Abstract
Background
Vector-/tick-borne pathogens (V/TBPs) pose a potential threat to human and animal health globally. Information regarding canine V/TBPs is scarce and no specific study has been conducted so far to explore the microbial diversity within ticks infesting dogs from Pakistan. Herein, this knowledge gap is addressed by assessing the genetic diversity and prevalence pattern of V/TBPs in ixodid ticks with special implications for public and canine health.
Methods
A total of 1150 hard ticks were collected from 300 dogs across central Khyber Pakhtunkhwa (KP), Pakistan. After morpho-molecular identification, 120 tick samples were screened for the presence of V/TBPs by amplifying 16S rRNA/gltA (Rickettsia/Ehrlichia and Wolbachia sp.), 18S rRNA (Theileria sp.) and cox1 (Dirofilaria sp.) genes through PCR followed by sequencing and phylogenetic study.
Results
In toto, 50 ixodid ticks (50/120, 41.7%) were found positive for V/TBPs DNA. The detected V/TBPs were categorized into five genera and eight species, viz. Ehrlichia (E. canis and Ehrlichia sp.), Rickettsia (R. massiliae, R. raoultii and Rickettsia sp.), Theileria (T. annulata), Dirofilaria (D. immitis) and Wolbachia (Wolbachia sp.). The pathogen prevalence patterns showed that R. massiliae was the most prevalent zoonotic V/TBP (19.5%), followed by E. canis (10.8%), Rickettsia sp. (7.5%), R. raoultii (6.7%), T. annulata (5.8%), D. immitis (5.8%), Wolbachia sp. (4.2%) and Ehrlichia sp. (3.3%), respectively. Among the screened tick species, most Rhipicephalus sanguineus sensu lato samples were found positive for V/TBP DNA (20/20,100%) followed by Rh. turanicus sensu stricto (13/20, 65%), Hyalomma dromedarii (8/20, 40%), Rh. haemaphysaloides (6/20, 30%), Hy. excavatum (2/20, 10%) and Rh. microplus (1/20, 5%). Co-occurrence of V/TBP was also detected in tick specimens (single V/TBP infection: 32 ticks; double and triple: 13 and 5 tick samples). The detected pathogens shared a phylogenetic relationship with similar isolates published in NCBI GenBank from Old and New World countries.
Conclusion
Ixodid ticks infesting dogs harbor a diverse array of V/TBPs including zoonotic agents from Pakistan. Furthermore, the presence of D. immitis in ticks that infest dogs raises the possibility that this parasite has either attained its dead-end host (i.e. the tick) while feeding on dogs or has expanded its range of intermediate/paratenic hosts. Further research work is needed to investigate the epidemiology and confirm the vector competence of screened tick species for these pathogens from Pakistan.
Graphical Abstract

Similar content being viewed by others
Background
Ticks are hematophagous arthropods infesting livestock/companion animals including dogs and are responsible for transmitting life-threatening pathogens infecting humans and animals worldwide [1, 2]. Research on tick and tick-borne diseases/pathogens (TBPs) has received considerable attention in the recent era of changing climatic scenarios which have resulted in global average temperature rise and annual rainfall shift, thus creating suitable biotopes in different landscapes for the survival and development of disease vector species. Consequently, the zoogeographical range of disease vectors and their associated pathogens expands, putting the public and veterinary health workers at risk of contracting V/TBPs infection [3, 4]. Globally ticks are known as vectors and reservoirs for several pathogenic microbes, i.e. viruses, bacteria (including Rickettsia) and protozoans [5]. Ticks stand second to the mosquito as a potent vector for transmitting infectious diseases to humans and animals [6, 7]. Ixodid tick attachment to the host and prolonged hematophagy may facilitate the transfer of TBPs from an infected tick to the animal hosts which in turn ensures pathogen dispersal to other geographic localities through pet/livestock intercontinental transportation [8].
Among TBPs, ehrlichiosis (canine monocytic ehrlichiosis/tropical canine pancytopenia) adversely affects dog health. Its etiological agent, E. canis, is an obligate intracellular alpha‐proteobacterium (of the Rickettsiae group), which replicates within mononuclear cells of the host [9]. There have been reports of canine ehrlichiosis from subtropical and tropical regions of the world [10]. Ehrlichia species can also infect humans and several other animal species like cats and horses. Important species of the genus Ehrlichia are E. chaffeensis, E. canis, E. ewingii, E. muris, E. sennetsu, E. risticii, E. equi and E. ruminantium [11]. Ehrlichia canis is among the TBPs that can infect dogs [12]. Infected dogs present a wider spectrum of ehrlichiosis consisting of three stages: acute, subclinical and clinical or chronic infection characterized by pyrexia, lymphadenitis, respiratory distress, body weight loss, bleeding episodes (epistaxis), visual and neurological involvement (blindness and meningitis) [13]. Dogs, red foxes and yellow jackals serve as probable reservoir hosts of E. canis, while the brown dog tick, Rhipicephalus sanguineus s.l., is the major arthropod vector of E. canis [14]. Rhipicephalus sanguineus s.l. ticks have a broader zoogeographical range that typically infests canids (dogs) and is most frequently found on dogs in South Asia including Pakistan [15]. Ehrlichia canis has zoonotic importance and is horizontally transmitted to humans by these ticks [16,17,18].
The rickettsial bugs constitute an important group of TBPs responsible for a wide spectrum of infections in humans and animals from subclinical to severe life-threatening forms [19,20,21]. Rickettsial pathogens have a cosmopolitan distribution and are grouped into two classes infecting humans and animals including dogs: (i) the typhus group (TG: Rickettsia prowazekii and R. typhii) and (ii) the spotted fever group (SFG: R. rickettsii, R. slovaca, R. sibirica, R. raoultii, R. conorii, R. peacockii, R. honei, R. japonica, R. montanensis, R. massiliae, R. ripicephali, R. amblyommii, R. africae, R. parkeri, R. heilongjiangensis and R. phillipi). The typhus and SFG rickettsiae are vectored by ixodid ticks, lice, fleas and other blood-sucking arthropods [22,23,24].
The SFG rickettsial microbe R. raoultii is the etiological agent of human scalp eschar and neck lymphadenopathy (SENLAT) across the globe. Rickettsia raoultii was first detected in Dermacentor nuttalli and Rhipicephalus pumilio ticks (as novel rickettsial genotypes: DnS14, DnS28 and RpA4) from the Siberian region of Russia at the end of the twentieth century [25]. The principal arthropod vectors involved in its transmission are Dermacentor ticks. However other ixodid ticks of the genera Amblyomma, Haemaphysalis, Hyalomma and Rhipicephalus were also incriminated in R. raoultii transmission to humans. To date, R. raoultii's presence has been confirmed in many Asian and European countries in ixodid ticks, except Pakistan [26].
The first human infection with R. raoultii was reported in a Spanish resident around 2006 [27]. The clinical manifestations of human SENLAT are a history of a tick bite, skin lesion at the site of tick bite/inoculation eschar on the scalp (because Dermacentor ticks prefer hairy prey), draining neck lymph node inflammation and surrounding alopecia, rarely with fever and rashes [26, 28].
Dirofilaria immitis, commonly known as dog heartworm, is a nematode parasite that causes fatal illness in dogs, known as cardiopulmonary dirofilariasis or dog heartworm disease [29, 30]. Dirofilaria species can also infect other animals including cats, badgers, coyotes, jackals, wolves and horses [29]. However, D. immitis, D. repens, D. tenuis, D. striata, D. ursi and D. subdermata have anthropozoonotic potential and have been detected in human patients. Human heartworm infection is relatively uncommon and categorized as pulmonary dirofilariasis (caused by D. immitis) and subcutaneous dirofilariasis (caused by D. repens, D. tenuis and other associated species) [31]. Canine dirofilariasis is prevalent in tropical, subtropical and temperate regions of the world [32]. Dirofilarial nematodes are vectored by the hematophagous female culicid mosquito of the genera Aedes, Culex and Anopheles [29, 32]. Canine dirofilariasis in dogs involve respiratory and cardiac complications with a typical clinical picture of chronic non-productive cough, shortness of breath, reduction in body weight, asthenia, epistaxis, cyanosis and congestive heart failure [30].
Wolbachia is an endosymbiotic α-proteobacteria (Rickettsiales) that shares a close genome resemblance with Anaplasma and Ehrlichia species [33]. These endosymbionts are widely distributed and detected in almost all arthropods and other invertebrates including filarial nematodes. Five decades earlier, the first bacteria-like microorganism was detected in the filarial worm D. immitis. Later, molecular findings showed that these bacterial bugs belong to the genus Wolbachia [34]. Wolbachia sp. exists in symbiotic associations (mutualism and parasitism) with roundworm microfilariae and influences its reproductive biology [35]. Research advancement with a focus on the interaction between Wolbachia and D. immitis has shown that these Wolbachia play a vital role in the development (embryogenesis and molting) of nematode microfilariae [36] while growing microfilariae provide essential nutrients (amino acids) for Wolbachia survival in a mutualistic manner [37]. The Wolbachia strains prevalent across diverse host species are categorized into different subgroups (each subgroup constitutes a separate phylogenetic lineage). In the beginning, Wolbachia endosymbionts were placed in subgroups A and B (infecting arthropods) followed by the addition of two more subgroups, C and D, after Wolbachia strains were discovered in parasitic nematode filariae. The ongoing discovery of the Wolbachia strain has resulted in Wolbachia subgroup multiplication and further creation of ten subclades, E, G, H, I, K, M, N, O, P and Q, infecting arthropods, one subclade J infecting filariae, one subclade L discovered in the plant-associated parasitic nematode (Radopholus similis) and one subgroup F infecting both arthropods and filariae [33, 38].
Dogs have been domesticated by humans since prehistoric times and live as pets in close association with humans [15]. Owing to massive population growth and global pet mobility, dogs are prone to contracting fatal parasitic illnesses including vector-borne diseases at the same time or in succession to locations where canine vector-borne diseases are endemic [8, 39]. Dogs also serve as preferred hosts of numerous hard tick species harboring zoonotic pathogens, thus increasing the likelihood of human infestation by these ticks and the eventual transfer of TBPs [40]. To track the TBPs of canids in a particular geographic location, the availability of blood-sucking arthropods transmitting these TBPs is a reliable marker for tracking pathogens in that particular region [25].
Currently, there is no comprehensive scientific report available on V/TBPs circulating in the tick vector and dog populations from Pakistan with implications for public and canine health. To address this crucial knowledge gap and provide baseline support for designing effective vector control strategies in the future as well as to educate dog owners about the risk of contracting V/TBPs infection, this scientific work explored the genetic diversity of pathogens in ixodid ticks collected from dogs.
Methods
Ethical consideration
This study was approved by the ethics committee on animal care and use in scientific experimentations at the College of Veterinary Sciences and Animal Husbandry, Abdul Wali Khan University Mardan, Pakistan (CVSAH/FCLS/AWKUM/2021/228).
Study location and tick sampling
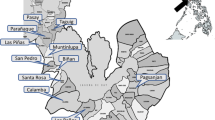
A total of 1150 ixodid ticks were collected from 300 pet dogs between January 1, 2022, and August 30, 2022. The dog population (n = 300, short-haired breed = 182, long-haired breed = 118) was examined for tick infestation and collection in 40 different towns of 5 districts located centrally in KP, Pakistan (Fig. 1). The collected ticks were stored in 70% ethanol and shipped to the public health laboratory at the City University of Hong Kong for further investigations. A comprehensive report has been published elsewhere that provides details about the present study area, its climatic conditions, dominant vegetation type, study design, tick sampling strategy as well as host demography [15].
Tick morphological identification and DNA extraction
The collected ticks were morphologically identified up to species level under a stereo zoom microscope (Olympus®, Tokyo, Japan) using standard morpho-taxonomic keys [41, 42] followed by molecular confirmation. The ixodid fauna was categorized into six species of hard ticks, viz. Rhipicephalus sanguineus s.l., Rh. turanicus s.s., Rh. haemaphysaloides, Rh. microplus, Hyalomma dromedarii and Hy. excavatum. Among the identified ticks, 120 hard ticks (20 specimens of each tick species) were selected for DNA isolation and pathogen screening. Genomic DNA was extracted from tick samples individually using a DNA extraction kit (QIAamp® DNA Mini Kit, Qiagen, Hilden, Germany) following the manufacturer's DNA extraction protocols. The DNA in the samples was analyzed qualitatively and quantitatively using a spectrophotometer (NanoDrop™ Thermo Scientific™, Waltham, MA, USA). All the DNA samples were preserved in a deep freezer at − 80 °C till further analysis.
Molecular screening of V/TBPs through PCR
For pathogen detection in ixodid tick species, three sets of PCRs were performed, a first PCR for the detection of Anaplasma/Ehrlichia/Rickettsia species using 16S rRNA/gltA gene primers sets, a second PCR for the detection of Theileria/Babesia species using 18S rRNA gene primer set and a third PCR for the detection of Dirofilaria species using cox1 gene primer set (Table 1). All PCR reactions were performed in a total reaction volume of 35 µl containing 17.5 µl Master Mix, 3 µl genomic DNA, 1 µl of each primer (forward and reverse, 10 pmol) and 12.5 µl PCR-grade water. The PCR conditions for Anaplasma/Ehrlichia/Rickettsia 16S rRNA/gltA were: initial denaturation of template DNA samples at 95 °C for 5 min followed by 35 cycles of denaturation at 95 °C for 30 s, annealing at 60 °C/55 °C (16S rRNA/gltA) for 30 s, extension at 72 °C for 30 s and a final extension at 72 °C for 10 min [43,44,45]. The thermodynamic conditions set in a thermocycler for Piroplasms 18S rRNA amplification were: an initial denaturation at 95 °C for 3 min followed by 35 cycles of denaturation at 94 °C for 1 min, annealing at 54 °C for 30 s, extension at 72 °C for 30 s and final extension at 72 °C for 10 min [46]. The Dirofilaria cox1 gene was amplified with the following thermocycling parameters: an initial denaturation at 95 °C for 3 min followed by 35 cycles of denaturation at 95 °C for 1 min, annealing at 40 °C for 1 min, extension at 72 °C for 1.5 min and final extension at 72 °C for 7 min [47]. The amplified PCR products were confirmed by submarine gel electrophoresis using 2% agarose gels stained with ethidium bromide and visualized under UV radiation in a Gel-Doc machine (Bio-Rad Laboratories, Hercules, CA, USA).
PCR product purification and sequencing
All the PCR products of desired genes were sent to a genome sequencing facility (BGI Tech Solutions, Hong Kong Co. Ltd., SAR China) for purification and unidirectional DNA sequencing. The query sequence dataset of V/TBPs was edited for trimming unnecessary nucleotides at termini and aligned in MEGA7 [48]. An online BLAST analysis of the query dataset was performed on the NCBI GenBank server to identify and characterize the sequenced pathogens. The reference sequences with query coverage of 99%–100% were downloaded as separate datasets for phylogenetic analysis of sequenced microorganisms. Finally, the 16S rRNA, gltA, 18S rRNA and partial cox1 nucleotide sequences of V/TBPs were submitted to the NCBI GenBank Nucleotide repository, and accession numbers were assigned.
Phylogenetic analyses of V/TBPs
Molecular phylogenies of V/TBPs were inferred from the partial nucleotide sequences of amplified genes using MEGA-7 software [48]. The maximum likelihood (ML) algorithm-based phylogenetic trees were constructed for Rickettsia, Ehrlichia, and Wolbachia sp. using 16S rRNA nucleotide sequences. Additionally, rickettsial isolates were further characterized by using partial gltA nucleotide sequence-based phylogram. Similarly, the same algorithm (ML) was applied to infer the evolutionary histories of Theileria and Dirofilaria isolates by using their target genes' partial nucleotide sequences (18S rRNA and cox1 genes). The whole dataset was resampled 1000 times for bootstrap value generation with Kimura two-parameter model for 16S rRNA- and 18S rRNA-based phylogenetic trees and Tamura three-parameter model for rickettsial 16S rRNA, gltA and dirofilarial cox1 isolates. These best-fit models of sequence evolution/phylogram construction were chosen based on the lowest Bayesian information criterion scores (BIC), corrected Akaike information criterion value (AIC) and maximum likelihood value (Inl) [49].
Statistical analyses
The dataset generated from this research work was analyzed statistically. The prevalence/infection rate of detected pathogens was calculated as the number of infected ticks (individual tick species) by V/TBP species divided by the total number of molecularly screened tick specimens (n = 120). Differences in the prevalence of pathogens in various tick species were assessed by Chi-square statistic in GraphPad software (Version 9.1.0, Boston, MA, USA). The significance level was set at P < 0.05.
Results
Detection, diversity and prevalence of V/TBPs in ixodid ticks
Out of 120 molecularly screened ixodid ticks, V/TBPs were detected in 50 (41.7%) tick specimens. The characterized pathogens were categorized into five genera and eight species, viz. Ehrlichia (E. canis, and Ehrlichia sp.); Rickettsia (R. massiliae, R. raoultii, and Rickettsia sp.); Theileria (T. annulata); Dirofilaria (D. immitis) and Wolbachia (Wolbachia sp.). Genus-based analysis showed that Rickettsial pathogens were most common (33.7%) followed by Ehrlichial microorganisms (14.1%), Theileria (5.8%), parasitic Dirofilaria of a dog (5.8%) and Wolbachia (4.2%). The species-wise prevalence pattern showed that R. massiliae was the most prevalent zoonotic V/TBP (19.5%), followed by E. canis (10.8%), Rickettsia sp. (7.5%), R. raoultii (6.7%), T. annulata (5.8%), D. immitis (5.8%), Wolbachia sp. (4.2%) and Ehrlichia sp. (3.3%), respectively (Table 2).
Among the molecularly screened hard tick species for pathogen detection, Rh. sanguineus s.l. was the most infected tick species by V/TBPs (20/20, 100%) followed by Rh. turanicus s.s. (13/20, 65%), Hy. dromedarii (8/20, 40%), Rh. haemaphysaloides (6/20, 30%), Hy. excavatum (2/20, 10%) and Rh. microplus (1/20, 5%). The occurrence of V/TBPs in ticks exhibits significant variations in different tick species (χ2 = 11.998, df = 5, P = 0.004). Rich pathogens diversity (number of species) was reported in Rh. sanguineus s.l. and Rh. turanicus s.s. (7/8 V/TBP species) followed by Rh. haemaphysaloides (5/8), Hy. dromedarii (3/8), Hy. excavatum (2/8) and Rh. microplus (1/8) (Table 2) (Fig. 2).
Single and multiple V/TBP infection in ixodid ticks
Single and mixed infections (co-infection: the presence of multiple V/TBP species within individual ticks) were observed within screened tick species. Out of 50 pathogen-positive ticks, 32 (64%) were found positive for a single V/TBP infection. Single pathogen prevalence pattern showed that R. massiliae was a highly prevalent zoonotic pathogen (22%) in ixodid ticks followed by R. raoultii (8%), Rickettsia sp. (8%), E. canis (6%), Ehrlichia sp. (6%), T. annulata (6%), D. immitis (2%) and Wolbachia sp. (2%). However, mixed/co-infection patterns revealed that 13 (26%) ticks were infected with two V/TBPs. Double co-infection in hard ticks showed that the most prevalent V/TBPs, i.e. R. massiliae + Rickettsia sp. (8%) detected in Rh. sanguineus s.l., Rh. turanicus s.s., Rh. haemaphysaloides and Hy. dromedarii ticks followed by E. canis + R. massiliae (6%) double coinfection in Rh. sanguineus s.l. and Rh. turanicus s.s.; R. massiliae + R. raoultii (4%) double co-infection in Rh. sanguineus s.l. and Rh. turanicus s.s.; E. canis + D. immitis (4%) double co-infection in Rh. sanguineus s.l. and Rh. turanicus s.s. ticks; D. immitis + Wolbachia sp. (2%) and E. canis + Ehrlichia sp. (2%) double co-infection in Rh. sanguineus s.l. ticks, respectively. DNA of three V/TBPs was detected in five (10%) tick samples. Triple co-infection was observed for E. canis + R. massiliae + R. raoultii (4%) in Rh. sanguineus s.l. and Rh. turanicus s.s. ticks followed by E. canis + D. immitis + Wolbachia sp. (4%) in Rh. sanguineus s.l. and R. massiliae + Rickettsia sp. + T. annulata (2%) triple co-infection in Hy. dromedarii. The associations among the different tick species and patterns of infections, viz. single (χ2 = 11.890, df = 5, P = 0.014), double (χ2 = 11.11, df = 3, P = 0.011), and triple (χ2 = 8.26, df = 2, P = 0.017) were found statistically significant (P < 0.05) (Table 3).
Molecular features of V/TBPs gene sequences
The resultant partial nucleotide sequence size of the detected pathogen's genes was: Ehrlichia canis 16S rRNA = 281 bp, Ehrlichia sp. 16S rRNA = 334 bp; R. massiliae 16S rRNA/gltA = 305/348 bps, R. raoultii 16S rRNA/gltA = 316/353 bps, Rickettsia sp. 16S rRNA/gltA = 713/344 bps; Wolbachia sp. 16S rRNA = 305 bp; T. annulata 18S rRNA = 980 bp and D. immitis cox1 = 166 bp, respectively (Additional file 1: Table S1). The characterized gene sequences can be retrieved from NCBI GenBank with the following accession numbers (E. canis 16S rRNA: OP605546-OP605547; Ehrlichia sp. 16S rRNA: ON926911-ON926912; R. massiliae 16S rRNA: ON909181-ON909182, gltA: ON952468-ON952469; R. raoultii 16S rRNA: ON909183-ON909184, gltA: ON952466-ON952467; Rickettsia sp. 16S rRNA: ON909179-ON909180, gltA: ON952470-ON952471; T. annulata 18S rRNA: ON982513-ON982514; D. immitis cox1:OQ600716, OQ600720-OQ603322 OQ600728 OQ600729 OQ603256 OQ603257 OQ601560 OQ601563 OQ600765 OQ600766; Wolbachia sp. 16S rRNA: OQ379166-OQ379169).
Online BLAST analysis of 16S rRNA isolates of E. canis and Ehrlichia sp. shared 100% and 99–100% homology with the same isolates published from Asia, Europe, South Africa and the USA. Likewise, partial 16S rRNA/gltA nucleotide isolates showed 98–100% sequence similarity with identical published isolates in NCBI GenBank from Asian, European and African countries. Similarly, partial 18S rRNA isolates of T. annulata, partial cox1 nucleotide sequences of D. immitis and partial 16S rRNA amplicons of Wolbachia sp. shared up to 99.90%, 100% and 99.01–100% sequence similarity with the same species isolates reported from Asia, Europe and South America, respectively (Additional file 1: Tables S2 and S3).
Phylogenetic profile of detected V/TPBs
The 16S rRNA-based phylograms were computed to infer the evolutionary relatedness of the species belonging to genera Ehrlichia, Rickettsia and Wolbachia species. The detected rickettsiae 16S rRNA isolate-based phylogenies were further supported by gltA isolate-based phylogram. Among the Ehrlichial microorganisms, E. canis specimens under this study clustered with the similar specimen on the 16S rRNA phylogram published from Europe (Greece MN922610, Spain AY394465), Asia including the Far East (China AF162860, Indonesia MT499360, Iraq MN227484, Thailand EF139458), North America (USA M73221) and South America (Brazil EF195134, Venezuela AF373612) with bootstrap support of 88%. The 16S rRNA-based ML phylogram showed that E. canis isolates from the present study clustered together with isolates reported from Greece and Indonesia as distinct sub-clades with bootstrap support of 95%, 99% and 100%, suggesting that E. canis isolates were past descendant to the same isolates reported from the aforementioned countries (Fig. 3). On the other hand, unidentified Ehrlichia sp. detected in the current study were closely grouped with identical isolates from China (KJ410254) with a maximum bootstrapping of 100% (Fig. 3).
Phylogenetic analysis of Ehrlichia canis and Ehrlichia sp. isolated from ixodid ticks (this study: colored) with reference Ehrlichia sp. isolates (NCBI GenBank) based on partial 16S rRNA gene sequences. The phylogram was constructed using the maximum likelihood algorithm based on the Kimura two-parameter model. The dataset was resampled 1000 times for bootstrap value generation. Borrelia miyamotoi (D45192) was used as the outgroup
Among the Rickettsial bugs, zoonotic partial R. massiliae 16S rRNA nucleotide sequences shared an evolutionary relationship with similar isolates available in the NCBI GenBank repository from a neighboring Asian country (India MZ851176, MZ851177) with a maximum bootstrap value of 100%. Anthropozoonotic R. raoultii 16S rRNA specimens from the present study grouped with the same isolates reported from Asia (China KJ410260, Russia MK304546) and Europe (Netherlands JN242190, Ukraine MZ093134, Poland KX024760) with a bootstrap support of 99%. The unclassified Rickettsia sp. 16S rRNA isolates from the present study showed evolutionary relatedness to the same pathogen’s sequences published from Asia (Thailand KF318168) and Africa (South Africa KX944386) with considerable bootstrap support of 89% (Fig. 4). The rickettsial gltA sequence-based phylogenetic tree showed that R. massiliae gltA isolates shared a phylogenetic relationship with identical specimens published from Asia (China KY069259, MF002497) and Europe (France U59719) with bootstrap support of 100%. Similarly, partial R. raoultii gltA nucleotide sequences shared evolutionary relatedness with the same gene sequences reported from a neighboring country (China MN046861, MN450398) with considerable branch support of 90%. However, the unclassified Rickettsia sp. gltA isolates clustered together with the identical isolates deposited in NCBI GenBank from Asia (Malaysia KU948238) and South America (Brazil MF175748) along with known Rickettsia vini isolates from Germany (MW864098 and MW864099) (Fig. 5).
Phylogenetic analysis of R. massiliae, Rickettsia raoultii and Rickettsia sp. isolated from ixodid ticks (this study: colored) with reference Rickettsia sp. isolates (NCBI GenBank) based on partial 16S rRNA gene sequences. The phylogram was constructed using the maximum likelihood algorithm based on the Kimura two-parameter model. The dataset was resampled 1000 times for bootstrap value generation. Anaplasma phagocytophilum (KJ782381) was used as the outgroup
Phylogenetic analysis of Rickettsia massiliae, R. raoultii, and Rickettsia sp. isolated from ixodid ticks (this study: colored) with reference Rickettsia sp. isolates (NCBI GenBank) based on partial gltA gene sequences. The phylogram was constructed using the maximum likelihood algorithm based on the Tamura three-parameter model. The dataset was resampled 1000 times for bootstrap value generation. Ehrlichia canis (AY647155) was used as the outgroup
The T. annulata 18S rRNA isolates from the present study shared a phylogenetic relationship with similar gene sequences published from Asia (China MK415058, KT959231; India T736498; UAE MW537791) and Europe (Turkey MK918607; Italy MT341858) with 99% bootstrap support (Fig. 6).
Phylogenetic analysis of Theileria annulata isolated from ixodid ticks (this study: colored) with reference Theileria sp. isolates (NCBI GenBank) based on partial 18S rRNA gene sequences. The phylogram was constructed using the maximum likelihood algorithm based on the Kimura two-parameter model. The dataset was resampled 1000 times for bootstrap value generation. Babesia bovis (L19077) was used as the outgroup
The partial D. immitis cox1 nucleotide sequences showed that the present study's dog heartworm samples clustered with the same species sequences separated from diverse host species (human, dog, cat and fox) across Asia (China EU159111; Iran MZ540219; Japan AB973226; Myanmar OL721650; Thailand MW577348; South Korea OM491241; Uzbekistan MN650648), Europe (France KP760184; Italy FN391553; Spain LC107816; Slovenia OP494255;), North America (USA MN945948, ON062406, MN945948) and South America (Colombia OQ179618) with maximum bootstrap support (100%). However, some D. immitis isolates detected during the present study clustered as separate sub-clades with considerable bootstrapping (95% and 99%) (Fig. 7).
Phylogenetic analysis of Dirofilaria immitis isolated from ixodid ticks (this study: colored) with reference Dirofilaria sp. isolates (NCBI GenBank) based on partial cox1 gene sequences. The phylogram was constructed using the maximum likelihood algorithm based on the Tamura three-parameter model. The dataset was resampled 1000 times for bootstrap value generation. Gongylonema pulchrum (NC026687) was used as the outgroup
The partial Wolbachia sp. 16S rRNA nucleotide sequences from the present study grouped with Wolbachia endosymbiont of filarial worm isolates (Wolbachia subclade C: infecting parasitic filarial nematodes) published in NCBI GenBank nucleotide repository from Asian and European countries with bootstrap statistical support of 93%. However, the present study's Wolbachia specimens shared a separate subclade within subgroup C along with unclassified Wolbachia sp. detected in dog blood samples in Portugal with considerable bootstrap statistic (97%). Further large-scale phylogenetic analysis of these unclassified Wolbachia sp. isolates is important to establish their accurate phylogenetic lineage (subgroup) (Fig. 8).
Phylogenetic analysis of Wolbachia sp. isolated from ixodid ticks (this study: purple colored) with reference Wolbachia sp. isolates (NCBI GenBank) based on partial 16S rRNA gene sequences. The phylogram was constructed using the maximum likelihood algorithm based on the Tamura three-parameter model. The dataset was resampled 1000 times for bootstrap value generation. Rickettsia sp. (ON952471) was used as the outgroup
Discussion
Climate change and global warming have facilitated disease vector spreading into new geographic locations and put the public and veterinary health workers at risk of contracting V/TBP infection in non-endemic areas [50]. Therefore, robust surveillance of disease vectors and associated pathogens in a particular geographic area is of utmost importance in devising appropriate vector and pathogen control interventions [15]. To the best of our knowledge, no comprehensive study has been conducted to explore the genetic diversity of V/TBPs in hard tick species infesting dog populations across Pakistan so far, with the exception of one mechanistic study (focused on the genetic diversity of E. canis in co-infected brown dog ticks) and two epidemiological investigations on pathogens detection in dog blood samples from Punjab province of Pakistan [51,52,53]. This is the first molecular report documenting a diverse array of zoonotic V/TBPs in ixodid ticks collected from dogs with implications for canine and public health from KP, Pakistan.
Vector-/tick-borne pathogens detected during the present study were grouped into five genera comprising eight species.
This study identified two species belonging to the genus Ehrlichia in ixodid ticks, viz. E. canis and Ehrlichia species. The zoonotic E. canis (causative agent of canine ehrlichiosis) was detected in Rh. sanguineus s.l., Rh. turanicus s.s. and Rh. haemaphysaloides ticks associated with dogs for the first time from KP, Pakistan. A significantly higher prevalence of the same pathogen was reported in dog blood samples and kennel tick Rh. sanguineus s.l. in Punjab province of Pakistan [51,52,53]. This fluctuation in the prevalence rate of E. canis across different agroecological zones of the country may be linked to the differences in sample size, diversity of infesting ticks and availability of competent tick vectors and host population that influence pathogen survival and in turn its prevalence rate. However, our findings are generally consistent with published literature that confirmed the presence of E. canis in the ixodid tick species and dog hosts from China, India, Korea, Philippines, Russia, Thailand, Portugal and Southeast Asia [3, 54,55,56,57,58,59,60]. The 16S rRNA consensus sequences of E. canis shared homology and evolutionary history with identical isolates from Asia, Europe, North America and South America, suggesting that E. canis' identical genotype is circulating in different tick species globally without geographic restrictions [60,61,62,63,64]. Presence of E. canis in Rh. sanguineus s.l. ticks required special attention from veterinary health specialists in the study area as it is a competent vector and can transmit E. canis to a dog host [14]. So, proper vector control strategies should be adopted against this tick species.
The uncharacterized Ehrlichia sp. DNA was detected in four ixodid ticks during the present study. Our results regarding Ehrlichia sp. detection in hard ticks are parallel and support findings on Ehrlichia sp. detection in hard tick/dog blood samples from other agroecological zones of the country and elsewhere globally [55, 65,66,67,68]. Ehrlichia sp. phylogeny was established during the present study using 16S rRNA nucleotide isolates. Similarly, multiple scientific reports inferred Ehrlichia sp. phylogeny successfully by using a 16S rRNA phylogenetic marker [69,70,71]. These unclassified isolates need further large-scale phylogenetic studies to establish/evaluate their taxonomic position and pathogenic potential for human and animal.
The present study detected and characterized two species of SFG Rickettsia, namely R. massiliae (causative agent of febrile illness with skin rash, lymphadenopathy and inoculation eschar) [72] and R. raoultii (causative agent of SENLAT) [27], and one unclassified Rickettsia sp. in hard ticks.
The SFG R. massiliae was recovered from all reported tick species involved in this study. These findings are in accordance with published reports that confirmed R. massiliae's presence in Rh. sanguineus s.l. collected from dogs across Punjab, Pakistan [52], and other multiple ixodid tick species infesting dogs worldwide [73,74,75]. Additionally, the presence of R. massiliae in Rh. sanguineus s.l. and Rh. turanicus s.s. ticks is predictive of increased transmission risk to dog owners as these tick species act as its competent vectors [76, 77]. The phylogenetic position of R. massiliae was successfully established based on 16S rRNA and rickettsial gltA gene isolates. Similar approaches were adopted elsewhere to infer the evolutionary history of R. massiliae using the same phylogenetic markers [73, 74, 78, 79]. Our study is supportive of their findings regarding rickettsial species detection and phylogenetic studies.
The SFG R. raoultii was identified in Rh. sanguineus s.l. and Rh. turanicus s.s. ticks, which frequently infest dogs. Rickettsia raoultii in a small number of dog blood samples (n = 2) has been reported by a comparative study from Pakistan and Iran so far [53]. This research work is the initial report on R. raoultii detection and circulation in ixodid ticks associated with dogs in different agroecological zones of KP, Pakistan. This scientific report is parallel and validates published literature on R. raoultii detection and isolation from ticks infesting animals and humans [80,81,82,83,84,85]. The presence of R. raoultii in Rh. sanguineus s.l. and Rh. turanicus s.s. ticks required special attention of public and veterinary health experts as their vector competence for R. raoultii has been confirmed. The presence of R. raoultii in competent vectors intensifies the likelihood of its transmission to those who are in frequent contact with pet dogs [76, 77]. The R. raoultii 16S rRNA and gltA isolates shared phylogeny with identical globally reported isolates. A parallel/similar approach was adopted to investigate and infer the molecular phylogeny of R. raoultii elsewhere [80, 83, 84].
Rickettsia sp. was detected in four ixodid tick species during this study. Phylogenetic analysis clustered Rickettsia sp. isolates with unidentified Rickettsia sp. available in the NCBI GenBank Nucleotide database. Our findings follow global literature that reported unidentified Rickettsia sp. from several ixodid ticks associated with livestock including companion animals [60, 86, 87]. The molecular phylogeny of detected Rickettsia sp. was successfully inferred by using 16S rRNA and gltA isolates. These findings support the published data elsewhere that had successfully inferred unclassified Rickettsia sp. phylogeny [60]. Further molecular work is needed to establish the taxonomic profile of unclassified Rickettsia sp. along with the prediction of its zoonotic potential for public and veterinary health.
No canine-associated Theileria sp. was detected in ixodid ticks analyzed by this study except T. annulata (causing bovine tropical theileriosis) in three species of hard ticks. Similarly, numerous studies reported the presence of Theileria sp. in dogs and hard ticks collected from them [87,88,89,90]. The phylogenetic profile of Theileria sp. was established by grouping with other T. annulata isolates on an 18S rRNA phylogram. Our results support scientific reports that detected T. annulata and inferred its phylogeny based on the 18S rRNA marker in ixodid ticks collected from diverse host species including dogs [89, 90].
The dog heartworm D. immitis (causative agent of canine cardiopulmonary dirofilariasis) was detected in two ixodid tick species. To our knowledge, Dirofilaria immitis detection in ixodid tick species by this study is the first to science, and no report has been published on D. immitis microfilariae detection in tick species so far, as this roundworm parasite is vectored by culicid mosquitos to canine hosts. These findings suggest that D. immitis microfilariae may have reached their dead-end host (i.e. the tick) during the feeding of infected ticks on dogs or that this parasite has extended its range of intermediate/paratenic hosts. To determine the role of kennel ticks in the dissemination of this parasite, their vector competence for D. immitis microfilariae should be investigated. Additionally, the presence of D. immitis in ixodid ticks indicates that the dog population in the study area is likely infected with this roundworm and that dog owners may be at risk of anthropozoonotic D. immitis infection if bitten by an infected tick. The D. immitis was successfully detected, and its phylogenetic position was determined using the cox1 universal barcode. Our report is consistent with published global literature that detected and inferred the evolutionary relationship of D. immitis recovered from diverse canine hosts including dogs using the same gene isolates [91,92,93,94,95].
The endosymbiotic Wolbachia sp. DNA was detected in Rh. sanguineus s.l. and Rh. turanicus s.s. ticks for the first time from Pakistan. Our findings follow the globally published data that detected uncharacterized Wolbachia sp. or Wolbachia endosymbiont of D. immitis in heartworm-positive dog blood samples [96, 97]. Research work has demonstrated that Wolbachia endosymbiont facilitates D. immitis microfilariae development (embryogenesis and molting) in the definitive host population [36]. Thus, the presence of Wolbachia endosymbiont potentiates D. immitis microfilariae's survival to exploit canine hosts effectively. Phylogenetic analysis of 16S rRNA isolates of Wolbachia sp. from the present study clustered with subclade C, which includes Wolbachia endosymbiont of filarial nematodes [33], and confirmed the close resemblance of these isolates with Wolbachia endosymbiont of D. immitis.
The co-existence of V/TBPs was found in molecularly screened hard tick species. Double and triple co-infections of pathogens were detected in 3/10 and 1/10 of the analyzed tick samples. The dog-associated ticks, i.e. Rh. sanguineus s.l. and Rh. turanicus s.s., were found positive for a maximum of V/TBP species and indicative of the high-level risk of pathogen transmission to humans and animals upon exposure to co-infected tick bite/infestation. A plethora of previously published data on vector or host-microbiome characterization revealed the co-occurrence of V/TBP and endosymbiont in ticks and canids [52, 73, 98, 99]. However, the composition and pattern (double/triple and so on) of co-infection varies regarding pathogen, host and vector species involved in time and space and is influenced by climatic factors, in particular landscape/agroecological zone. Additionally, levels of infection may differ among unfed, partially fed and completely engorged ticks [100]. To date, no human case has been reported involving co-infection of V/TBPs in Pakistan. The co-existence of V/TBPs in hard tick fauna across the study area is of public health interest as most of the pathogens detected by this study share common reservoir hosts and vector ticks, which assure co-infection transmission to dog owners.
Conclusion
This scientific work is a comprehensive report on V/TBPs detection, their genetic diversity and prevalence patterns in ixodid ticks collected from dogs across KP, Pakistan. Five genera of V/TBPs were detected in molecularly screened hard ticks, i.e. Ehrlichia, Rickettsia, Theileria, Dirofilaria and Wolbachia. To our knowledge, detection of D. immitis in ixodid ticks is first to science while E. canis and R. raoultii's presence in hard ticks is first from Pakistan. Phylogenetic analysis showed that V/TBP isolates shared homology and evolutionary histories with identical isolates published in the NCBI GenBank nucleotide repository from Old and New World countries. Among the reported pathogens, E. canis, R. raoultii, R. massiliae, and D. immitis were zoonotic V/TBPs of public and canine health concerns. The brown dog ticks Rh. sanguineus s.l. and Rh. turanicus s.s. carried a maximum of the detected pathogens, and for some of them Rh. sanguineus s.l. and/or Rh. turanicus s.s. is also a competent vector. During this study, there was evidence of the co-occurrence of various vector- and tick-borne pathogens. This raises the possibility of co-infections being transmitted to humans who have frequent contact with pet dogs. Physicians in Pakistan should be aware of the detection of V/TBPs in competent vector ticks and consider these pathogens in the diagnosis of patients with a history of tick bites from the study area. Moreover, it is recommended that further large-scale scientific investigations be conducted to explore the exclusive microbiome (pathobiome) of ixodid ticks associated with diverse host species in Pakistan. Such research would have significant implications for both public and veterinary health, including canid pets.
Availability of data and materials
The dataset and related information generated during the present study have been included in this manuscript in the best interest of the public and veterinary health.
Abbreviations
- SFG:
-
Spotted fever group
- TG:
-
Typhus group
- SENLAT:
-
Scalp eschar and neck lymphadenopathy
- PCR:
-
Polymerase chain reaction
- 16S rRNA :
-
16S ribosomal RNA gene
- 18S rRNA :
-
18S ribosomal RNA gene
- gltA :
-
Rickettsial citrate synthase-encoding gene
- cox1 :
-
Cytochrome c oxidases subunit 1 gene
- KP:
-
Khyber Pakhtunkhwa
- V/TBPs:
-
Vector/tick-borne pathogens
- sp.:
-
Species
References
Chomel B. Tick-borne infections in dogs—an emerging infectious threat. Vet Parasitol. 2011;179:294–301.
Dantas-Torres F, Otranto D. Best practices for preventing vector-borne diseases in dogs and humans. Trends Parasitol. 2016;32:43–55.
Zhang J, Liu Q, Wang D, Li W, Beugnet F, Zhou J. Epidemiological survey of ticks and tick-borne pathogens in pet dogs in south-eastern China. Parasite. 2017;24:1.
Shaw SE, Day MJ, Birtles RJ, Breitschwerdt EB. Tick-borne infectious diseases of dogs. Trends Parasitol. 2001;17:74–80.
Estrada-Peña A. Ticks as vectors: taxonomy, biology and ecology. Rev Sci Tech. 2015;34:53–65.
Hillyard P. Diseases carried by ticks in NW Europe: their medical and veterinary importance. Ticks of North-West Europe. Synopsis of the British Fauna (New Series). London: FSC Publications; 1996;22–23.
Elelu N, Bankole AA, Daphne HP, Rabiu M, Ola-Fadunsin SD, Ambali HM, et al. Molecular characterisation of Rhipicephalus sanguineus sensu lato ticks from domestic dogs in Nigeria. Vet Med Sci. 2022;8:454–9.
Otranto D, Dantas-Torres F, Breitschwerdt EB. Managing canine vector-borne diseases of zoonotic concern: part one. Trends Parasitol. 2009;25:157–63.
Harrus S, Waner T. Diagnosis of canine monocytotropic ehrlichiosis (Ehrlichia canis): an overview. Vet J. 2011;187:292–6.
Abd Rani PA, Irwin PJ, Coleman GT, Gatne M, Traub RJ. A survey of canine tick-borne diseases in India. Parasit Vectors. 2011;4:1–8.
Dumler JS. Anaplasma and Ehrlichia infection. Ann N Y Acad Sci. 2005;1063:361–73.
Carvalho FS, Wenceslau AA, Carlos RSA, Albuquerque GR. Epidemiological and molecular study of Ehrlichia canis in dogs in Bahia. Brazil Genet Mol Res. 2008;7:657–62.
Cardoso L, Mendão C, Madeira De Carvalho L. Prevalence of Dirofilaria immitis, Ehrlichia canis, Borrelia burgdorferi sensu lato, Anaplasma spp. and Leishmania infantum in apparently healthy and CVBD-suspect dogs in Portugal—a national serological study. Parasit Vectors. 2012;5, 1–9.
Masala G, Chisu V, Foxi C, Socolovschi C, Raoult D, Parola P. First detection of Ehrlichia canis in Rhipicephalus bursa ticks in Sardinia. Italy Ticks Tick Borne Dis. 2012;3:396–7.
Zeb J, Song B, Senbill H, Aziz MU, Hussain S, Khan MA, et al. Ticks infesting dogs in Khyber Pakhtunkhwa, Pakistan: detailed epidemiological and molecular report. Pathogens. 2023;12:98.
Rahbari S, Nabian S, Shayan P. Primary report on distribution of tick fauna in Iran. Parasitol Res. 2007;101:175–7.
Dantas-Torres F. Biology and ecology of the brown dog tick, Rhipicephalus sanguineus. Parasit Vectors. 2010;3:1–11.
Khazeni A, Telmadarraiy Z, Oshaghi MA, Mohebali M, Zarei Z, Abtahi SM, et al. Molecular detection of Ehrlichia canis in ticks’ population collected on dogs in Meshkin-Shahr, Ardebil Province, Iran. J Biomed Sci Eng. 2013;6:1–5.
Raoult D, Roux V. Rickettsioses as paradigms of new or emerging infectious diseases. Clin Microbiol Rev. 1997;10:694.
Parola P, Paddock CD, Raoult D. Tick-borne rickettsioses around the world: emerging diseases challenging old concepts. Clin Microbiol Rev. 2005;18:719.
Perlman SJ, Hunter MS, Zchori-Fein E. The emerging diversity of Rickettsia. Proc Biol Sci. 2006;273:2097–106.
Fournier PE, Raoult D. Current knowledge on phylogeny and taxonomy of Rickettsia spp. Ann N Y Acad Sci. 2009;1166:1–11.
Murray GGR, Weinert LA, Rhule EL, Welch JJ. The phylogeny of Rickettsia using different evolutionary signatures: How tree-like is bacterial evolution? Syst Biol. 2016;65:265–79.
Merhej V, Angelakis E, Socolovschi C, Raoult D. Genotyping, evolution and epidemiological findings of Rickettsia species. Infect Genet Evol. 2014;25:122–37.
Rydkina E, Roux V, Fetisova N, Rudakov N, Gafarova M, Tarasevich I, et al. New Rickettsiae in ticks collected in territories of the former soviet union. Emerg Infect Dis. 1999;5:811–4.
Li H, Zhang P-H, Huang Y, Du J, Cui N, Yang Z-D, et al. Isolation and identification of Rickettsia raoultii in human cases: a surveillance study in 3 medical centers in China. Clin Infect Dis. 2018;66:1109–15.
Ibarra V, Oteo JA, Portillo A, Santibanez S, Blanco JR, Metola L, et al. Rickettsia slovaca infection: DEBONEL/TIBOLA. Ann N Y Acad Sci. 2006;1078:206–14.
Angelakis E, Pulcini C, Waton J, Imbert P, Socolovschi C, Edouard S, et al. Scalp eschar and neck lymphadenopathy caused by Bartonella henselae after tick bite. Clin Infect Dis. 2010;50:549–51.
McCall JW, Genchi C, Kramer LH, Guerrero J, Venco L. Heartworm disease in animals and humans. Adv Parasitol. 2008;66:193–285.
Dantas-Torres F. Canine vector-borne diseases in Brazil. Parasit Vectors. 2008;1:1–17.
Lee ACY, Montgomery SP, Theis JH, Blagburn BL, Eberhard ML. Public health issues concerning the widespread distribution of canine heartworm disease. Trends Parasitol. 2010;26:168–73.
Morchón R, Carretón E, González-Miguel J, Mellado-Hernández I. Heartworm disease (Dirofilaria immitis) and their vectors in Europe—new distribution trends. Front Physiol. 2012;3:196.
Lefoulon E, Bain O, Makepeace BL, D’Haese C, Uni S, Martin C, et al. Breakdown of coevolution between symbiotic bacteria Wolbachia and their filarial hosts. PeerJ. 2016;4:e1840.
Simón F, Siles-Lucas M, Morchón R, González-Miguel J, Mellado I, Carretón E, et al. Human and animal dirofilariasis: the emergence of a zoonotic mosaic. Clin Microbiol Rev. 2012;25:507–44.
Zug R, Hammerstein P. Bad guys turned nice? A critical assessment of Wolbachia mutualisms in arthropod hosts. Biol Rev Camb Philos Soc. 2015;90:89–111.
Bandi C, Dunn AM, Hurst GDD, Rigaud T. Inherited microorganisms, sex-specific virulence and reproductive parasitism. Trends Parasitol. 200;17:88–94.
Foster J, Ganatra M, Kamal I, Ware J, Makarova K, Ivanova N, et al. The Wolbachia genome of Brugia malayi: endosymbiont evolution within a human pathogenic nematode. PLoS Biol. 2005;3:0599–614.
Wang Z, Su XM, Wen J, Jiang LY, Qiao GX. Widespread infection and diverse infection patterns of Wolbachia in Chinese aphids. Insect Sci. 2014;21:313–25.
Cardoso L, Yisaschar-Mekuzas Y, Rodrigues FT, Costa Á, MacHado J, Diz-Lopes D, et al. Canine babesiosis in northern Portugal and molecular characterization of vector-borne co-infections. Parasit Vectors. 2010;3:1–10.
Guglielmone AA, Petney TN, Robbins RG. Ixodidae (Acari: Ixodoidea): descriptions and redescriptions of all known species from 1758 to December 31, 2019. Zootaxa. 2020;4871.
Estrada-Peña A, Bouattour A, Camicas JL, Walker AR. Ticks of domestic animals in the Mediterranean region. University of Zaragoza: Zaragoza, Spain. 2004; p. 131.
Walker JB, Keirans JE, Horak IG. The genus Rhipicephalus (Acari, Ixodidae): a guide to the brown ticks of the world. Cambridge: Cambridge University Press; 2005.
Parola P, Inokuma H, Camicas JL, Brouqui P, Raoult D. Detection and identification of spotted fever group Rickettsiae and Ehrlichiae in African ticks. Emerg Infect Dis. 2001;7:1014–7.
Nijhof AM, Bodaan C, Postigo M, Nieuwenhuijs H, Opsteegh M, Franssen L, et al. Ticks and associated pathogens collected from domestic animals in the Netherlands. Mary Ann Liebert, Inc. 2 Madison Avenue Larchmont, NY 10538 USA; 2008;7:585–95.
Roux V, Rydkina E, Eremeeva M, Raoult D. Citrate synthase gene comparison, a new tool for phylogenetic analysis, and its application for the rickettsiae. Int J Syst Bacteriol. 1997;47:252–61.
Casati S, Sager H, Gern L, Piffaretti JC. Presence of potentially pathogenic Babesia sp. for human in Ixodes ricinus in Switzerland. Ann Agric Environ Med. 2006;13:65–70.
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3:294–9.
Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–4.
Nei M, Kumar S. Molecular evolution and phylogenetics. Oxford: Oxford University Press; 2000.
Dantas-Torres F. Climate change, biodiversity, ticks and tick-borne diseases: the butterfly effect. Int J Parasitol Parasites Wildl. 2015;4:452–61.
Malik MI, Qamar M, Ain Q, Hussain MF, Dahmani M, Ayaz M, et al. Molecular detection of Ehrlichia canis in dogs from three districts in Punjab (Pakistan). Vet Med Sci. 2018;4:126–32.
Cabezas-Cruz A, Allain E, Ahmad AS, Saeed MA, Rashid I, Ashraf K, et al. Low genetic diversity of Ehrlichia canis associated with high co-infection rates in Rhipicephalus sanguineus (s.l.). Parasit Vectors. 2019;12:1–3.
Iatta R, Sazmand A, Nguyen VL, Nemati F, Ayaz MM, Bahiraei Z, et al. Vector-borne pathogens in dogs of different regions of Iran and Pakistan. Parasitol Res. 2021120:4219–28.
Behera SK, Dimri U, Banerjee P, Garg R, Dandapat S, Sharma B. Molecular detection and assessment of hemato-biochemistry, oxidant/antioxidant status in natural canine monocytic ehrlichiosis cases from northern India. Proc Natl Acad Sci India Sect B Biol Sci. 2017;87:361–8.
Suh GH, Ahn KS, Ahn JH, Kim HJ, Leutenegger C, Shin SS. Serological and molecular prevalence of canine vector-borne diseases (CVBDs) in Korea. Parasit Vectors. 2017;10:1–8.
Galay RL, Manalo AAL, Dolores SLD, Aguilar IPM, Sandalo KAC, Cruz KB, et al. Molecular detection of tick-borne pathogens in canine population and Rhipicephalus sanguineus (sensu lato) ticks from southern Metro Manila and Laguna, Philippines. Parasit Vectors. 2018;11:643.
Livanova NN, Fomenko NV, Akimov IA, Ivanov MJ, Tikunova NV, Armstrong R, et al. Dog survey in Russian veterinary hospitals: tick identification and molecular detection of tick-borne pathogens. Parasit Vectors. 2018;11:1–10.
Do T, Inpankaew T, Duong DH, Bui KL. First molecular evidence of pathogens in fleas collected from dogs in Northern Vietnam. Pathogens. 2021;10:1185.
Maia C, Almeida B, Coimbra M, Fernandes MC, Cristóvão JM, Ramos C, et al. Bacterial and protozoal agents of canine vector-borne diseases in the blood of domestic and stray dogs from southern Portugal. Parasit Vectors. 2015;8:1–7.
Nguyen VL, Colella V, Greco G, Fang F, Nurcahyo W, Hadi UK, et al. Molecular detection of pathogens in ticks and fleas collected from companion dogs and cats in East and Southeast Asia. Parasit Vectors. 2020;13:1–11.
Anderson BE, Dawson JE, Jones DC, Wilson KH. Ehrlichia chaffeensis, a new species associated with human ehrlichiosis. J Clin Microbiol. 1991;29:2838–42.
Unver A, Perez M, Orellana N, Huang H, Rikihisa Y. Molecular and antigenic comparison of Ehrlichia canis isolates from dogs, ticks, and a human in Venezuela. J Clin Microbiol. 2001;39:2788–93.
Wen B, Cao W, Pan H. Ehrlichiae and ehrlichial diseases in china. Ann N Y Acad Sci. 2003;990:45–53.
Kostopoulou D, Gizzarelli M, Ligda P, Foglia Manzillo V, Saratsi K, Montagnaro S, et al. Mapping the canine vector-borne disease risk in a Mediterranean area. Parasit Vectors. 2020;13:1–9.
Suto Y, Suto A, Inokuma H, Obayashi H, Hayashi T. First confirmed canine case of Ehrlichia canis infection in Japan. Vet Rec. 2001;148:809–11.
Lee SO, Na DK, Kim CM, Li YH, Cho YH, Park JH, et al. Identification and prevalence of Ehrlichia Chaffeensis infection in Haemaphysalis Longicornis ticks from Korea by PCR, sequencing and phylogenetic analysis based on 16S RRNA gene. J Vet Sci. 2005;6:151–5.
Talukder M, Matsuu A, Iguchi A, Roy B, Nishii N, Hikasa Y. PCR-based survey of vector-borne pathogens in dogs in Dhaka, Bangladesh. J Bangladesh Agric Univ. 2013;10:249–54.
Chen Z, Liu Q, Liu JQ, Xu BL, Lv S, Xia S, et al. Tick-borne pathogens and associated co-infections in ticks collected from domestic animals in central China. Parasit Vectors. 2014;7:1–8.
Parola P, Cornet JP, Sanogo YO, Miller RS, Van Thien H, Gonzalez JP, et al. Detection of Ehrlichia spp., Anaplasma spp., Rickettsia spp., and other eubacteria in ticks from the Thai-Myanmar border and Vietnam. J Clin Microbiol. 2003;41:1600–8.
Kang YJ, Diao XN, Zhao GY, Chen MH, Xiong Y, Shi M, et al. Extensive diversity of Rickettsiales bacteria in two species of ticks from China and the evolution of the Rickettsiales. BMC Evol Biol. 2014;14:1–12.
Ledger KJ, Beati L, Wisely SM. Survey of ticks and tick-borne Rickettsial and protozoan pathogens in Eswatini. Pathogens. 2021;10:1043.
Vitale G, Mansueto S, Rolain JM, Raoult D. Rickettsia massiliae human isolation. Emerg Infect Dis. 2006;12:174–5.
Psaroulaki A, Ragiadakou D, Kouris G, Papadopoulos B, Chaniotis B, Tselentis Y. Ticks, tick-borne rickettsiae, and Coxiella burnetii in the Greek Island of Cephalonia. Ann N Y Acad Sci. 2006;1078:389–99.
Maia C, Ferreira A, Nunes M, Vieira ML, Campino L, Cardoso L. Molecular detection of bacterial and parasitic pathogens in hard ticks from Portugal. Ticks Tick Borne Dis. 2014;5:409–14.
Barradas PF, Mesquita JR, Ferreira P, Amorim I, Gärtner F. Detection of tick-borne pathogens in Rhipicephalus sanguineus sensu lato and dogs from different districts of Portugal. Ticks Tick Borne Dis. 2020;11:101536.
Matsumoto K, Ogawa M, Brouqui P, Raoult D, Parola P. Transmission of Rickettsia massiliae in the tick, Rhipicephalus turanicus. Med Vet Entomol. 2005;19:263–70.
Olivieri E, Wijnveld M, Bonga M, Berger L, Manfredi MT, Veronesi F, et al. Transmission of Rickettsia raoultii and Rickettsia massiliae DNA by Dermacentor reticulatus and Rhipicephalus sanguineus (s.l.) ticks during artificial feeding. Parasit Vectors. 2018;11, 1–7.
Moraga-Fernández A, Chaligiannis Ι, Cabezas-Cruz A, Papa A, Sotiraki S, de la Fuente J, et al. Molecular identification of spotted fever group Rickettsia in ticks collected from dogs and small ruminants in Greece. Exp Appl Acarol. 2019;78:421–30.
Song S, Chen C, Yang M, Zhao S, Wang B, Hornok S, et al. Diversity of Rickettsia species in border regions of northwestern China. Parasit Vectors. 2018;11:1–7.
Jia N, Zheng YC, Ma L, Huo QB, Ni XB, Jiang BG, et al. Human infections with Rickettsia raoultii. China Emerg Infect Dis. 2014;20:866–8.
Wijnveld M, Schötta AM, Pintér A, Stockinger H, Stanek G. Novel Rickettsia raoultii strain isolated and propagated from Austrian Dermacentor reticulatus ticks. Parasit Vectors. 2016;9:1–7.
Chisu V, Foxi C, Masala G. First molecular detection of the human pathogen Rickettsia raoultii and other spotted fever group rickettsiae in Ixodid ticks from wild and domestic mammals. Parasitol Res. 2018;117:3421–9.
Turebekov N, Abdiyeva K, Yegemberdiyeva R, Dmitrovsky A, Yeraliyeva L, Shapiyeva Z, et al. Prevalence of Rickettsia species in ticks including identification of unknown species in two regions in Kazakhstan. Parasit Vectors. 2019;12:1–16.
Tariq M, Seo JW, Kim DY, Panchali MJL, Yun NR, Lee YM, et al. First report of the molecular detection of human pathogen Rickettsia raoultii in ticks from the Republic of Korea. Parasit Vectors. 2021;14:1–5.
Chang QC, Hu Y, Wu TT, Ma XX, Jiang BG, Jia N, et al. The role of ranged horses in eco-epidemiology of Rickettsia raoultii infection in China. Front Microbiol. 2022;12:4382.
Melo ALT, Witter R, Martins TF, Pacheco TA, Alves AS, Chitarra CS, et al. A survey of tick-borne pathogens in dogs and their ticks in the Pantanal biome, Brazil. Med Vet Entomol. 2016;30:112–6.
Chaligiannis Ι, Fernández de Mera IG, Papa A, Sotiraki S, de la Fuente J. Molecular identification of tick-borne pathogens in ticks collected from dogs and small ruminants from Greece. Exp Appl Acarol. 2018;74:443–53.
Bigdeli M, Rafie SM, Namavari MM, Jamshidi S. Report of Theileria annulata and Babesia canis infections in dogs. Comp Clin Path. 2012;21:375–7.
Rjeibi MR, Amairia S, Rouatbi M, Ben Salem F, Mabrouk M, Gharbi M. Molecular prevalence and genetic characterization of piroplasms in dogs from Tunisia. Parasitol. 2016;143:1622–8.
Habibi G, Imani A, Afshari A, Bozorgi S. Detection and molecular characterization of Babesia canis vogeli and Theileria annulata in free-ranging dogs and ticks from Shahriar County, Tehran Province, Iran. Iran J Parasitol. 2020;15:321.
Víchová B, Miterpáková M, Iglódyová A. Molecular detection of co-infections with Anaplasma phagocytophilum and/or Babesia canis canis in Dirofilaria-positive dogs from Slovakia. Vet Parasitol. 2014;203:167–72.
Heidari Z, Kia EB, Arzamani K, Sharifdini M, Mobedi I, Zarei Z, et al. Morphological and molecular identification of Dirofilaria immitis from Jackal (Canis aureus) in North Khorasan, northeast Iran. J Vector Borne Dis. 2015;52:329–33.
Khanmohammadi M, Akhlaghi L, Razmjou E, Falak R, Zolfaghari Emameh R, Mokhtarian K, et al. Morphological description, phylogenetic and molecular analysis of Dirofilaria immitis Isolated from dogs in the northwest of Iran. Iran J Parasitol. 2020;15:57–66.
Sharifdini M, Karimi M, Ashrafi K, Soleimani M, Mirjalali H. Prevalence and molecular characterization of Dirofilaria immitis in road killed canids of northern Iran. BMC Vet Res. 2022;18:161.
Brianti E, Panarese R, Napoli E, De Benedetto G, Gaglio G, Bezerra-Santos MA, et al. Dirofilaria immitis infection in the Pelagie archipelago: the southernmost hyperendemic focus in Europe. Transbound Emerg Dis. 2022;69:1274–80.
Wong SSY, Teng JLL, Poon RWS, Choi GKY, Chan KH, Yeung ML, et al. Comparative evaluation of a point-of-care immunochromatographic test SNAP 4Dx with molecular detection tests for vector-borne canine pathogens in Hong Kong. Vector Borne Zoonotic Dis. 2011;11:1269–77.
Landum M, Ferreira CC, Calado M, Alho AM, Maurício IL, Meireles JS, et al. Detection of Wolbachia in Dirofilaria infected dogs in Portugal. Vet Parasitol. 2014;204:407–10.
Manoj RRS, Iatta R, Latrofa MS, Capozzi L, Raman M, Colella V, et al. Canine vector-borne pathogens from dogs and ticks from Tamil Nadu, India. Acta Trop. 2020;203:105308.
Qiu Y, Simuunza M, Kajihara M, Ndebe J, Saasa N, Kapila P, et al. Detection of tick-borne bacterial and protozoan pathogens in ticks from the Zambia-Angola Border. Pathogens. 2022;11:566.
Estrada-Peña A, Gray JS, Kahl O, Lane RS, Nijhof AM. Research on the ecology of ticks and tick-borne pathogens–methodological principles and caveats. Front Cell Infect Microbiol. 2013;3:29.
Acknowledgements
Not applicable.
Funding
This research work was funded by the scientific research project at the Department of Infectious Diseases and Public Health, the City University of Hong Kong (PI: Professor Olivier Andre Sparagano, Project No. 9380108).
Author information
Authors and Affiliations
Contributions
JZ and OAS designed the study. JZ experimented and drafted the initial manuscript. JZ, BS, MAK, MUA, SH and AW conducted a formal analysis. JZ and MAK prepared the study area map and graphical abstract. MAK, HS, OAS and AE-T provided intellectual inputs and critically revised the manuscript. JZ OAS and MAK edited, revised and proofread the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The study was performed according to the guidelines of the Declaration of Abdul Wali Khan University Mardan (AWKUM), Pakistan, and approved by the Ethics Committee of the College of Veterinary Sciences and Animal Husbandry, AWKUM (CVSAH/FCLS/AWKUM/2021/228).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1. Table S1
: Vector/tick-borne pathogen target gene amplicon size and nucleotide composition. Table S2: Vector/tick-borne pathogen target gene consensus sequences. Table S3: Vector/tick-borne pathogen target gene sequenceNCBI GenBank BLAST summary.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Zeb, J., Song, B., Khan, M.A. et al. Genetic diversity of vector-borne pathogens in ixodid ticks infesting dogs from Pakistan with notes on Ehrlichia canis, Rickettsia raoultii and Dirofilaria immitis detection. Parasites Vectors 16, 214 (2023). https://doi.org/10.1186/s13071-023-05804-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-023-05804-2