Abstract
Background
Breast cancer (BC) is a great clinical challenge because of its aggressiveness and poor prognosis. Zinc Finger Protein 64 (ZFP64), as a transcriptional factor, is responsible for the development and progression of cancers. This study aims to investigate whether ZFP64 regulates stem cell-like properties and tumorigenesis in BC by the glycolytic pathway.
Results
It was demonstrated that ZFP64 was overexpressed in BC specimens compared to adjacent normal tissues, and patients with high ZFP64 expression had shorter overall survival and disease-free survival. The analysis of the association of ZFP64 expression with clinicopathological characteristics showed that high ZFP64 expression is closely associated with N stage, TNM stage, and progesterone receptor status. Knockdown of ZFP64 suppressed the viability and colony formation capacity of BC cells by CCK8 and colony formation assays. The subcutaneous xenograft models revealed that ZFP64 knockdown reduced the volume of formatted tumors, and decreased Ki67 expression in tumors. The opposite effects on cell proliferation and tumorigenesis were demonstrated by ZFP64 overexpression. Furthermore, we suggested that the stem cell-like properties of BC cells were inhibited by ZFP64 depletion, as evidenced by the decreased size and number of formatted mammospheres, the downregulated expressions of OCT4, Nanog, and SOX2 proteins, as well as the reduced proportion of CD44+/CD24− subpopulations. Mechanistically, glycolysis was revealed to mediate the effect of ZFP64 using mRNA-seq analysis. Results showed that ZFP64 knockdown blocked the glycolytic process, as indicated by decreasing glycolytic metabolites, inhibiting glucose consumption, and reducing lactate and ATP production. As a transcription factor, we identified that ZFP64 was directly bound to the promoters of glycolysis-related genes (ALDOC, ENO2, HK2, and SPAG4), and induced the transcription of these genes by ChIP and dual-luciferase reporter assays. Blocking the glycolytic pathway by the inhibition of glycolytic enzymes ENO2/HK2 suppressed the high proliferation and stem cell-like properties of BC cells induced by ZFP64 overexpression.
Conclusions
These data support that ZFP64 promotes stem cell-like properties and tumorigenesis of BC by activating glycolysis in a transcriptional mechanism.
Similar content being viewed by others
Introduction
Breast cancer (BC) is a substantial worldwide health burden in women with high mortality rates [1, 2]. Given its heterogeneous characteristics and limited therapy options, BC patients remain to have poor prognosis [2]. Therefore, it is extremely urgent to understand the mechanisms and identify novel therapeutic targets for BC.
Cancer stem cells, a small population of tumor cells with stem cell-like properties, are characterized to be capable of self-renewal and pluripotent differentiation [3]. It is known that cancer stem cells are identified in numerous solid tumors, such as BC [4]. The presence of cancer stem cells is the major cause of tumor initiation, relapse, metastasis, and therapy resistance [5, 6]. However, the underlying mechanisms of maintaining stem cell-like properties are still poorly illustrated.
To our knowledge, metabolic reprogramming is an emerging hallmark of tumorigenesis, which includes multiple metabolic pathways, such as glycolysis, tricarboxylic acid cycle (TCA), and oxidative phosphorylation (OXPHOS) [7, 8]. Cancer cells preferentially undergo the glycolysis pathway, also named the Warburg effect, to support cell growth and malignancies [9]. For example, Park et al. demonstrate that inhibiting glycolysis genetically or pharmacologically suppresses BC tumorigenesis and metastasis [10]. It also highlights the significant role of glycolysis in maintaining stem cell-like properties, self-renew, and chemoresistance [11]. The work by Prokakis et al. reports that the glycolytic program supports the maintenance of stem cell-like features in BC [12]. Thus, it indicates that targeting glycolytic metabolism is an important mechanism for tumorigenesis and stem cell-like properties in BC.
Zinc Finger Protein 64 (ZFP64), also known as ZNF338, is a member of the Kruppel-like C2H2-type zinc finger family that functions as a transcription activator [13]. The previous study suggests that ZFP64 activates the mixed lineage leukemia (MLL) gene expression by binding to its promoter, implying the potential role of ZFP64 in MLL development [14]. In gallbladder cancer, it is shown that ZFP64 is required for proliferation, anti-apoptosis, migration, and invasion [15]. Wei et al. propose that ZFP64 promotes tumor progression through driving immune escape in hepatocellular carcinoma. Furthermore, evidence supports that ZFP64 plays an important role in facilitating stem cell-like properties and immunosuppression in gastric cancer [16]. However, the role of ZFP64 in BC remains to be clarified.
Based on the publicly available datasets [17, 18], we found a significantly high expression of ZFP64 in BC. The glycolysis pathway was demonstrated to be associated with the biological function of ZFP64 in BC via our mRNA-seq. Therefore, we speculated that ZFP64 might support stem cell-like properties and tumorigenesis in BC via the glycolysis program.
Materials and methods
Clinical samples
This work was performed based on the Declaration of Helsinki, and approved by the ethical committee of Dalian Municipal Central Hospital. All BC patients had written the informed consent. The fresh tumor tissues (n = 43) and adjacent normal tissues (n = 43), were collected for mRNA assessment. The paraffin-embedded tumor tissues from 126 BC patients were used for immunohistochemistry (IHC). ZFP64 expression level was scored semi-quantitatively as previously described [19]. The IHC score ≤ 4 was defined as a low ZFP64 expression, while the IHC score > 4 was defined as a high ZFP64 expression.
Cell culture and transfection
Human BC cell lines MCF-7 and MDA-MB-468 were purchased from Icellbioscience (Shanghai, China), and these cell lines were authenticated by STR identification (data in Supplementary Material 2). MCF-7 cells were cultured in the Minimum Essential Medium (MEM; Cat# G4550, Servicebio, Wuhan, China) supplemented with 10% fetal bovine serum (FBS; Cat# 11011 − 8611, TianHang, Huzhou, China). MDA-MB-468 cells were cultured in an L15 medium (Cat# LA9510, Solarbio, Beijing, China) containing 10% FBS. All cell lines were incubated in a humidified atmosphere with 5% CO2 at 37 °C.
To explore the effect of ZFP64, the overexpression plasmids and shRNA plasmids targeting ZFP64 were prepared to be transfected into MCF-7 and MDA-MB-468 cell lines. G418 (Cat# BS150, Biosharp Life Sciences, Hefei, China) was performed to select stably transfected cells. To block the glycolytic process, shRNA plasmids targeting ENO2 or HK2 were transfected into MCF-7 cells. Lipofectamine 3000 (Cat# L3000015, Invitrogen, Carlsbad, CA, USA) was used to mediate plasmid transfections.
Animal studies
Six-week-old female BALB/c nude mice were purchased from Cavens (Changzhou, China). All animal experiments were done under the Guide for the Care and Use of Laboratory Animals and the approval of the ethics committee of Dalian Municipal Central Hospital. MCF-7 and MDA-MB-468 cells (1 × 106 cells/mice) were inoculated into the mammary fat pads subcutaneously. Tumor volume was measured every 5 days with calipers, and calculated as the formula Volume=(length×width×width)/2.
Immunohistochemistry (IHC)
IHC analysis was performed to determine ZFP64 levels in tumors of BC patients and Ki67 expression in xenograft tumors. Briefly, the sections were dewaxed in xylene and hydrated in ethanol. The endogenous peroxidase was quenched with the 3% H2O2 solution. After blocking with 1% BSA, the slides were incubated with anti-ZFP64 (Cat# DF15064, Affinity, Changzhou, China) or anti-Ki67 (Cat# 28074-1-AP, Proteintech, Wuhan, China) primary antibodies at 4 °C overnight. After washing, the tissues were incubated with HRP-conjugated goat anti-rabbit antibody (Cat# D110058, Sangon, Shanghai, China) for 60 min at room temperature. Finally, the sections were developed with DAB solution (Cat# DA1016, Solarbio) and counterstained with hematoxylin (Cat# H8070, Solarbio). The images were captured using the microscope (BX53, Olympus, Tokyo, Japan).
Cell proliferation and colony formation assays
Cell viability was examined using the Cell Counting Kit-8 (CCK-8; Cat# C0037, Beyotime, Shanghai, China) following the protocols provided by the manufacturer. In brief, cells were seeded in 96-well plates, and CCK-8 reagent was added to each well for 2 h. Then the absorbance at 450 nm was recorded on a microplate reader (Cat# 800TS, BioTek, Winooski, VT, USA). The cell viability analysis consisted of five technical replicates per sample and three independent biological replicates.
For colony formation assay, cells were plated and cultured for 2 weeks. After fixation in 4% paraformaldehyde, cells were stained with the Wright’s-Giemsa staining solution (Cat# KGA227, KeyGEN BioTech, Nanjing, China) for 5 min. Finally, the IX53 microscope (Olympus) was used to count colonies. The colony formation rate was calculated as the formula: colony formation rate = clone counts/seeded cell counts×100%. Three independent experiments were performed in the colony formation assay.
Sphere formation
For mammosphere assay, cells were seeded at ultralow-attachment plates supplemented with B27 (1:50, Cat# B917514, MACKLIN, Shanghai, China), 20 ng/mL EGF (Cat# H980384, MACKLIN) and 10 ng/mL bFGF (Cat# RP01042, ABclonal, Wuhan, China). After 10 days, the mammospheres were observed at the magnification of 100× using the microscope (IX53, Olympus), and harvested for flow cytometry and Western blot assays. The spheres above 50 μm in diameter were counted. Three independent experiments were used in the mammosphere assay.
Flow cytometry
To identify the mammosphere features, mammosphere cells were stained with anti-CD44 (Cat# F1104403, Multi Sciences, Hangzhou, China) or anti-CD24 (Cat# 12-0247-41, ThermoFisher Scientific, Pittsburgh, PA, USA) for 30 min at 4 °C. The NovoCyte flow cytometer (Agilent, Santa Clara, CA, USA) was used to measure CD44+/CD24− subpopulations, and the data were analyzed using NovoExpress software (1.4.1 version; Aligent). The flow cytometry analysis was carried out with three independent experiments.
mRNA-seq analysis
To profile the differentially expressed genes (DEGs), total RNA was extracted from MCF-7 cells transfected with ZFP64 overexpression or Empty plasmids. Sequencing libraries were generated and index codes were added to attribute sequences to each sample. The raw data were recorded in FASTQ format and data quality was evaluated. Genes with the thresholds of |log2FC| ≥ 1.0 and p < 0.05 were identified as DEGs. The DEGs list was shown in Supplementary Material 1-Table S1.
Detection of glycolytic metabolites
The concentrations of glycolytic metabolites, including glucose-6-phosphate (G-6-P; Cat# E-BC-K011-M, Elabscience, Houston, TX, USA), 2-phosphoglycerate (2-PG; Cat# Ab174097, Abcam, Cambridge, UK), pyruvate (Cat# R22024, YuanYe, Shanghai, China), and ATP (Cat# S0026, Beyotime) were determined using the commercial detection kits according to the manufacturer’s instructions. All assays were performed with three independent experiments.
Measurement of glucose consumption and lactate production
To determine glucose consumption and lactate production, MCF-7 and MDA-MB-468 cells were seeded into the culture plates. Forty-eight hours later, glucose and lactate contents were analyzed using the Glucose assay kit (Cat# F006, JianCheng, Nanjing, China) and the Lactate Colorimetric Assay kit (Cak# A019, JianCheng) following the manufacturer’s instructions, respectively. Glucose consumption was calculated by subtracting the measured amount of glucose in the medium from the original amount of glucose. The produced lactate was assessed by subtracting the original lactate content in the medium from the measured lactate content. Experiments were performed three times independently.
Chromatin immunoprecipitation (ChIP)
Cells were cross-linked with 1% formaldehyde and stopped with glycine. After washing with cold PBS containing PMSF, cells were lysed in SDS Lysis buffer from a ChIP Assay Kit (P2078, Beyotime) and sonicated to obtain chromatin fragments. Subsequently, the samples were incubated with anti-ZFP64 antibody (Cat# 17187-1-AP, Proteintech) or control IgG antibody (Cat# A7001, Beyotime) overnight at 4 °C, and were added with Protein A + G Agarose/Salmon Sperm DNA to immunoprecipitate DNA-protein complexes. Finally, the qRT-PCR reaction was performed to determine the immunoprecipitated DNA fragments. The primers employed in ChIP assay were listed in Table 1.
Dual-luciferase reporter assay
To determine the transcription activity of target genes, the pGL3 luciferase reporter plasmids containing the promoter (-1347 ~ + 42 bp) of potential targets (including ALDOC, ENO2, HK2, and SPAG4) were co-transfected into MCF-7 cells with ZFP64 overexpression or Empty plasmids, respectively. After 48 h of transfection, cells were harvested to detect the luciferase activity using the Dual-Luciferase Reporter Assay Kit (Cat# KGE3302, KeyGEN BioTech). The relative luciferase activity was calculated as the ratio of the firefly to the renilla luminescence activity. The analysis was performed with three independent experiments.
Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from cells using the TRIpure reagent (Cat# RP1001, BioTeke, Beijing, China), and RNA purity was checked using A260/A280 and A260/A230 ratios (mean A260/A280 = 1.94, mean A260/230 = 2.12). The RNA was reverse-transcribed into cDNA using the All-in-One First-Strand SuperMix reverse transcription kit (Cat# MD80101, Magen Biotech, Guanzhou, China). Then, the qRT-PCR reaction was conducted in a Real-Time PCR system instrument (Cat# Exicycler96, Bioneer, Daejeon, Korea) with the SYBR Green PCR reagent (Cat# SR4110, Solarbio). The melting curve analysis demonstrated a signal melting curve peak for all primers. The relative mRNA expression was calculated using the 2−∆∆Ct method. β-actin was used as an internal control. The primers used were shown in Table 2. All analyses were undergone for three technical replicates and three biological replicates.
Western blot
Total protein was extracted from tumor tissues and cell lines using the Cell Lysis Buffer (Cat# P0013, Beyotime), and the protein concentration was quantified in a BCA Kit (Cat# P0011, Beyotime). Then, the equal proteins were loaded onto homemade 10% or 12% separation SDS-PAGE gels. After electrophoresis, the proteins were transferred to the PVDF membrane (Cat# Ab133411, Abcam). Membranes were blocked with the Blocking Buffer (Cat# P0023, Beyotime) for 60 min at room temperature, and then incubated with primary antibodies overnight at 4 °C. The target proteins were detected using the HRP-conjugated secondary antibodies for 45 min at 37 °C and were visualized with the ECL kit (Cat# P0018, Beyotime), followed by film exposure. β-actin was used as an internal control. All antibodies used were listed in Table 3. Western blot analysis was performed with three independent experiments in vitro and four independent experiments in vivo.
Statistical analysis
All data were presented as mean ± SD and analyzed using Graph Pad Prism 9.0 software. Two-tailed t-test (Unpaired or paired, as appropriate) was used to evaluate the differences between the two groups. For comparisons among multiple groups, one-way ANOVA followed by the Bonferroni post hoc test was performed. The association between clinicopathological variables and ZFP64 expression was analyzed using χ2 test. P values less than 0.05 were identified to be significant.
Results
ZFP64 is highly expressed in BC
Based on the TIMER platform [17], ZFP64 expression was highly expressed across various tumors, such as bladder urothelial carcinoma (BLCA), breast invasive carcinoma ( BRCA ), and esophageal carcinoma (ESCA) (Fig. 1A). The TNMplot web tool [18] confirmed an increase in ZFP64 expression in BRCA tumors compared to normal tissues (Fig. 1B). Furthermore, the PrognoScan analysis (http://dna00.bio.kyutech.ac.jp/PrognoScan/index.html) revealed that high ZFP64 expression was correlated with shorter overall survival (OS) and disease-free survival (DFS) in BC patients (Fig. 1C-D). To verify the data, the tumor tissues and adjacent normal tissues from BC patients were collected for further experiments. Results showed that both the mRNA and protein levels of ZFP64 were upregulated in tumor tissues compared with adjacent normal tissues of BC patients (Fig. 1E-F). Then IHC analysis was performed to investigate the significance of ZFP64 expression in BC. Representative images with high or low ZFP64 expression were shown in Fig. 1G, and ZFP64 was found to be mainly localized to the cell nucleus in tumors. The χ2 test suggested that high ZFP64 expression was significantly associated with N stage, TNM stage, and progesterone receptor (PR) status, rather than age, T stage, estrogen receptor (ER) status, and human epidermal growth factor receptor-2 (HER-2) status (Fig. 1H). Taken together, these findings indicated high ZFP64 expression might be associated with poor prognosis in BC.
ZFP64 is highly expressed in BC. (A) Pan-cancer analysis of ZFP64 expression using the TIMER. *** p < 0.001. (B) ZFP64 expression in normal and tumor tissues of BRCA using the TNMplot. *** p < 0.001. (C-D) Kaplan-Meier plot showing the correlation of ZFP64 level with overall survival and disease-free survival of BC patients via the PrognoScan analysis. (E-F) The mRNA and protein expressions of ZFP64 in BC specimens and adjacent normal tissues were determined by qRT-PCR and Western blot assays. * p < 0.05. (G) Representative IHC staining for ZFP64 with low or high expression in tumor tissues. Scale bar: 100 μm. (H) Association of ZFP64 expression with clinicopathological features of BC patients. * p < 0.05
ZFP64 promotes BC cell proliferation and tumorigenesis
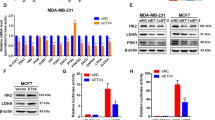
To investigate the effects of ZFP64 on BC, the gain-of-function and loss-of-function experiments were performed. As shown in Fig. 2A-B, both the mRNA and protein levels of ZFP64 were upregulated by a ZFP64-GFP fusion protein vector. The viability and colony formation ability of BC cells were increased by the ZFP64-overexpression plasmids fused with GFP (Fig. 2C-D). Similar biological functions of ZFP64 on BC cells were confirmed using the non-fusion ZFP64 overexpression vectors (Figure S1). In addition, results in Fig. 2E-F suggested the knockdown plasmids targeting ZFP64 significantly downregulated its expression. Cell viability and colonies were reduced by ZFP64 knockdown (Fig. 2G-H).
ZFP64 promotes BC cell proliferation in vitro. (A-B) Cells were transfected with GFP-fusion plasmids for ZFP64 overexpression or negative control. qRT-PCR and Western blot analyses were conducted to determine ZFP64 mRNA and protein levels. (C) CCK-8 analyzes the viability of BC cells. (D) Representative images of colony formation assay and quantification data. Scale bar: 10 mm. (E-F) Cells were transfected with ZFP64 inhibition plasmids or negative control. qRT-PCR and Western blot analyses were conducted to determine ZFP64 mRNA and protein levels. (G) CCK-8 analyzes the viability of BC cells. (H) Representative images of colony formation assay and quantification data. Scale bar: 10 mm. * p < 0.05, compared to Empty-GFP; # p < 0.05 compared to shNC
Then, the xenograft tumor models were employed using MCF-7 and MDA-MB-468 cell lines. As shown in Fig. 3A-B and Figure S2, ZFP64 overexpression increased the volume of formatted tumors, whereas ZFP64 knockdown decreased it. IHC staining showed that the expression of Ki67, a cellular marker for proliferation [20], was upregulated by ZFP64 overexpression but downregulated by ZFP64 inhibition in tumor tissues (Fig. 3C-D). Therefore, the in vitro and in vivo data demonstrated the promotive effect of ZFP64 on BC cell proliferation and tumorigenesis.
ZFP64 facilitates tumor growth in vivo. (A-B) MCF-7 or MDA-MB-468 cell lines were inoculated into the mammary fat pads of female BALB/c nude mice to establish the xenograft model. Tumor volume was recorded. (C-D) Representative IHC images of Ki67 expression in tumors. Scale bar: 50 μm. * p < 0.05, compared to Empty-GFP; # p < 0.05 compared to shNC
ZFP64 supports stem cell-like properties in BC
As tumor cells with stem cell-like properties promote tumor formation and progression, we further explore the role of ZFP64 in the maintenance of stem cell-like features. As shown in Fig. 4A, the size and number of formatted mammospheres were reduced by ZPF64 knockdown (Fig. 4A). The pluripotency factors, such as OCT4, Nanog, and SOX2, are critical regulators for stem-like cell phenotypes in cancers [21]. We identified the expression of these stem cell markers using Western blot, and results showed that ZFP64 depletion reduced the protein levels of OCT4, Nanog, and SOX2 in the mammospheres (Fig. 4B). It is well-known that CD44+/CD24− BC cells exhibited stem cell-like properties [22]. As shown in Fig. 4C, the proportion of the CD44+/CD24− subpopulations was reduced by ZFP64 knockdown. Altogether, the data suggested that ZFP64 played a positive role in the maintenance of stem cell-like characteristics of BC.
ZFP64 supports stem cell-like properties of BC cells. (A) Representative images of the formatted mammospheres and quantification of the number of mammospheres. Scale bar: 200 μm. (B) Western blot analysis of OCT4, Nanog, and SOX2 protein levels in mammospheres. (C) Flow cytometry analysis of the CD44+/CD24− cell populations in mammospheres. # p < 0.05 compared to shNC
Glycolysis is involved in the role of ZFP64 in BC through mRNA-seq analysis
To further elucidate the underlying mechanism of ZFP64 in BC, we performed mRNA-seq analysis in ZFP64-overexpression and control MCF-7 cells (Fig. 5A). As shown in Fig. 5B, 721 upregulated genes and 1006 downregulated genes were screened in cells with ZFP64 overexpression. The upregulated genes by ZFP64 were evaluated for GO, KEGG, and GSEA enrichment analysis, because ZFP64 functions as a transcription activator. Results in KEGG analysis showed that several pathways such as “MAPK signaling pathway”, “Glycolysis/Gluconeogenesis”, “Apoptosis”, and “Cell cycle” were enriched by the upregulated genes (Fig. 5C). The GO terms related to tumor development were enriched by the upregulated genes, such as “Response to hypoxia”, “Glycolytic process”, “Cell migration”, “Positive regulation of cell proliferation”, “Positive regulation of angiogenesis”, and “Gluconeogenesis” (Fig. 5D). Furthermore, based on the network cluster analysis using the GSEA of the GO dataset, we observed that ZFP64 dysregulated critical pathways in BC, such as “Canonical glycolysis”, “Response to hypoxia, “Positive regulation of T cell activation”, “ERK1 and ERK2 cascade”, and “Chemokine-mediated signaling pathway”(Fig. 5E). Importantly, we identified that the gene sets both in the GO Glycolytic process by GSEA and the KEGG Glycolysis/Gluconeogenesis by GSEA were upregulated by ZFP64 overexpression (Fig. 5F-G). It supported that the glycolytic process might be of great significance in the regulation of ZFP64 in BC.
Glycolysis is involved in the role of ZFP64 in BC through RNA-seq analysis. (A) Schematic diagram of MCF-7 cells subjected to mRNA-seq. (B) Volcano plot of the differentially expressed genes. (C) Column diagram showing the enriched KEGG pathway from the upregulated genes. (D) Chord diagram showing the enriched GO terms from the upregulated genes. (E) An enrichment network generated from GSEA combined with GO analysis showing biological processes regulated by ZFP64. The red nodes represent upregulated biological pathways, the blue nodes represent downregulated biological pathways, and the blue lines represent gene overlap between pathways. (F) GSEA combined with GO analysis showing ZFP64 related to the Glycolytic process. (G) GSEA combined with KEGG analysis showing ZFP64 related to Glycolysis/Gluconeogenesis
ZFP64 promotes the glycolytic process in BC
Given that glycolysis is a key metabolic program that often modulates stem cell-like phenotypes and tumorigenesis [9, 11], we then focused on the role of ZFP64 in the glycolytic process in BC cells. As shown in Fig. 6A-C, the master metabolites produced by glycolysis, like glucose-6-phosphate (G-6-P), 2-phosphoglycerate (2-PG), and pyruvate, were demonstrated to be inhibited by ZFP64 knockdown. We further found that ZFP64 depletion blocked the glucose consumption, lactate production, as well as ATP generation of BC cells (Fig. 6D-F). These results suggested an important effect of ZFP64 on facilitating the glycolytic process of BC cells.
ZFP64 directly activated glycolysis-related genes to promote cell proliferation and stem cell-like properties
Through the mRNA-seq analysis, we identified the expression alterations of several genes involved in glycolysis by ZFP64 overexpression in MCF-7 cells (Fig. 7A). To confirm it, qRT-PCR analysis results showed that ZFP64 knockdown decreased the mRNA levels of ALDOC, ENO2, HK2, PGK1, and SPAG4, whereas it did not alter the expressions of PDK1 and PFKFB4 (Fig. 7B). As ZFP64 is a Kruppel-like C2H2 transcriptional activator, we found that among the above-mentioned glycolysis-related genes, ALDOC, ENO2, HK2, and SPAG4 might be potential downstream targets of ZFP64 through the JASPER database (Fig. 7C) [23]. ChIP combined with qRT-PCR assay confirmed that ZFP64 was directly bound to the promoter of ALDOC, ENO2, HK2, or SPAG4 (Fig. 7D). The dual-luciferase reporter assay demonstrated that the transcription activity of ALDOC, ENO2, HK2, or SPAG4 was significantly enhanced by ZFP64 overexpression (Fig. 7E). These results indicated that the glycolysis-related genes, including ALDOC, ENO2, HK2, and SPAG4, were the direct targets of ZFP64.
ZFP64 directly activates glycolysis-related genes to promote cell proliferation and stem-like properties. (A) Heatmap showing the expression profiles of glycolysis-related genes regulated by ZFP64 overexpression via mRNA-seq. (B) qRT-PCR was performed to determine the relative mRNA levels of glycolysis-related genes in cells with ZFP64 knockdown compared to control cells. (C) Representation of ZFP64 binding motifs and predicted sequences at the promoters of ALDOC, ENO2, HK2, or SPAG4 by JASPER software. (D) Relative enrichment of ZFP64 on the indicated regions (rectangles with different colors) at the promoters of ALDOC, ENO2, HK2, or SPAG4 was detected in MCF-7 cells by ChIP assay combined with qRT-PCR analysis. (E) The transcription activity of ALDOC, ENO2, HK2, or SPAG4 regulated by ZFP64 was examined by the dual-luciferase reporter assay in MCF-7 cells. (F-G) MCF-7 cells with ZFP64 overexpression were transfected with ENO2/HK2 inhibition plasmids or negative control. Relative levels of glucose uptake and lactate production were measured. (H) CCK8 assay analyzes the viability of MCF-7 cells. (I) Representative images of the formatted mammospheres and quantification of the number of mammospheres in MCF-7 cells. Scale bar: 200 μm. (J) Schematic representation of the role of ZFP64 in BC. # p < 0.05 compared to shNC; * p < 0.05 compared to anti-IgG; $ p < 0.05 compared to Empty; & p < 0.05 compared to ZFP64-GFP + shNC
Finally, as ENO2 and HK2 are key glycolysis enzymes [24, 25], the inhibitory plasmids targeting ENO2 and HK2 were employed to block the glycolytic process in MCF-7 cells. As shown in Fig. 7F-G, the knockdown of ENO2 or HK2 caused a reduction of glucose consumption and lactate production in ZFP64-overexpression cells. ZFP64-induced cell growth and sphere formation of BC cells were impeded by the inhibition of ENO2 or HK2 (Fig. 7H-I). Thus, these results suggested that ZFP64 promoted the glycolytic process to support the growth of BC cells.
Discussion
The present study demonstrated that ZFP64 was overexpressed in BC specimens compared with adjacent normal specimens. Patients with high ZFP64 expression had shorter OS and DFS than patients with low ZFP64 expression. The in vitro and in vivo experiments showed that ZFP64 facilitated cell growth and stem cell-like properties of BC cells. Mechanistically, the glycolysis program was identified to be an underlying mechanism involved in the role of ZFP64 in BC using mRNA-seq. ZFP64 knockdown reduced the glycolytic metabolites by inhibiting the glycolysis-related gene transcription. Furthermore, ENO2 and HK2, the key glycolytic enzymes, were responsible for the effect of ZFP64 on cell proliferation and stem cell-like properties. These findings illustrated that ZFP64 played an important role in stem cell-like properties and tumorigenesis by regulating glycolysis (Fig. 7J).
Zinc finger proteins (ZFPs) are the most abundant family of transcription factors involved in BC development and progression. For example, ZNF451 is reported to recruit and activate tumor-associated macrophages by facilitating CCL5 transcription in triple-negative BC [26]. Li et al. show that ZNF32 transcriptionally activates GPER to promote BC stem cell-like properties [27]. ZFP64 is a newly discovered transcription factor among ZFPs. The previous work shows that ZFP64 acts as a transcription factor to activate the oncogene MLL expression in MLL-rearranged leukemia, implying that ZFP64 is a critical transcription factor in cancer [14]. The online tool analysis suggests that ZFP64 expression is overexpressed in numerous tumors, especially in BC [17, 18]. Here, ZFP64 was upregulated in BC samples compared with adjacent normal tissues. Patients with high ZFP64 expression had a shorter survival time and poorer clinicopathological characteristics. The results in clinical indicated that ZFP64 might be a biomarker of the diagnosis of BC. The previous studies describe that ZFP64 contributes to cell proliferation, migration, and invasion in gallbladder cancer [15]. In addition, the stem cell-like phenotype of tumor cells is the predominant cause of tumor development and progression [22]. Zhu et al. demonstrate that ZFP64 promotes the stem cell-like phenotypes of gastric cancer cells [16]. Consistent with these reports, our work suggested that ZFP64 facilitated cell growth and stem cell-like properties in BC cells.
The glycolysis program is one of the main metabolic pathways in cancer that provides adequate nutrients and energy to support cancer cell proliferation, metastasis and stem cell-like phenotypes [8, 28]. To our knowledge, cancer metabolism, especially glycolysis, has become the major research hotspot. The previous study demonstrated that glycolysis participates in promoting cell proliferation and metastasis of BC [10]. Inhibiting glycolysis using 2-DG (a competitive inhibitor of G-6-P produced from glucose) blocks the stem-like states of BC cells [29]. Furthermore, our present work showed that the glycolytic pathway was significantly upregulated by ZFP64 overexpression using the mRNA-seq analysis. The mRNA-seq results also suggested that a series of glycolysis-related genes were increased in ZFP64-overexpression cells, including ALDOC, ENO2, HK2, PGK1, and SPAG4. Therefore, we made great focus on the glycolytic process to explore the underlying mechanism regulated by ZFP64. The results indicated that ZFP64 knockdown decreased glycolytic metabolites, glucose consumption, and ATP production, as well as the expressions of glycolysis-related genes. It underlined that ZFP64 could promote the glycolytic process of BC cells.
The glycolysis-related genes, including ALDOC, ENO2, HK2, PGK1, and SPAG4, have been identified to play critical roles in various cancers. For instance, the overexpression of ALDOC and ENO2 glycolytic enzymes promotes sphere formation and viability of BC cells [30]. HK2 facilitates the maintenance and self-renewal of liver cancer stem cells and promotes tumor growth [31]. Similar effects of PGK1 on stem cell-like properties and tumorigenesis were described in oral squamous cell carcinoma [32]. SPAG4 is revealed to serve as a glycolytic marker and is associated with poor prognosis in pancreatic ductal adenocarcinoma and glioblastoma [33, 34]. Thus, we further determined the molecular mechanism by ZFP64 on the expressions of glycolysis-related genes. As a transcription factor, ZFP64 is involved in the transcriptional activation of NOTCH1 target genes Hes1 and Hey1 [35]. Zhu et al. suggest that ZFP64 promotes Galetin-1 transcription to induce chemoresistance and tumor immunosuppression in gastric cancer [16]. In hepatocellular carcinoma, ZFP64 induces the transcriptional activation of CSF1 to trigger immune invasion [36]. Using the JASPER analysis, we found that ZFP64 had potential binding sites on the promoters of ALDOC, ENO2, HK2, and SPAG4, but not PGK1. ChIP and dual-luciferase reporter assay confirmed that ZFP64 was directly bound to the promoters of ALDOC, ENO2, HK2, and SPAG4, and promoted the transcription of these genes. Blocking the glycolytic pathways by the knockdown of the glycolytic enzymes ENO2 and HK2 resulted in inhibitory effects on cell proliferation and stem cell-like phenotypes induced by ZFP64. Altogether, these data proposed that ZFP64 modulated stem cell-like properties and tumorigenesis of BC through the glycolytic process in a transcriptional-dependent mechanism.
In conclusion, ZFP64 drives glycolysis-mediated stem cell-like properties and tumorigenesis of BC. Mechanistically, ZFP64 induces the transcriptional activation of several glycolysis-related genes to accelerate the glycolytic process. The discovery of ZFP64’s function may offer new insights into the diagnostic or prognostic biomarkers in BC. However, several limitations exist in this present work. For example, the enriched “Response to hypoxia”, “Positive regulation of angiogenesis”, “Chemokine-mediated signaling pathway”, and “positive regulation of T cell activation” indicates that ZFP64 is associated with the regulation of the tumor microenvironment through mRNA-seq. Furthermore, besides glycolysis, a variety of oncogenic pathways may also mediate the role of ZFP64, like the MAPK signaling pathway, Wnt signaling pathway, and ERK1/2 signaling pathway. However, whether ZFP64 affects the tumor microenvironment and underlying mechanisms in BC is still unclear. It will be illustrated in more depth for our follow-up studies.
Data availability
No datasets were generated or analysed during the current study.
References
Harbeck N, Penault-Llorca F, Cortes J, Gnant M, Houssami N, Poortmans P, et al. Breast cancer. Nat Reviews Disease Primers. 2019;5(1):66.
Siegel RL, Miller KD, Fuchs HE, Jemal A, Cancer Statistics. 2021. CA: a cancer journal for clinicians. 2021;71(1):7–33.
Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414(6859):105–11.
Najafi M, Farhood B, Mortezaee K. Cancer stem cells (CSCs) in cancer progression and therapy. J Cell Physiol. 2019;234(6):8381–95.
Fan M, Shi Y, Zhao J, Li L. Cancer stem cell fate determination: mito-nuclear communication. Cell Communication Signaling: CCS. 2023;21(1):159.
Clevers H. The cancer stem cell: premises, promises and challenges. Nat Med. 2011;17(3):313–9.
Pavlova NN, Thompson CB. The emerging Hallmarks of Cancer Metabolism. Cell Metabol. 2016;23(1):27–47.
Cantor JR, Sabatini DM. Cancer cell metabolism: one hallmark, many faces. Cancer Discov. 2012;2(10):881–98.
Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Sci (New York NY). 2009;324(5930):1029–33.
Park MK, Zhang L, Min KW, Cho JH, Yeh CC, Moon H, et al. NEAT1 is essential for metabolic changes that promote breast cancer growth and metastasis. Cell Metabol. 2021;33(12):2380–e979.
Jang H, Yang J, Lee E, Cheong JH. Metabolism in embryonic and cancer stemness. Arch Pharm Res. 2015;38(3):381–8.
Prokakis E, Jansari S, Boshnakovska A, Wiese M, Kusch K, Kramm C, et al. RNF40 epigenetically modulates glycolysis to support the aggressiveness of basal-like breast cancer. Cell Death Dis. 2023;14(9):641.
Mack HG, Beck F, Bowtell DD. A search for a mammalian homologue of the Drosophila photoreceptor development gene glass yields Zfp64, a zinc finger encoding gene which maps to the distal end of mouse chromosome 2. Gene. 1997;185(1):11–7.
Lu B, Klingbeil O, Tarumoto Y, Somerville TDD, Huang YH, Wei Y, et al. A transcription factor addiction in Leukemia imposed by the MLL promoter sequence. Cancer Cell. 2018;34(6):970–e818.
He Z, Zhong Y, Hu H, Li F. ZFP64 promotes Gallbladder Cancer Progression through recruiting HDAC1 to Activate NOTCH1 Signaling Pathway. Cancers. 2023;15:18.
Zhu M, Zhang P, Yu S, Tang C, Wang Y, Shen Z, et al. Targeting ZFP64/GAL-1 axis promotes therapeutic effect of nab-paclitaxel and reverses immunosuppressive microenvironment in gastric cancer. J Experimental Clin cancer Research: CR. 2022;41(1):14.
Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, et al. TIMER: a web server for Comprehensive Analysis of Tumor-infiltrating Immune cells. Cancer Res. 2017;77(21):e108–10.
Bartha Á, Győrffy B. TNMplot.com: a web Tool for the comparison of Gene expression in normal, Tumor and metastatic tissues. Int J Mol Sci. 2021;22(5).
Lai YH, Chen J, Wang XP, Wu YQ, Peng HT, Lin XH, et al. Collagen triple helix repeat containing-1 negatively regulated by microRNA-30c promotes cell proliferation and metastasis and indicates poor prognosis in breast cancer. J Experimental Clin cancer Research: CR. 2017;36(1):92.
Kayaselçuk F, Zorludemir S, Gümürdühü D, Zeren H, Erman T. PCNA and Ki-67 in central nervous system tumors: correlation with the histological type and grade. J Neurooncol. 2002;57(2):115–21.
Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40(5):499–507.
Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100(7):3983–8.
Rauluseviciute I, Riudavets-Puig R, Blanc-Mathieu R, Castro-Mondragon JA, Ferenc K, Kumar V, et al. JASPAR 2024: 20th anniversary of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2024;52(D1):D174–82.
Wolf A, Agnihotri S, Micallef J, Mukherjee J, Sabha N, Cairns R, et al. Hexokinase 2 is a key mediator of aerobic glycolysis and promotes tumor growth in human glioblastoma multiforme. J Exp Med. 2011;208(2):313–26.
Leonard PG, Satani N, Maxwell D, Lin YH, Hammoudi N, Peng Z, et al. SF2312 is a natural phosphonate inhibitor of enolase. Nat Chem Biol. 2016;12(12):1053–8.
Zhang Y, Wang W, Min J, Liu S, Wang Q, Wang Y, et al. ZNF451 favors triple-negative breast cancer progression by enhancing SLUG-mediated CCL5 transcriptional expression. Cell Rep. 2023;42(6):112654.
Li Y, Gong D, Zhang L, Li H, Zhang S, Zhang J, et al. Zinc finger protein 32 promotes breast cancer stem cell-like properties through directly promoting GPER transcription. Cell Death Dis. 2018;9(12):1162.
Ciavardelli D, Rossi C, Barcaroli D, Volpe S, Consalvo A, Zucchelli M, et al. Breast cancer stem cells rely on fermentative glycolysis and are sensitive to 2-deoxyglucose treatment. Cell Death Dis. 2014;5(7):e1336.
Gao R, Li D, Xun J, Zhou W, Li J, Wang J, et al. CD44ICD promotes breast cancer stemness via PFKFB4-mediated glucose metabolism. Theranostics. 2018;8(22):6248–62.
De Vitis C, Battaglia AM, Pallocca M, Santamaria G, Mimmi MC, Sacco A, et al. ALDOC- and ENO2- driven glucose metabolism sustains 3D tumor spheroids growth regardless of nutrient environmental conditions: a multi-omics analysis. J Experimental Clin cancer Research: CR. 2023;42(1):69.
Li H, Song J, He Y, Liu Y, Liu Z, Sun W, et al., et al. CRISPR/Cas9 screens reveal that Hexokinase 2 enhances Cancer Stemness and Tumorigenicity by activating the ACSL4-Fatty acid β-Oxidation pathway. Adv Sci (Weinheim Baden-Wurttemberg Germany). 2022;9(21):e2105126.
Zhang Y, Cai H, Liao Y, Zhu Y, Wang F, Hou J. Activation of PGK1 under hypoxic conditions promotes glycolysis and increases stem cell–like properties and the epithelial–mesenchymal transition in oral squamous cell carcinoma cells via the AKT signalling pathway. Int J Oncol. 2020;57(3):743–55.
Tian G, Li G, Liu P, Wang Z, Li N. Glycolysis-based genes Associated with the clinical outcome of pancreatic ductal adenocarcinoma identified by the Cancer Genome Atlas Data Analysis. DNA Cell Biol. 2020;39(3):417–27.
Zhang C, Wang M, Ji F, Peng Y, Wang B, Zhao J, et al. A novel glucose metabolism-related gene signature for overall survival prediction in patients with Glioblastoma. Biomed Res Int. 2021;2021:8872977.
Sakamoto K, Tamamura Y, Katsube K, Yamaguchi A. Zfp64 participates in notch signaling and regulates differentiation in mesenchymal cells. J Cell Sci. 2008;121(Pt 10):1613–23.
Wei CY, Zhu MX, Zhang PF, Huang XY, Wan JK, Yao XZ, et al. PKCα/ZFP64/CSF1 axis resets the tumor microenvironment and fuels anti-PD1 resistance in hepatocellular carcinoma. J Hepatol. 2022;77(1):163–76.
Acknowledgements
Not applicable.
Funding
No funding.
Author information
Authors and Affiliations
Contributions
Conceptualization, Jiayi Sun, and Jintao Liu; methodology, Jiayi Sun, Jinquan Liu, Yudong Hou, Jianheng Bao, and Teng Wang; investigation, Jiayi Sun; visualization, Longbi Liu, Yidan Zhang, Rui Zhong, and Zhenxuan Sun; project administration, Jintao Liu; supervision, Jintao Liu; writing manuscript, Jiayi Sun, Yan Ye, and Jintao Liu.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Sun, J., Liu, J., Hou, Y. et al. ZFP64 drives glycolysis-mediated stem cell-like properties and tumorigenesis in breast cancer. Biol Direct 19, 83 (2024). https://doi.org/10.1186/s13062-024-00533-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13062-024-00533-7