, Tan J, Zhao J, Kass DJ, Zhao Y. De-ubiquitinating enzyme, USP11, promotes transforming growth factor beta-1 signaling through stabilization of transforming growth factor beta receptor II. Cell Death Dis. 2016;7(11):e2474.
Ke JY, Dai CJ, Wu WL, Gao JH, Xia AJ, Liu GP, Lv KS, Wu CL. USP11 regulates p53 stability by deubiquitinating p53. J Zhejiang Univ Sci B. 2014;15(12):1032–8.
Deng T, Xie L, Xiaofang C, Zhang Z, Xiao Y, Peng Y, et al. ATM-Mediated translocation of RanBPM regulates DNA damage response by stabilizing p21 in non-small cell lung cancer cells. Cell Oncol (Dordr). 2023;47(1):245–8.
Jin Q, Gutierrez Diaz B, Pieters T, Zhou Y, Narang S, Fijalkwoski I, Borin C, Van Laere J, Payton M, Cho BK, et al. Oncogenic deubiquitination controls tyrosine kinase signaling and therapy response in acute lymphoblastic leukemia. Sci Adv. 2022;8(49):eabq8437.
Sun H, Ou B, Zhao S, Liu X, Song L, Liu X, Wang R, Peng Z. USP11 promotes growth and metastasis of colorectal cancer via PPP1CA-mediated activation of ERK/MAPK signaling pathway. EBioMedicine. 2019;48:236–47.
Zhang S, Xie C, Li H, Zhang K, Li J, Wang X, Yin Z. Ubiquitin-specific protease 11 serves as a marker of poor prognosis and promotes metastasis in hepatocellular carcinoma. Lab Invest. 2018;98(7):883–94.
Zhu X, Zhang Y, Luo Q, Wu X, Huang F, Shu T, Wan Y, Chen H, Liu Z. The deubiquitinase USP11 promotes ovarian cancer chemoresistance by stabilizing BIP. Signal Transduct Target Ther. 2021;6(1):264.
Liu H, Liu M, He B, Li Q. Inhibition of USP11 sensitizes gastric cancer to chemotherapy via suppressing RhoA and ras-mediated signaling pathways. Clin Res Hepatol Gastroenterol. 2022;46(1):101779.
Li L, Deng T, Zhang L, Wang Y, Zhou Y, Liu Y, Li H, Dai J, Yang Y, Ling N, et al. ERK-mediated cytoplasmic Retention of USP11 contributes to breast Cancer cell proliferation by stabilizing cytoplasmic p21. Int J Biol Sci. 2022;18(6):2568–82.
Funding
This work was supported by grants from the Research Project of Maternal and Child Health of Jiangsu Province (F201953), the Science and Technology Project of Nantong (JC2020067), the Research project of Nantong Municipal Health Commission (MS2023023) and Preventive medicine and research projects on blood transfusion prevention (X202354) to ZH; the Research project of Nantong Municipal Health Commission (MS2023001) to FW; the Research project of Nantong Municipal Health Commission (MS2023009) to QW;
Author information
Authors and Affiliations
Contributions
(I) Conception and design: F Wang, N Zhang; (II) Administrative support: Z He; (III) Provision of study materials or patients: N Zhang; (IV) Collection and assembly of data: Q Wang; (V) Data analysis and interpretation: Y Lu; (VI) Manuscript writing: All authors; (VII) Final approval of manuscript: All authors.
Corresponding author
Ethics declarations
Ethical approval
The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the institutional ethics committee of the Affiliated Hospital of Nantong University (ID: 2023-L084). Informed consent was obtained from each participant. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The animal research protocol was approved by the Animal Ethics Committee of Nantong University (ID: P20230222-001), in compliance with institutional guidelines for the care and use of animals.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, N., Wang, Q., Lu, Y. et al. The deubiquitinating enzyme USP11 regulates breast cancer progression by stabilizing PGAM5. Breast Cancer Res 26, 135 (2024). https://doi.org/10.1186/s13058-024-01892-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13058-024-01892-9