Abstract
Background
Excavation of key molecules can help identify therapeutic targets and improve the prognosis of pancreatic cancer. This study evaluated the roles of SUMO3 in cell viability, glycolysis, gemcitabine (GEM) sensitivity, and the antitumor activity of butyric acid (BA) in pancreatic cancer.
Methods
The mRNA and protein levels of SUMO3 were detected by qRT-PCR, Western blot, and immunohistochemical assay. SUMO3 was silenced or overexpressed in pancreatic cancer cells with or without Wnt/β-catenin pathway inhibitor, glycolysis inhibitor, GEM, or BA treatment. Cell viability was measured using the Cell Counting Kit-8 assay. Glycolysis was measured by determining the extracellular acidification rate, ATP level, and lactate content. Apoptosis was measured by flow cytometry, and TUNEL staining was used to examine in vitro and in vivo sensitivity to GEM chemotherapy. Luciferase reporter and chromatin immunoprecipitation assays were conducted to detect the binding of the SUMO3 promoter and NF-κB p65.
Results
SUMO3 was increased and associated with poor survival in pancreatic cancer. SUMO3 knockdown decreased cell viability and glycolysis in vitro and inhibited tumor growth in vivo. SUMO3 overexpression increased cell viability and glycolysis in vitro through the β-catenin pathway. SUMO3 knockdown increased GEM sensitivity, whereas SUMO3 overexpression decreased GEM sensitivity and inhibited the antitumor activity of BA. BA promoted histone acetylation and p-IκBα expression to inhibit NF-κB p65-mediated SUMO3 transcription.
Conclusion
SUMO3 acted as an active molecule in cell survival and growth by enhancing glycolysis in response to either GEM or BA. The mechanism was related to the constitutive IκBα/NF-κB/SUMO3/β-catenin signaling pathway.
Similar content being viewed by others
Introduction
Pancreatic cancer is a type of malignancy, which has been shown to have poor survival rate and high morbidity in clinical practice in recent years, and is a major cause of death [1]. The treatment strategies for pancreatic cancer include surgery and chemotherapy; however, the latter is more widely used, because most patients are initially diagnosed at an advanced stage. Thus, rapid acquisition of chemotherapy resistance has been the main reason for the poor prognosis of pancreatic cancer [2]. Targeted cancer therapy, which is based on an understanding of the molecular and genetic changes of a disease, has been shown to have good potential. Therefore, improved understanding of the functional molecules involved in cancer cell homeostasis can help identify therapeutic targets and improve the treatment strategy for pancreatic cancer.
The small ubiquitin-like modifier (SUMO) family of proteins is covalently linked with their target proteins through a multistep enzyme-linked reaction. This SUMOylation regulates protein stabilization, subcellular localization, innate immunity, signal transduction, and antiviral defense [3] and is recognized as a key biological regulator of various pathological and physiological processes [4, 5], such as cancer pathogenesis [6]. SUMO pathway enzyme levels are typically elevated in various cancer types and are associated with cancer progression and poor clinical outcomes. In pancreatic cancer, SUMOylation level was found to be significantly increased, and inhibition of SUMOylation blocked cancer cell cycle progression [7]. Moreover, SUMOylation is involved in lymphangiogenesis, lymph node metastasis [8], and chemosensitivity in pancreatic cancer [9]. SUMO3 is one of the five SUMO members responsible for stabilizing various proteins to promote the progression of solid cancers, including gastric, prostate, and breast cancer [6, 10, 11]. In pancreatic cancer, SUMO3 levels appear to be upregulated in tumor tissues [7], but its specific function in cancer progression is unknown.
In this study, we aimed to determine the level of SUMO3 expression in pancreatic cancer tissues and cell lines and to define the correlation between SUMO3 expression and patient survival and cancer cells. We found that SUMO3 actively promoted cancer cell growth and survival, even in the presence of the chemotherapy drug gemcitabine (GEM) and the antitumor agent butyric acid (BA), by enhancing glycolysis, which is important for energy metabolism for cancer cell survival and metastasis [12]. Overall, our data revealed the potential clinical utility of targeted SUMO3 inhibition by enhancing chemotherapy sensitivity and tumor eradication.
Materials and methods
Bioinformatics analysis of the association between SUMO3 expression and pancreatic cancer
Using the RNAseq data from The Cancer Genome Atlas (TCGA) dataset, Gene Expression Profiling Interactive Analysis was performed to determine the SUMO3 level, which was compared between the pancreatic cancer and normal control groups. The Kaplan–Meier plot was used to estimate the correlation between SUMO3 and patient survival. The gene set enrichment analysis (GSEA) algorithm was used to discriminate the functional clustering associated high and low SUMO3 expression.
Patient samples
SUMO3 expression was determined using quantitative RT-PCR in 30 pancreatic cancer tissues obtained from patients who were recruited from October 2021 to March 2023 in the hospital. All patients provided written informed consent before tissue collection. Human pancreatic cancer tissue microarrays (Shanghai Outdo Biotech, China), which comprised 40 tumor tissues and 27 adjacent normal tissues, were used to determine SUMO3 protein expression on immunohistochemistry (IHC). Among the 40 patients with pancreatic cancer, SUMO3 expression was compared between 7 patients who were sensitive to chemotherapy (CR + PR) and 4 patients who were not (SD + PD). The experimental procedures involving human samples were approved by the medical ethics committee of Fudan University Shanghai Cancer Center (approval number 050432-4-2108) and were performed in accordance with the Declaration of Helsinki.
Immunohistochemistry
Paraffin-embedded tissues were sectioned to detect SUMO3 expression by IHC. Histological sections were sequentially reacted with anti-SUMO3 antibody (4971; Cell Signaling Technology, USA) and anti-IgG antibody (D-3004; Long Island Biotech, China). The H-score system, which ranged from 0 to 12, was applied to evaluate the immunoreactivity of the target protein by calculating the ratio of positive-staining cells to the color intensity. Scoring was performed independently by two investigators.
Cell lines and cell culture
The human pancreatic cancer cell lines (PANC-1, AsPC-1, SW1990, HPAC, and BXPC3) and control human pancreatic ductal epithelial (HPDE) cell line were purchased from the American Type Culture Collection. DMEM containing 10% fetal bovine serum, 1% streptomycin, and penicillin and a 5% CO2 cell incubator provided the nutrient and environmental conditions for cell growth.
For the experimental protocols, HPAC cells were transduced by a SUMO3 expression vector and stimulated by 10 µM of XAV939, which is an inhibitor of the Wnt/beta-catenin pathway, or 10 and 20 µM of GEM (Selleck, USA) for 48 h. SW1990 cells were transduced with the SUMO3 short hairpin RNA (shRNA) vector and treated with 10 and 20 µM of GEM and/or 5 mM of 2-DG (Selleck, USA) or different concentrations of BA (Selleck, USA) for 48 h.
Cell transfection
To achieve gene overexpression or knockdown, pancreatic cancer cells were transduced by pLVX-Puro-SUMO3 (oeSUMO3), pLKO.1-SUMO3 shRNA (shSUMO3), or pLVX-Puro-NF-κBp65 (oep65) vectors, which were constructed by GENERAL BIO (Chuzhou, China) using Lipofectamine 2000 (Invitrogen), according to the manufacturer’s instructions. The shRNA sequences targeting the human SUMO3 gene were as follows: shSUMO3#1 GCTCCGTGGTGCAGTTCAAGA, shSUMO3#2 TGGTGCAGTTCAAGATCAAGA, shSUMO3#3 GTGGTGCAGTTCAAGATCAAG, and scramble shRNA GGACGAGCTGTACAAGTAA. Cells transduced by blank pLVX-Puro vector (oeNC) or pLKO.1-scrambled shRNA served as the negative control.
Cell counting Kit-8 assay
Cell viability was measured using a Cell Counting Kit-8 (CCK-8) kit (Signaling Antibody, USA) according to the manufacturer’s protocols. Briefly, 3 × 103 cells were cultured in a 96-well plate overnight. At 12, 24, and 48 h after treatment, cells in each well were incubated with 10 µL CCK-8 solution for 1 h. Subsequently, the absorbance value at OD450 nm was detected on a microplate reader and translated into cell viability.
Cell apoptosis assay by flow cytometer
Cells were stained and incubated in Annexin V-FITC at 4 °C for 20 min, followed by propidium iodide for 20 min. Cell apoptosis was analyzed using an Accuri™ C6 flow cytometer.
Extracellular acidification rate
Cells were maintained in an XF-24 culture plate (Agilent Technologies, USA) at a density of 1 × 104/well for 24 h and then recultured in XF Base Medium (Agilent Technologies) 1 h before testing. Extracellular acidification rate (ECAR) was determined using an XF24 Extracellular Flux Analyzer (USA). During the test, glucose (1 µM), the ATP synthase inhibitor oligomycin (1 µM), and the glycolytic inhibitor 2-DG (0.5 µM) were used to stimulate cells at the test node.
Measurement of lactic acid and ATP
After normal cell culture for 12 h and another 24 h of reculture with fresh DMEM (50 µL/well), a culture supernatant of 3.5 × 103 cells/well in a 96-well plate was acquired and assessed for lactic acid content. ATP content was determined in the lysate that were acquired from cells cultured for 12–24 h in a 6-well plate. The lactic acid and ATP assay test kits used in this experiment were obtained from Nanjing Jiancheng Bioengineering Institute (China), and the protocols were followed according to the manufacturer’s instructions.
Quantitative RT-PCR analysis
Total RNA was extracted using TRIzol and used as the template for reverse transcription using Superscript II (Invitrogen, Shanghai). The cDNA product was amplified by quantitative RT-PCR (qRT-PCR) using SYBR master mix (Bio-Rad, USA). The primers used were as follows: SUMO3-F: 5′-ATGTCCGAGGAGAAGCCCAA-3′; SUMO3-R: 5′-ATTGACAAGCCCTGCCTCTCG-3′; β-actin-F: 5′-AGGATTCCTATGTGGGCGAC-3′; and β-actin-R: 5′-ATAGCACAGCCTGGATAGCAA-3′. Using β-actin as the reference gene, relative mRNA expression levels were calculated using 2−△△Ct.
DNA copy analysis of fecal samples from patients with pancreatic cancer
DNA was extracted using Stool Genomic DNA Extraction Kit (D2700; Solarbio Beijing, China) according to the manufacturer’s procedures. The DNA copy numbers of Faecalibacterium prausnitzii and Bifidobacterium were determined using qRT-PCR with Hieff® qPCR SYBR Green Master Mix (11203ES03; Yeasen Biotechnology Co., Ltd., Shanghai, China). The primer sequences were ACGCACATAGAAACAGTAG (F), TGTAGCCCAAGTCATAAAG (R) for Faecalibacterium prausnitzii and ATCGGGCTTTGCTTGGTGG (F), AGTCTGGGCCGTATCTCAGTC (R) for Bifidobacterium. pGM-T plasmids containing the bacterial DNA sequence were constructed by GENERAL BIO (Chuzhou, China) and used as the standard for qRT-PCR reactions.
Western blot
Cells were lysed using radioimmunoprecipitation assay lysis buffer (Beyotime, Shanghai, China). Nuclear and cytoplasmic fraction extracts were obtained using the NE-PER™ Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Scientific, USA). The total protein in the sample was evaluated using the Bicinchoninic Acid Protein Assay Kit (Beyotime, China). The same content of the protein sample (20 µg) was subjected to SDS-PAGE to separate proteins of different weights and then transferred onto polyvinylidene fluoride membranes (Millipore, USA). Nonspecific proteins were blocked by 5% fat-free milk, and the target proteins were subsequently reacted with the primary antibodies anti-SUMO3 (ab203570, Abcam); anti-β-catenin (8480T, Cell Signaling Technology); anti-lactate dehydrogenase (LDHA) (3582T, Cell Signaling Technology); cleaved PARP (ab32064, Abcam); cleaved caspase 3 (PA5-114687, Invitrogen); anti-H3 (ab1791, Abcam); and anti-β-actin (60066-1-AP; Proteintech) and a secondary antibody (AB-2301, ZB-2305; ZSGB-BIO, Beijing, China). Blots of target proteins were visualized by chemiluminescence (Enhanced Chemiluminescence Detection Kit; Pierce Biotechnology, USA).
Luciferase reporter assay
Luciferase reporter assay was performed to identify the transcription activity of p65 on the SUMO3 promoter. The pGL3 wild-type SUMO3 promoter (WT) vector or the pGL3 mutant-type SUMO3 promoter (MUT), which was acquired by amplification and cloning of the promoter fragment into pGL3 vectors (Promega, USA), was combined with empty pGL3 or p65 overexpression vector-transduced cells (Lipofectamine 3000; Invitrogen, USA). At 48 h after transfection, the cells were collected and evaluated for luciferase activity using the Dual-Luciferase Reporter Assay System (Promega), as per the procedures provided by the manufacturer.
Chromatin immunoprecipitation assays
The interactions of specific proteins with gene promoters were evaluated using a chromatin immunoprecipitation (ChIP) assay (Magnetic ChIP kit; Millipore, USA). Briefly, cells immobilized with 1% formaldehyde were subjected to DNA fragmentation by sonication in the presence of MNase. Immunoprecipitation assays were conducted using the antibodies H3K9ac (ab32129, Abcam); H3K14ac (ab203952, Abcam); H3K27ac (ab4729, Abcam); or negative control IgG (30000-0-AP, Proteintech). The immunoprecipitate was eluted and subsequently subjected to reverse crosslinking. The precipitated DNA was amplified by PCR, and the target DNA was normalized to the input.
In vivo tumor-bearing mice model
Male BALB/c nude mice (5 weeks old) (Shanghai Lab. Animal Co., China) were used to establish a tumor-bearing model by subcutaneous injection of SUMO3 shRNA-transduced SW1990 cells or SUMO3 expression vector-transduced HPAC cells (n = 6 in each group). To examine the GEM sensitivity of the tumors, the mice were administered intraperitoneal GEM at 50 mg/kg once every 2 days. Tumor size was monitored every 3 days, and xenografts were collected for Ki67 immunofluorescence staining (27309-I-AP; Proteintech, USA) and TUNEL assay (Roche, USA). The animal studies were approved by the Ethics Committee of Fudan University (approval number 2020 Shanghai Cancer Center JS-213).
Data processing and analysis
Data were shown as mean ± standard deviation. Significant differences between groups were assessed using one-way analysis of variance or Student’s t-test. Statistical analyses were conducted using GraphPad Prism 8.4.2 software (USA). P values < 0.05 were considered significant.
Results
SUMO3 was increased and associated with poor survival in pancreatic cancer
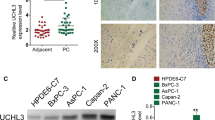
Figure 1 presents SUMO3 expression in patients and cell lines and its relationship with survival in pancreatic cancer. In the TCGA dataset, the SUMO3 level in pancreatic cancer tissue was significantly upregulated (Fig. 1A). Based on the SUMO3 expression, patients were classified into the high (n = 45) and low (n = 132) SUMO3 groups. Survival time was short in the high SUMO3 group (Fig. 1B). Consistent with the TCGA dataset, the pancreatic cancer tissue microarray was found to have significantly increased SUMO3 expression, and high SUMO3 expression predicted poor survival rate (Fig. 1C and E). Analysis of the clinicopathological characteristics of patients with pancreatic cancer revealed that high SUMO3 expression was associated with tumor T stage and pathological stage (Table 1). In addition, SUMO3 expression was high in the pancreatic cancer cell lines AsPC-1, BXPC3, HPAC, SW1990, and PANC-1 (Fig. 1F and G). SW1990 cells had the highest expression level and were used to construct a SUMO3 knockdown cell line, whereas HPAC cells had the lowest expression level and were used to construct a SUMO3-overexpressing cell line for the subsequent experiments. GSEA showed that SUMO3 was related with the HALLMARK_GLYCOLYSIS, FEVR_CTNNB1_TARGETS_UP, and HONMA_DOCETAXEL_RESISTANCE pathways (Fig. 1H), suggesting the involvement of SUMO3 in glycolysis and chemosensitivity of pancreatic cancer cells.
Increased SUMO3 expression in patients and cell lines indicates poor survival in pancreatic cancer. The TCGA dataset is used to analyze SUMO3 expression (A) in pancreatic cancer tissues and assess its correlation with prognosis (B). Immunohistochemical image (C) shows increased SUMO3 expression, as evaluated by the IHC score (D) and its correlation with low survival rate (E) in clinical pancreatic cancer tissues. SUMO3 expression in cancer cell lines is assessed by qRT-PCR (F) and Western blot (G). GSEA shows that SUMO3 is related with the HALLMARK_GLYCOLYSIS, FEVR_CTNNB1_TARGETS_UP, HONMA_DOCETAXEL_RESISTANCE pathways (H). Scale bar: 100 μm. *P < 0.05, **P < 0.01, ***P < 0.001 vs. normal or HPDE
SUMO3 knockdown inhibited glycolysis in vitro and tumor growth in vivo
Figure 2 summarizes the effects of SUMO3 knockdown on glycolysis in vitro and tumor growth in vivo. After transduction of three specific shRNAs (shSUMO3#1, shSUMO3#2, and shSUMO3#3), SUMO3 was significantly knocked down in SW1990 cells (Fig. 2A and B). Observation of shSUMO3#1 and shSUMO3#2 knockdown cell line revealed reduced cell viability and suppressed ECAR and production of ATP and lactic acid. These results suggested that SUMO3 was indispensable for glycolysis in SW1990 cells (Fig. 2C and F). The shSUMO3#1 knockdown SW1990 cell line was transplanted into nude mice to observe subcutaneous xenograft growth. Tumor growth was weaker in the shSUMO3#1 knockdown SW1990 cell xenografts than in the negative control cell xenografts (Fig. 2G). This observation was supported by the findings on tumor size (Fig. 2H) and weight (Fig. 2I) of the transplanted xenograft. The immunofluorescence of Ki67-labeled cells was significantly less in the shSUMO3#1 knockdown xenograft than in the negative control cell xenograft (Fig. 2J and K), suggesting the indispensability of SUMO3 for SW1990 cell proliferation. Moreover, SUMO3 expression was significantly decreased in the shSUMO3#1 knockdown xenograft (Fig. 2L).
SUMO3 knockdown inhibits glycolysis in vitro and tumor growth in vivo. SUMO3 knockdown SW1990 cell line is constructed by gene interference lentivirus transduction, and SUMO3 expression is evaluated by qRT-PCR (A) and Western blot (B). The SUMO3 knockdown SW1990 cells are assessed for cell viability using the CCK8 assay (C), ECAR using a Seahorse energy analyzer (D), ATP production (E), and lactate level in the supernatant (F). In the tumor-bearing model, which is built by subcutaneous transplantation of SUMO3 knockdown SW1990 cells into nude mice, tumor growth is evaluated by tumor growth curve (G), tumor size (H), tumor weight (I), cell proliferation (J), immunofluorescence of Ki67-positive cells (K), and SUMO3 protein expression (L). Scale bar: 100 μm. *P < 0.05, **P < 0.01, ***P < 0.001 vs. shNC
SUMO3 overexpression increased glycolysis in vitro through the β-catenin signaling pathway
Figure 3 shows the effects and mechanisms of SUMO3 overexpression on glycolysis. After overexpression vector (oeSUMO3) transduction, SUMO3 was significantly overexpressed in HPAC cells (Fig. 3A and B). β-catenin, which is a crucial functional molecule downstream of the classical Wnt signaling pathway, plays a critical role in tumorigenesis [13]. We found that SUMO3 knockdown suppressed nuclear β-catenin and promoted cytoplasmic β-catenin expression in SW1990 cells, whereas SUMO3 overexpression had the opposite effects on β-catenin expression in HPAC cells (Fig. 3C). The changes in LDHA expression were consistent with the changes in nuclear β-catenin in the SUMO3 knockdown SW1990 cells and HPAC cells with SUMO3 overexpression (Fig. 3C). To verify the involvement of β-catenin in promoting cell viability and the glycolysis effects of SUMO3, the β-catenin inhibitor XAV939 was used to stimulate HPAC cells with SUMO3 overexpression. We observed that compared with SUMO3 overexpression alone, combined XAV939 treatment reduced cell viability (Fig. 3D), ECAR (Fig. 3E), ATP production, (Fig. 3F) and lactic acid production (Fig. 3G). These results suggested that XAV939 attenuated cell viability and glycolysis in HPAC cells with SUMO3 overexpression.
SUMO3 overexpression enhances glycolysis in vitro through the β-catenin signaling pathway. SUMO3 overexpression in HPAC cells is constructed by overexpression lentivirus transduction, and the SUMO3 expression is evaluated by qRT-PCR (A) and Western blot (B). Western blot shows the expressions of nuclear and cytoplasmic β-catenin and LDHA in SUMO3 knockdown SW1990 cells and HPAC cells with SUMO3 overexpression (C). HPAC cells with SUMO3 overexpression that are treated alone or in combination with 10 µmol/L of the β-catenin inhibitor XAV939 for 48 h are collected for analysis of cell viability (D), ECAR (E), ATP (F), and cell supernatant lactic acid level (G). *P < 0.05, **P < 0.01, ***P < 0.001 vs. oeNC or oeNC + Vehicle. #P < 0.05, ##P < 0.01, ###P < 0.001 vs. oeSUMO3 + Vehicle
SUMO3 regulated the antitumor activity of GEM
Figure 4 illustrates the role of SUMO3 in the sensitivity of cancer cells to GEM chemotherapy. SUMO3 was expressed at different levels in pancreatic cancer tissue microarray solid tumors under different chemotherapy efficacy (Fig. 4A). In particular, pancreatic cancer tissue microarray with high sensitivity (CR + PR) exhibited low SUMO3 expression (Fig. 4B). In the in vitro GEM exposure experiment, the viability of SW1990 and HPAC cells gradually decreased with increasing doses of GEM and decreased to < 50% when the GEM concentration reached 100 µM (Fig. 4C). After exposure to 10 and 20 µM of GEM, apoptosis was significantly increased in the shSUMO3#1 knockdown SW1990 cells (Fig. 4D and E) and decreased in the HPAC cells with SUMO3 overexpression (Fig. 4F and G). Moreover, in the GEM-treated SW1990 cells, the cleaved PARP and cleaved caspase 3 levels were increased and further enhanced by shSUMO3#1 knockdown (Fig. 4H). Meanwhile, in the GEM-treated HPAC cells, the cleaved PARP and cleaved caspase 3 levels were also increased, but these changes were reversed by SUMO3 overexpression (Fig. 4I)
SUMO3 regulates the antitumor activity of GEM in vitro. SUMO3 is positive on IHC staining of a pancreatic cancer tissue microarray (A) and is low in patients who are sensitive to chemotherapy (B). The viability of SW1990 cells treated with different doses of GEM (0, 2, 5, 10, 20, 50, and 100 µM) is shown (C). Apoptosis of SW1990 cells treated with SUMO3 gene interference alone or in combination with 10 and 20 µM of GEM is shown (D, E). Apoptosis of HPAC cells treated with SUMO3 overexpression alone or in combination with 10 and 20 µM of GEM is shown (F, G). Expression levels of cleaved PARP and cleaved caspase 3 in SW1990 cells treated with SUMO3 gene interference alone or in combination with 10 and 20 µM of GEM are shown (H). Expressions of cleaved PARP and cleaved caspase 3 in HPAC cells treated with SUMO3 overexpression alone or in combination with 10 and 20 µM of GEM are shown (I). *P < 0.05, ***P < 0.001 vs. CR + PR, shNC + Vehicle or oeNC + Vehicle. ###P < 0.001 vs. oeSUMO3 + Vehicle.
In vivo xenograft experiments showed that the tumor growth, size, and weight of GEM-treated HPAC cell xenografts with SUMO3 overexpression were superior to those of GEM-treated xenografts but inferior to those of HPAC cell xenografts with SUMO3 overexpression alone (Fig. 5A C), suggesting that SUMO3 overexpression reduced the sensitivity of cells to GEM. On the xenograft tissue, the cleaved PARP and cleaved caspase 3 levels and the TUNEL-positive cells induced by GEM were attenuated by SUMO3 overexpression (Fig. 5D F). Moreover, we found that inhibition of glycolysis by 2-DG treatment increased the antitumor activity of GEM in SW1990 cells (Figure S1). These data suggested that SUMO3 enhanced the resistance of pancreatic cancer cells to GEM, and the mechanism was likely related to glycolysis.
SUMO3 regulates the antitumor activity of GEM in vivo. HPAC cells with SUMO3 overexpression are transplanted subcutaneously into nude mice, and 50 mg/kg of GEM is injected intraperitoneally after tumor formation. Tumor growth is evaluated by the tumor growth curve (A), tumor size (B), tumor weight (C), expressions of cleaved PARP and cleaved caspase 3 (D), and TUNEL staining (E, F). Scale bar: 100 μm. **P < 0.01, ***P < 0.001 vs. oeNC + Vehicle. ###P < 0.001 vs. oeSUMO3 + Vehicle
SUMO3 overexpression inhibited BA antitumor activity in vitro
Figure 6 illustrates the effects of SUMO3 overexpression on the antitumor activity of BA in vitro. Using the median SUMO3 expression as the cutoff, 30 pancreatic cancer tissues were divided into the low and high SUMO3 expression groups; the intestinal probiotic Faecalibacterium prausnitzii and Bifidobacterium contents were lower in the latter (Fig. 6A and C). BA is a short-chain fatty acid that is generated by gut microbiota and has been proven to function as an antitumor agent [14]. We observed that BA inhibited cell viability (Fig. 6D) and SUMO3 expression (Fig. 6E and F) in SW1990 cells in a dose-dependent manner. At a dose of 10 mM, BA inhibited the viability of SW1990 cells in a time-dependent manner and attenuated the protective effect of SUMO3 overexpression on cell viability (Fig. 6G). Moreover, 10 mM BA reduced ECAR (Fig. 6H), ATP content (Fig. 6I), and lactic acid content (Fig. 6J) in the SW1990 cells with SUMO3 overexpression.
SUMO3 overexpression inhibits BA antitumor activity in vitro. SUMO3 expression in 30 pancreatic cancer tumor tissues is detected by qRT-PCR (A). The presence of Faecalibacterium Prausnitzii (B) and Bifidobacterium (C) in the feces of SUMO3-high and SUMO3-low groups of patients with pancreatic cancer is detected by qRT-PCR. SW1990 cells exposed to different BA doses (0, 2, 5, 10, 20, and 50 mM) are assessed for cell viability (D), SUMO3 mRNA expression (E), and SUMO3 protein expression (F). SW1990 cells treated with SUMO3 overexpression alone or in combination with 10 mM BA treatment for 48 h were collected for analysis of cell viability (G), ECAR (H), ATP (I), and cell supernatant lactate level (J). *P < 0.05, **P < 0.01, ***P < 0.001 vs. oeNC + Vehicle. #P < 0.05, ##P < 0.01, ###P < 0.001 vs. oeNC + BA
BA inhibited SUMO3 expression by inactivating the IκBα/NF-κB p65 signaling pathway
The transcriptional activity of NF-κB can be stimulated by phosphorylated p65, which is a subunit of NF-κB, and suppressed by IκBα, which is a membrane of the IκB inhibitor family that binds to and suppresses NF-κB transcriptional activity in the cytoplasm. We examined the influence of BA on the transcriptional activity of NF-κB. Figure 7 shows the effect and mechanism of BA on SUMO3 expression. Western blot assay suggested that BA promoted total and phosphorylated IκBα expression but suppressed phosphorylated NF-κB p65 expression (Fig. 7A), suggesting that BA inhibited NF-κB activity by increasing IκBα expression. Histones are structural elements of chromatin, and their acetylation is essential for gene transcription initiation. In the BA-treated SW1990 cells, we observed increased histone acetylation levels of H3K9, H3K14, and H3K27 (Fig. 7B). To study the effects of H3 acetylation on histone binding to the IκBα chromatin, we performed ChIP assay using three antibodies against H3K9ac, H3K14ac, and H3K27ac. Analysis of the data demonstrated significant enrichment of acetylated H3 in the IκBα promoter (Fig. 7C-6E), indicating that hyperacetylation promoted histone H3 binding to the IκBα promoter. Luciferase assay revealed that NF-κB p65 overexpression increased SUMO3 promoter activity (Fig. 7F). Moreover, the ChIP assay revealed that the SUMO3 promoter was enriched after NF-κB p65 immunoprecipitation (Fig. 7G). These data demonstrated that activation of the IκBα/NF-κB p65 signaling pathway modulated SUMO3 expression in BA-treated pancreatic cancer cells.
BA promotes SUMO3 expression through the IκBα/NF-κB p65 signaling pathway. Western blot of SW1990 cells treated with 10 mM BA shows the IκBα and NF-κB p65 expressions (A) and histone acetylation levels (B). ChIP-PCR shows the binding of (C) H3K9ac, (D) H3K14ac, and (E) H3K27ac with the IκBα promoter. SUMO3 promoter activity is evaluated using a luciferase reporter assay through cotransfection of the WT or mutant SUMO3 promoter vectors with the NF-κB p65 overexpression plasmid in the SW1990 cell line (F). NF-κB p65 binding and the SUMO3 promoter are evaluated by ChIP-PCR (G). ***P < 0.001 vs. oeNC or anti-IgG
Discussion
SUMOylation mediates several processes responsible for cancer cell adaptation and survival, but the specific molecular mechanism is unclear. Elucidation of the molecular functions of the SUMOylation pathway will help to understand the mechanism of SUMOylation in tumorigenesis and identify new therapeutic targets. In this study, we comprehensively demonstrated the potential of SUMO3 to act as a therapeutic target molecule, owing to its positive effects on cancer cell survival and growth, even after exposure of cancer cells to GEM and BA.
Glycolysis is the dominant metabolic profile in cancer cells that have undergone metabolic reprogramming and affords a survival advantage for cancer cells, making the tumor environment more conducive to carcinogenesis [15]. Increased glycolysis promotes cancer progression, chemotherapy resistance to GEM, and metastasis [16, 17]. Therefore, targeting glycolysis is a promising strategy for tumor suppression. However, only few strategies targeting cancer cell glycolysis have been effective in enhancing treatment outcomes. Based on our results on the changes in ECAR, ATP, LDHA, and lactic acid levels in response to SUMO3 overexpression or knockdown, SUMO3 is a potential target for controlling glycolysis in cancer cells. Lactic acid, which had been traditionally considered a metabolic waste product of glycolysis, has regained scientific interest since the discovery of the Warburg effect in tumor cells [18]. LDHA is the rate-limiting enzyme that converts glucose-derived pyruvate to lactate and NAD + in the final step of glycolysis. Considering that LDHA is a crucial biological molecule involved in glucose metabolism, its inhibition may help reverse glucose metabolism from glycolysis to oxidative phosphorylation in the cytoplasm, thereby, inducing cancer cell death [19]. Given its critical role in tumor cell survival, growth, and glycolysis, SUMO3 is a potential therapeutic target.
Nuclear β-catenin serves as a coactivator of the TCF/LEF family of transcription factors in the nucleus and functions on cell proliferation by encoding cycle proteins. Abnormal β-catenin activation represents tumor progression or chemotherapy resistance, which is reflected in various solid cancers, such as prostate, ovarian, and colon [20,21,22], as well as pancreatic cancer [23]. The nuclear β-catenin protein level is closely regulated at multiple levels, including its stability, subcellular localization, and transcriptional activity. SUMOylation has been shown to be involved in regulating the intracellular level and activity of β-catenin by directly modifying β-catenin or indirectly modifying molecules that regulate the β-catenin pathway [24,25,26]. In this study, changes in nuclear β-catenin expression with SUMO3 upregulation or downregulation indicated that the upregulated activity of the Wnt/β-catenin pathway mediated the cancer-promoting effect of SUMO3 in pancreatic cancer.
GEM resistance has been reported to be a poor prognostic factor in pancreatic cancer, and some studies have attempted to identify new targets to enhance the sensitivity of pancreatic cancer cells to GEM [27, 28]. Notably, SUMOylation has been shown to contribute to drug resistance, based on the profound changes in the SUMOylation profiles of GEM-resistant pancreatic cancer cells [2]. However, the specific mechanism underlying the association between GEM resistance and SUMOylation in pancreatic cancer cells is unclear. In our study, patients with high SUMO3 expression exhibited resistance to chemotherapy. Moreover, the effects of the changes in SUMO3 expression on cancer cell viability and growth suggested that SUMO3 was a potential target to regulate the sensitivity of pancreatic cancer cells to GEM.
Gut microbiota is involved in cancer progression by affecting metabolism and immunity, and this effect has been reported in pancreatic cancer [29]. Among them, probiotics appear to have anticancer effects. In fact, some reports have confirmed that Faecalibacterium Prausnitzii and Bifidobacterium suppressed tumor progression and reduced chemotherapy resistance and intestinal adverse effects by enhancing immunity [30, 31]. BA is a metabolite of the gut microbiota, such as Bifidobacterium, and plays a key role in maintaining intestinal health. It is an active antitumor molecule that restricts the growth and metastasis of pancreatic cancer [14, 32]. In this study, the reduced Faecalibacterium Prausnitzii and Bifidobacterium content in patients with high SUMO3 expression and inhibition of SUMO3 expression by BA suggested that SUMO3 was involved in gut microbiota dysbiosis in pancreatic cancer. Previous studies have observed that gut microbiota and its products regulate host inflammatory responses by influencing SUMOylation [33, 34]. The correlation between gut microbiota and SUMOylation in cancer progression are preliminary results that need to be further explored.
A significant finding of our study was that NF-κB p65 was enriched and promoted the transcriptional activity of the SUMO3 promoter. NF-κB acts as a critical transcription factor for controlling the immune microenvironment and cell survival and contributes to cancer progression and chemotherapy resistance [35]. Inducible phosphorylation of NF-κB p65 is considered a key mechanism in the positive regulation of NF-κB activity in cancer [36, 37]. Our experiments showed that BA treatment inhibited NF-κB p65 phosphorylation by promoting IκBα expression, implying that the antitumor activity of BA was mediated by inhibition of IκBα/NF-κB transcriptional activity. This phenomenon has also been demonstrated in other tumors [38, 39]. SUMOylation has been commonly found to promote or inhibit NF-κB pathway activation by acting indirectly or directly on different NF-κB transcription factors under different physiological and pathological conditions [40, 41]. However, little is known about the effects of NF-κB transcriptional activity on SUMO3 expression. Overall, understanding the relevance of NF-κB and SUMO3 expression can help deepen our comprehension of the pathological mechanisms of cancer progression and chemotherapy resistance in pancreatic cancer.
In conclusion, the present study highlighted the role of SUMO3 in regulating glycolysis, GEM sensitivity, and the antitumor activity of BA in pancreatic cancer cells through the NF-κB/SUMO3/β-catenin signaling axis. Our findings showed the potential application of SUMO3 as a therapeutic target for pancreatic cancer.
Data availability
No datasets were generated or analysed during the current study.
References
Klein AP. Pancreatic cancer epidemiology: understanding the role of lifestyle and inherited risk factors. Nat Rev Gastroenterol Hepatol. 2021;18(7):493–502. https://doi.org/10.1038/s41575-021-00457-x. Epub 2021/05/19.
Swayden M, Alzeeb G, Masoud R, Berthois Y, Audebert S, Camoin L, et al. PML hyposumoylation is responsible for the resistance of pancreatic cancer. FASEB J. 2019;33(11):12447–63. https://doi.org/10.1096/fj.201901091. Epub 2019/09/27.
Sheng Z, Zhu J, Deng YN, Gao S, Liang S. SUMOylation modification-mediated cell death. Open Biol. 2021;11(7):210050. https://doi.org/10.1098/rsob.210050. Epub 2021/07/14.
Queiroz LY, Kageyama R, Cimarosti HI. SUMOylation effects on neural stem cells self-renewal, differentiation, and survival. Neurosci Res. 2023. https://doi.org/10.1016/j.neures.2023.09. Epub 2023/09/25.
Sahin U, de The H, Lallemand-Breitenbach V. Sumoylation in Physiology, Pathology and Therapy. Cells. 2022;11(5). Epub 2022/03/11. https://doi.org/10.3390/cells11050814. PubMed PMID: 35269436; PubMed Central PMCID: PMCPMC8909597.
Hu Y, Chen C, Tong X, Chen S, Hu X, Pan B, et al. NSUN2 modified by SUMO-2/3 promotes gastric cancer progression and regulates mRNA m5C methylation. Cell Death Dis. 2021;12(9):842. https://doi.org/10.1038/s41419-021-04127-3. Epub 2021/09/11.
Kumar S, Schoonderwoerd MJA, Kroonen JS, de Graaf IJ, Sluijter M, Ruano D, et al. Targeting pancreatic cancer by TAK-981: a SUMOylation inhibitor that activates the immune system and blocks cancer cell cycle progression in a preclinical model. Gut. 2022;71(11):2266–83. https://doi.org/10.1136/gutjnl-2021-324834. Epub 2022/01/26.
Luo Y, Li Z, Kong Y, He W, Zheng H, An M, et al. KRAS mutant-driven SUMOylation controls extracellular vesicle transmission to trigger lymphangiogenesis in pancreatic cancer. J Clin Invest. 2022;132(14). https://doi.org/10.1172/JCI157644. PubMed PMID: 35579947; PubMed Central PMCID: PMCPMC9282935. Epub 2022/05/18.
Biederstadt A, Hassan Z, Schneeweis C, Schick M, Schneider L, Muckenhuber A, et al. SUMO pathway inhibition targets an aggressive pancreatic cancer subtype. Gut. 2020;69(8):1472–82. https://doi.org/10.1136/gutjnl-2018-317856. Epub 2020/02/01.
Yang N, Liu S, Qin T, Liu X, Watanabe N, Mayo KH, et al. SUMO3 modification by PIAS1 modulates androgen receptor cellular distribution and stability. Cell Commun Signal. 2019;17(1):153. https://doi.org/10.1186/s12964-019-0457-9. Epub 2019/11/23.
Wu R, Fang J, Liu M, Liu AJ, Chen J. SUMOylation of the transcription factor ZFHX3 at Lys-2806 requires SAE1, UBC9, and PIAS2 and enhances its stability and function in cell proliferation. J Biol Chem. 2020;295(19):6741–53. https://doi.org/10.1074/jbc.RA119.012338. Epub 2020/04/07.
Cao L, Wu J, Qu X, Sheng J, Cui M, Liu S, et al. Glycometabolic rearrangements–aerobic glycolysis in pancreatic cancer: causes, characteristics and clinical applications. J Exp Clin Cancer Res. 2020;39(1):267. https://doi.org/10.1186/s13046-020-01765-x. Epub 2020/12/02.
Dev A Jr., Vachher M, Prasad CP. beta-catenin inhibitors in cancer therapeutics: intricacies and way forward. Bioengineered. 2023;14(1):2251696. https://doi.org/10.1080/21655979.2023.2251696. Epub 2023/09/01.
Panebianco C, Villani A, Pisati F, Orsenigo F, Ulaszewska M, Latiano TP, et al. Butyrate, a postbiotic of intestinal bacteria, affects pancreatic cancer and gemcitabine response in in vitro and in vivo models. Biomed Pharmacother. 2022;151:113163. https://doi.org/10.1016/j.biopha.2022.113163. Epub 2022/05/27.
Chelakkot C, Chelakkot VS, Shin Y, Song K. Modulating Glycolysis to Improve Cancer Therapy. Int J Mol Sci. 2023;24(3). Epub 2023/02/12. https://doi.org/10.3390/ijms24032606. PubMed PMID: 36768924; PubMed Central PMCID: PMCPMC9916680.
Dai S, Peng Y, Zhu Y, Xu D, Zhu F, Xu W, et al. Glycolysis promotes the progression of pancreatic cancer and reduces cancer cell sensitivity to gemcitabine. Biomed Pharmacother. 2020;121:109521. https://doi.org/10.1016/j.biopha.2019.109521. Epub 2019/11/07.
Yang J, Ren B, Yang G, Wang H, Chen G, You L et al. The enhancement of glycolysis regulates pancreatic cancer metastasis. Cell Mol Life Sci. 2020;77(2):305 – 21. Epub 2019/08/23. https://doi.org/10.1007/s00018-019-03278-z. PubMed PMID: 31432232.
Hu Y, He Z, Li Z, Wang Y, Wu N, Sun H, et al. Lactylation: the novel histone modification influence on gene expression, protein function, and disease. Clin Epigenetics. 2024;16(1):72. https://doi.org/10.1186/s13148-024-01682-2. Epub 2024/05/30.
Xiang S, Huang D, He Q, Li J, Tam KY, Zhang SL, et al. Development of dual inhibitors targeting pyruvate dehydrogenase kinases and human lactate dehydrogenase A: high-throughput virtual screening, synthesis and biological validation. Eur J Med Chem. 2020;203:112579. https://doi.org/10.1016/j.ejmech.2020.112579. Epub 2020/07/21.
Atawia IM, Kushwaha PP, Verma S, Lin S, Shankar E, Abdel-Gawad O, et al. Inhibition of Wnt/beta-catenin pathway overcomes therapeutic resistance to abiraterone in castration-resistant prostate cancer. Mol Carcinog. 2023;62(9):1312–24. https://doi.org/10.1002/mc.23565. Epub 2023/05/26.
Xie H, Chen J, Ma C, Zhao J, Cui M. UBP43 promotes epithelial ovarian carcinogenesis via activation of beta-catenin signaling pathway. Cell Biol Int. 2023;47(8):1427–40. https://doi.org/10.1002/cbin.12028. Epub 2023/05/15.
Iloki Assanga SB, Lewis Lujan LM, McCarty MF. Targeting beta-catenin signaling for prevention of colorectal cancer - nutraceutical, drug, and dietary options. Eur J Pharmacol. 2023;956:175898. https://doi.org/10.1016/j.ejphar.2023.175898. Epub 2023/07/23.
Chen L, Xu Z, Li Q, Feng Q, Zheng C, Du Y, et al. USP28 facilitates pancreatic cancer progression through activation of Wnt/beta-catenin pathway via stabilising FOXM1. Cell Death Dis. 2021;12(10):887. https://doi.org/10.1038/s41419-021-04163-z. Epub 2021/09/30.
Fan L, Yang X, Zheng M, Yang X, Ning Y, Gao M, et al. Regulation of SUMOylation targets Associated with Wnt/beta-Catenin pathway. Front Oncol. 2022;12:943683. https://doi.org/10.3389/fonc.2022.943683. Epub 2022/07/19.
Liang S, Zhou Z, Zhou Z, Liang J, Lin W, Zhang C et al. Blockade of CBX4-mediated beta-catenin SUMOylation attenuates airway epithelial barrier dysfunction in asthma. Int Immunopharmacol. 2022;113(Pt A):109333. Epub 2022/10/29. https://doi.org/10.1016/j.intimp.2022.109333. PubMed PMID: 36306558.
Li W, Han Q, Zhu Y, Zhou Y, Zhang J, Wu W, et al. SUMOylation of RNF146 results in Axin degradation and activation of Wnt/beta-catenin signaling to promote the progression of hepatocellular carcinoma. Oncogene. 2023;42(21):1728–40. https://doi.org/10.1038/s41388-023-02689-4. Epub 2023/04/08.
Jiang X, Ma Y, Wang T, Zhou H, Wang K, Shi W, et al. Targeting UBE2T potentiates Gemcitabine Efficacy in Pancreatic Cancer by regulating pyrimidine metabolism and replication stress. Gastroenterology. 2023;164(7):1232–47. https://doi.org/10.1053/j.gastro.2023.02.025. Epub 2023/02/27.
Yang G, Guan W, Cao Z, Guo W, Xiong G, Zhao F, et al. Integrative Genomic Analysis of Gemcitabine Resistance in Pancreatic Cancer by patient-derived xenograft models. Clin Cancer Res. 2021;27(12):3383–96. https://doi.org/10.1158/1078-0432.CCR-19-3975. Epub 2021/03/07.
Li Q, Jin M, Liu Y, Jin L. Gut microbiota: its potential roles in pancreatic Cancer. Front Cell Infect Microbiol. 2020;10:572492. https://doi.org/10.3389/fcimb.2020.572492. Epub 2020/10/30.
Dikeocha IJ, Al-Kabsi AM, Chiu HT, Alshawsh MA. Faecalibacterium prausnitzii Ameliorates Colorectal Tumorigenesis and Suppresses Proliferation of HCT116 Colorectal Cancer Cells. Biomedicines. 2022;10(5). Epub 2022/05/29. https://doi.org/10.3390/biomedicines10051128. PubMed PMID: 35625865; PubMed Central PMCID: PMCPMC9138996.
Sobocki BK, Kazmierczak-Siedlecka K, Folwarski M, Hawrylkowicz V, Makarewicz W, Stachowska E. Pancreatic Cancer and Gut Microbiome-Related Aspects: A Comprehensive Review and Dietary Recommendations. Nutrients. 2021;13(12). Epub 2021/12/29. https://doi.org/10.3390/nu13124425. PubMed PMID: 34959977; PubMed Central PMCID: PMCPMC8709322.
Farrow B, Rychahou P, O’Connor KL, Evers BM. Butyrate inhibits pancreatic cancer invasion. J Gastrointest Surg. 2003;7(7):864 – 70. Epub 2003/11/01. https://doi.org/10.1007/s11605-003-0031-y. PubMed PMID: 14592659.
Ezzine C, Loison L, Montbrion N, Bole-Feysot C, Dechelotte P, Coeffier M, et al. Fatty acids produced by the gut microbiota dampen host inflammatory responses by modulating intestinal SUMOylation. Gut Microbes. 2022;14(1):2108280. PubMed PMID: 35978476; PubMed Central PMCID: PMCPMC9466625.
Dalmasso G, Cougnoux A, Delmas J, Darfeuille-Michaud A, Bonnet R. The bacterial genotoxin colibactin promotes colon tumor growth by modifying the tumor microenvironment. Gut Microbes. 2014;5(5):675–80. https://doi.org/10.4161/19490976.2014.969989. Epub 2014/12/09.
Deka K, Li Y. Transcriptional Regulation during Aberrant Activation of NF-kappaB Signalling in Cancer. Cells. 2023;12(5). Epub 2023/03/12. https://doi.org/10.3390/cells12050788. PubMed PMID: 36899924; PubMed Central PMCID: PMCPMC10001244.
Valovka T, Hottiger MO. p65 controls NF-kappaB activity by regulating cellular localization of IkappaBbeta. Biochem J. 2011;434(2):253–63. https://doi.org/10.1042/BJ20101220. Epub 2010/12/17. PubMed PMID: 21158742.
Mirzaei S, Saghari S, Bassiri F, Raesi R, Zarrabi A, Hushmandi K, et al. NF-kappaB as a regulator of cancer metastasis and therapy response: a focus on epithelial-mesenchymal transition. J Cell Physiol. 2022;237(7):2770–95. https://doi.org/10.1002/jcp.30759. Epub 2022/05/14.
Schwab M, Reynders V, Loitsch S, Steinhilber D, Stein J, Schroder O. Involvement of different nuclear hormone receptors in butyrate-mediated inhibition of inducible NF kappa B signalling. Mol Immunol. 2007;44(15):3625–32. https://doi.org/10.1016/j.molimm.2007.04.010. Epub 2007/05/25.
Luhrs H, Gerke T, Schauber J, Dusel G, Melcher R, Scheppach W, et al. Cytokine-activated degradation of inhibitory kappaB protein alpha is inhibited by the short-chain fatty acid butyrate. Int J Colorectal Dis. 2001;16(4):195–201. https://doi.org/10.1007/s003840100295. Epub 2001/08/23.
Li W, Qiao J, You Q, Zong S, Peng Q, Liu Y, et al. SARS-CoV-2 Nsp5 activates NF-kappaB pathway by upregulating SUMOylation of MAVS. Front Immunol. 2021;12:750969. https://doi.org/10.3389/fimmu.2021.750969. Epub 2021/12/04.
Hegde S, Sreejan A, Gadgil CJ, Ratnaparkhi GS. SUMOylation of Dorsal attenuates Toll/NF-kappaB signaling. Genetics. 2022;221(3). Epub 2022/05/15. https://doi.org/10.1093/genetics/iyac081. PubMed PMID: 35567478; PubMed Central PMCID: PMCPMC9252280.
Acknowledgements
Not applicable.
Funding
This study was supported by the National Natural Science Foundation of China (82074204).
Author information
Authors and Affiliations
Contributions
LZ, GC and YW organized the data and drafted the manuscript. CH, HG, YW and YS: performed experiments and analyzed the data. LZ and YS revised manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Experimental procedures related to human samples had been given permission by the medical ethics committee of the Fudan University Shanghai Cancer Center (approval number 050432-4-2108) and were operated in accordance with the Declaration of Helsinki. Animal studies were authorized by the Ethics Committee of Fudan University (approval number 2020 Shanghai Cancer Center JS-213).
Consent for publication
All authors consent to submit the manuscript for publication.
Conflict of interest
The authors have no conflict of interest to declare.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhu, L., Chen, G., Huang, C. et al. SUMO3 inhibition by butyric acid suppresses cell viability and glycolysis and promotes gemcitabine antitumor activity in pancreatic cancer. Biol Direct 19, 74 (2024). https://doi.org/10.1186/s13062-024-00513-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13062-024-00513-x