Abstract
Background
Bartonella spp. infect a variety of vertebrates throughout the world, with generally high prevalence. Several Bartonella spp. are known to cause diverse clinical manifestations in humans and have been recognized as emerging pathogens. These bacteria are mainly transmitted by blood-sucking arthropods, such as fleas and lice. The role of ticks in the transmission of Bartonella spp. is unclear.
Methods
A recently developed quadruplex polymerase chain reaction (PCR) amplicon next-generation sequencing approach that targets Bartonella-specific fragments on gltA, ssrA, rpoB, and groEL was applied to test host-seeking Ixodes scapularis ticks (n = 1641; consisting of 886 nymphs and 755 adults) collected in 23 states of the eastern half of the United States and Ixodes pacificus ticks (n = 966; all nymphs) collected in California in the western United States for the presence of Bartonella DNA. These species were selected because they are common human biters and serve as vectors of pathogens causing the greatest number of vector-borne diseases in the United States.
Results
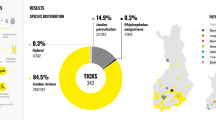
No Bartonella DNA was detected in any of the ticks tested by any target.
Conclusions
Owing to the lack of Bartonella detection in a large number of host-seeking Ixodes spp. ticks tested across a broad geographical region, our results strongly suggest that I. scapularis and I. pacificus are unlikely to contribute more than minimally, if at all, to the transmission of Bartonella spp.
Graphical Abstract

Similar content being viewed by others
Background
Bartonella is a diverse genus of gram-negative bacteria that are highly adapted to intracellular persistence in a wide range of vertebrates. Many mammalian species are natural reservoirs for Bartonella spp. and experience chronic, asymptomatic, intraerythrocytic bacteremia when infected [1]. Over the last two decades, more than 40 species belonging to the genus Bartonella have been described from different mammalian hosts, with more than a dozen having been associated with diverse clinical manifestations of diseases in humans and recognized as emerging pathogens [2,3,4,5,6,7,8,9]. Many Bartonella spp. are host-specific, suggesting maintenance of individual species in independent enzootic cycles [10].
Bartonella spp. are typically vector-borne, transmitted between mammalian hosts by hematophagous arthropods. To date, several arthropods have been proven to be vectors for Bartonella spp. transmission. Sand flies (Lutzomyia verrucarum), human body lice (Pediculus humanis corporis), and cat fleas (Ctenocephalides felis) are well-known vectors that transmit Bartonella bacilliformis (Carrión’s disease) [11], B. quintana (trench fever) [12, 13], and B. henselae (cat scratch disease) [14], respectively. Additionally, the Oriental rat flea (Xenopsylla cheopis) and other rodent fleas have been implicated as potential vectors transmitting rodent-associated Bartonella spp., such as B. elizabethae, B. grahamii, and B. taylorii [15, 16].
In the United States, the majority of reported vector-borne disease cases are caused by pathogens spread by ticks [17]. There has been considerable interest in ticks as potential vectors for Bartonella species. Numerous molecular surveys to detect Bartonella DNA in various tick species from around the world have been conducted in recent years [18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39]. Studies focused on host-seeking ticks are particularly intriguing because detection of Bartonella in unfed ticks would suggest that the bacterium can survive the transstadial molt from larvae to nymphs or from nymphs to adults; demonstration of transstadial transmission is one component in demonstrating vector competence [40]. The results from these studies were discordant. Some studies reported detection of Bartonella DNA with high prevalence in host-seeking ticks. For example, Chang et al. [19] detected B. quintana and several other Bartonella species by gltA in 19.2% of host-seeking Ixodes pacificus ticks in California, USA; Adelson et al. [21] reported that 34.5% of host-seeking Ixodes scapularis ticks collected in New Jersey, USA, were infected with Bartonella spp. using 16S primers; Morozova et al. [22] identified B. henselae by groEL in as much as 44% of host-seeking Ixodes persulcatus ticks from Russia. By contrast, other studies reported very low prevalence of Bartonella in ticks. For example, only 0.9% of host-seeking I. scapularis ticks from Maryland, USA [27], and 0.2% of host-seeking I. ricinus ticks from France [29] harbored Bartonella DNA; both studies were based on gltA results. Moreover, absence of Bartonella in host-seeking ticks was reported in studies from Hungary, Korea, and Finland based on the results of gltA, groEL, or ssrA [25, 28, 41].
With these varied results, it has been debated whether ticks can serve as vectors of Bartonella spp. In the USA, the blacklegged tick (I. scapularis) and the western blacklegged tick (I. pacificus) are frequent human-biters in the eastern and far western USA, respectively [42]. Both species serve as vectors of the Lyme disease spirochete, as well as several other human pathogens [43,44,45]. In this study, we tested host-seeking I. scapularis and I. pacificus nymphs and adults collected across 24 states for the presence of Bartonella DNA. Previous surveys mainly used traditional polymerase chain reaction (PCR) with a single target [18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38]. Because of the genetic diversity of Bartonella spp., however, a single target may not be able to detect all species, and in some cases, single targets might not be sufficiently specific to accurately detect Bartonella, yielding false-positive results. In our study, we applied a recently described quadruplex PCR amplicon next-generation sequencing approach which amplifies and sequences four Bartonella-specific gene targets (gltA, ssrA, rpoB, and groEL) [46]. The use of multiple target sequences increases detection success and also improves the accuracy of species identification compared to single-target methods. Our objective was to elucidate whether Bartonella DNA is present in the host-seeking Ixodes ticks using this powerful detection method and to infer whether the ticks can serve as vectors for transmission of Bartonella spp. based on the prevalence of infection in ticks collected over a broad geographical area in the USA.
Methods
Tick collections and DNA templates
The ticks tested in this study were from our archived DNA samples from ticks collected as part of national tick surveillance [47] or research efforts. Host-seeking I. scapularis were collected in 23 states in the eastern half of the USA by dragging or flagging during 2013–2023. Ticks included in this study were chosen based on life stage, sex, and collection site. We were aiming to include ~100 ticks from each state, but the actual number of ticks per state varied between 13 and 153 due to low availability in some states or inclusion of more collection sites in other states. As a result, a total of 1641 I. scapularis ticks consisting of 886 nymphs and 755 adults (330 males and 425 females) were tested for the presence of Bartonella spp. (Table 1). DNA was extracted previously using the KingFisher DNA extraction system (Thermo Fisher Scientific, Waltham, MA, USA) and the MagMAX™ CORE Kit (Thermo Fisher Scientific) [48] for tick surveillance testing and stored at −80 °C afterwards. Residual DNA was used for the present study.
Host-seeking I. pacificus ticks were collected from different woodland habitat types in highly climatically and ecologically diverse Mendocino County, CA, by dragging in 2004 as previously described [49]. A total of 966 I. pacificus nymphs with representatives of all woodland habitat types described in the county were tested for the presence of Bartonella spp. (Table 2). Total DNA had been extracted from individual nymphs previously using the DNeasy Blood & Tissue Kit (Qiagen, Valencia, CA, USA) [50], and archived DNA was used for the present study.
PCR amplification, library preparation, and next-generation sequencing
Ticks were tested for the presence of Bartonella DNA using a quadruplex PCR amplicon next-generation sequencing assay that targets gltA, ssrA, rpoB, and groEL using Bartonella-specific primers [46]. Additionally, an internal control using 16S ribosomal RNA (rRNA) primer (forward 5′-CTGCTCAATGATTTTTTAAATTGCTGTGG-3′ and reverse 5′-CCGGTCTGAACTCAGATCAAGT-3′) [51] was included to ensure the presence of tick DNA and to confirm the original morphological identification of tick species.
Detailed procedures follow those described in Bai et al. [46]. Briefly, a primary PCR reaction containing 12.5 μl of TEMPase 2× master mix (AMPLICON, Denmark), the four pairs of Bartonella-specific primers and the 16S rRNA tick-specific primer (final concentration of 300 nM each), and 5 μl of tick DNA was first amplified. Positive and negative controls were always included in each PCR run to evaluate performance and detect contamination. Upon completion of amplification, the PCR products were purified with AMPure XP magnetic beads (Beckman Coulter, Brea, CA, USA), followed by index PCR using dual unique barcode indices (Nextera XT Index Kit V2, Illumina, San Diego, CA, USA), then purified with MagSi-DNA allround magnetic beads (BOCA Scientific, Westwood, MA, USA). The purified products were then pooled, quantified, normalized, and denatured to generate the final library to be loaded into a MiSeq Nano v2 (500 cycles) reagent cassette (Illumina, San Diego, CA, USA) to start sequencing on an Illumina MiSeq instrument (Illumina, San Diego, CA, USA).
Tick DNA interference testing
The inhibitory effect is a commonly observed issue during PCR amplification, which sometimes causes false-negative results for pathogen detection [52]. Before applying the assay for field tick testing, we evaluated the assay performance to check whether tick DNA would cause any inhibition or interference during PCR amplification. Clean tick DNA was extracted from I. scapularis ticks raised at the Centers for Disease Control and Prevention (CDC) colony using the KingFisher DNA extraction system; Bartonella DNA from pure culture of B. henselae, B. koehlerae, and B. grahamii (collections at CDC Division of Bacterial Diseases branch in Fort Collins, CO, USA) was prepared by heating a heavy suspension of microorganisms for 15 min at 95 °C followed by centrifugation of the lysed cells for 1 min at 8000 rpm. The supernatant was transferred to a clean centrifuge tube to be used as the DNA template.
The DNA concentration was measured using the Invitrogen™ Qubit™ 4 Fluorometer dsDNA [double-stranded DNA] HS Assay (Fisher Scientific, Pittsburgh, PA, USA). Then a high concentration of the clean tick DNA (5 ng/μl) was mixed with DNA of B. henselae (10 pg/μl), B. koehlerae (10 pg/μl), and B. grahamii (10 pg/μl), respectively. Five microliters of each mixture was then used for the interference testing following the procedures described in the preceding section. Triplicates of each mixture were tested.
Bioinformatics analysis
After sequencing was completed, the raw sequences were analyzed with a custom Nextflow bioinformatics pipeline described by Osikowicz et al. [53]. Briefly, quality control analysis and primer trimming were first performed followed by error correction, paired read merging, and amplicon sequence variant (ASV) grouping. The observed ASVs were then aligned to reference sequences with the nucleotide Basic Local Alignment Search Tool (BLASTn) [54, 55]. The minimum read cut-off for a sample to be considered positive was set to 50 reads. A 95% sequence similarity and 90% minimum sequence alignment length were used to align the observed ASVs to the reference sequences. Sequences that represent different Bartonella species for each target were obtained from GenBank and used as reference sequences.
Results
Tick DNA interference testing
Spiked DNA from B. henselae, B. koehlerae, and B. grahamii was successfully detected and identified by each of the four targets used in the quadruplex sequencing in all triplicates of each mixture of tick DNA and Bartonella DNA, showing no interference from tick DNA.
Host-seeking tick testing
The internal control with the 16S rRNA tick primer showed that tick DNA was present in all samples. All ticks from the eastern USA were confirmed to be I. scapularis ticks, and all ticks from Mendocino County, CA, were confirmed to be I. pacificus.
No Bartonella DNA was detected in any ticks, nymph or adult, by any target (Tables 1 and 2).
Discussion
Ixodes scapularis and I. pacificus ticks are important vectors in the USA that are responsible for transmission of Borrelia burgdorferi (the Lyme disease agent) and several other human pathogens [43,44,45]. However, the role of these ticks in Bartonella transmission was not clear. In the present study, we tested more than 2600 host-seeking I. scapularis and I. pacificus ticks for the presence of Bartonella DNA using a sensitive and specific quadruplex PCR amplicon sequencing approach. No Bartonella DNA was detected in any ticks, nymphs, or adults, by any target. Similar results have been reported by other investigators who tested host-seeking Ixodes ticks across more limited spatial scales but found no detectable Bartonella DNA [25, 28, 41]. These results demonstrate that host-seeking Ixodes ticks are unlikely to contribute to the transmission of Bartonella spp.
Previous studies that tested blood-fed Ixodes ticks collected from different animals were able to detect Bartonella infections [56, 57], demonstrating that Ixodes ticks are exposed to Bartonella through blood-feeding on infected hosts. Bartonella infections are prevalent in rodent species that commonly serve as blood meal sources for I. scapularis and I. pacificus [58, 59]. However, the lack of detection of Bartonella in unfed Ixodes ticks, which take only a single blood meal per life stage, suggests that the bacteria seldom, if ever, survive the transstadial molt from larva to nymph or nymph to adult. The lack of Bartonella DNA in adult ticks is particularly compelling to support the conclusion that Bartonella does not survive the transstadial molt, because adult ticks could have been exposed to any potential infections twice by taking two blood meals.
Bartonella spp. typically cause long-lasting hemotrophic bacteremia in their mammalian reservoir hosts. The reservoir hosts, however, may clear the infection after being bacteremic for several months, but later may acquire the infection again, with the same or a different strain [58, 60]. The temporal dynamics observed in mammals may apply to the ticks as well. Both I. scapularis and I. pacificus ticks go through four life stages (egg, larva, nymph, and adult) to complete their life cycles. After a blood meal, the ticks take months to a year to digest and molt into the next life stage. During the long molting process, Bartonella infection might have been cleared, assuming the ticks were infected during the previous life stage. This may explain the absence of Bartonella DNA in host-seeking ticks. Alternatively, Bartonella could have deleterious effects on tick survival, resulting in very low pathogen prevalence in surviving host-seeking ticks.
Notably, molecular detection of Bartonella DNA has been reported in ticks (primarily Ixodes spp.) collected at various locations in the USA, Europe, and other parts of the world, with high or low infection rates. It is worth noting that high Bartonella prevalence in ticks (> 30%) were mostly from studies using 16S rRNA [21, 30]. Such results are questionable due to the highly conserved 16S rRNA target used, which had little genetic diversity and shared homology with non-target microbes [61, 62]. Studies utilizing non-specific targets could yield a falsely high prevalence of Bartonella infections in ticks. On the other hand, lower Bartonella prevalence in host-seeking Ixodes ticks (< 1%) has been reported, mostly in studies using much more specific genes, for example, gltA [27, 29, 37]. Such low prevalence is consistent with our findings. Although we tested over 2600 ticks in our study, we cannot rule out the possibility that Bartonella infections could occur at very low prevalence in host-seeking Ixodes spp. ticks. However, it is important to emphasize that the mere presence of Bartonella DNA within a tick does not imply that the bacteria are viable or that the tick could transmit it during the course of blood-feeding [61].
Although our field investigation suggests that I. scapularis and I. pacificus are unlikely to contribute to transmission of Bartonella spp., laboratory experiments demonstrated the vector competence of other Ixodes ticks, in particular I. ricinus, which could acquire B. henselae and B. birtlesii through blood-feeding, maintain the infections throughout the molt, and transmit B. henselae/B. birtlesii during a subsequent blood meal [63, 64]. Bartonella is a very diverse taxon, and it is possible that vector competence and efficiency differ across Ixodes and Bartonella species combinations. However, in those studies, the I. ricinus ticks were continuously fed on blood with a very high bacteremia load (108–109 CFU) [63, 64], which is rarely seen in natural infections. Thus, the experimental results may not be relevant to establishing vector competence under natural conditions. Nevertheless, vector competence has not been demonstrated for I. scapularis or I. pacificus and any Bartonella species that naturally occur in the USA. Laboratory transmission studies on I. scapularis and I. pacificus are needed to elucidate acquisition, survival, and transmission rates.
Conclusions
Although sample sizes were low for many states, our testing spanned a broad geographical region, and cumulatively a large number of ticks were tested. Using a highly sensitive and specific next-generation sequencing approach, we tested more than 2600 host-seeking I. scapularis and I. pacificus ticks from 24 states. No Bartonella DNA was detected in the ticks. Our data, together with previous studies from more limited geographical regions [27, 38, 39], strongly suggest that I. scapularis and I. pacificus are unlikely to contribute more than minimally, if at all, to the transmission of Bartonella spp. in the USA.
Availability of data and materials
All data of this study are presented in the manuscript.
References
Deng H, Le Rhun D, Buffet JP, Cotté V, Read A, Birtles RJ, et al. Strategies of exploitation of mammalian reservoirs by Bartonella species. Vet Res. 2012;43:15.
Daly JS, Worthington MG, Brenner DJ, Moss CW, Hollis DG, Weyant RS, et al. Rochalimaea elizabethae sp. nov. isolated from a patient with endocarditis. J Clin Microbiol. 1993;31:872–81.
Drancourt M, Mainardi JL, Brouqui P, Vandenesch F, Carta A, Lehnert F, et al. Bartonella (Rochalimaea) quintana endocarditis in three homeless men. N Engl J Med. 1995;332:419–23.
Anderson B, Neuman M. Bartonella spp. as emerging human pathogens. Clin Microbiol Rev. 1997;10:203–19.
Kerkhoff FT, Bergmans AM, van Der Zee A, Rothova A. Demonstration of Bartonella grahamii DNA in ocular fluids of a patient with neuroretinitis. J Clin Microbiol. 1999;37:4034–8.
Roux V, Eykyn SJ, Wyllie S, Raoult D. Bartonella vinsonii subsp. berkhoffii as an agent of afebrile blood culture-negative endocarditis in a human. J Clin Microbiol. 2000;38:1698–700.
Eremeeva ME, Gerns HL, Lydy SL, Goo JS, Ryan ET, Mathew SS, et al. Bacteremia, fever, and splenomegaly caused by a newly recognized Bartonella species. N Engl J Med. 2007;356:2381–7.
Kosoy M, Morway C, Sheff KW, Bai Y, Colborn J, Chalcraft L, et al. Bartonella tamiae sp. nov., a newly recognized pathogen isolated from three human patients from Thailand. J Clin Microbiol. 2008;46:772–5.
Chomel BB, Kasten RW. Bartonellosis, an increasingly recognized zoonosis. J Appl Microbiol. 2010;109:743–50.
Vayssier-Taussat M, Moutailler S, Michelet L, Devillers E, Bonnet S, Cheval J, et al. Next generation sequencing uncovers unexpected bacterial pathogens in ticks in western Europe. PLoS ONE. 2013;8:e81439.
Garcia-Caceres U, Garcia FU. Bartonellosis: an immunodepressive disease and the life of Daniel Alcides Carrion. J Clin Pathol. 1991;95:58–66.
Byam W. Trench fever: a louse-borne disease. London: Oxford University Press; 1919. p. 196.
Roux V, Raoult D. Body lice as tools for diagnosis and surveillance of reemerging diseases. J Clin Microbiol. 1999;37:596–9.
Higgins JA, Radulovic S, Jaworski DC, Azad AF. Acquisition of the cat scratch disease agent Bartonella henselae by cat fleas (Siphonaptera: Pulicidae). J Med Entomol. 1996;33:490–5.
Bown KJ, Bennet M, Begon M. Flea-borne Bartonella grahamii and Bartonella taylorii in bank voles. Emerg Infect Dis. 2004;10:684–7.
McKee CD, Osikowicz LM, Schwedhelm TR, Maes SE, Enscore RE, Gage KL, et al. Acquisition of Bartonella elizabethae by experimentally exposed Oriental rat fleas (Xenopsylla cheopis; Siphonaptera, Pulicidae) and excretion of Bartonella DNA in flea feces. J Med Entomol. 2018;55:1292–8.
Eisen RJ, Kugeler KJ, Eisen L, Beard CB, Paddock CD. Tick-borne zoonoses in the United States: persistent and emerging threats to human health. ILAR J. 2017;58:319–35.
Schouls LM, van de Pol I, Rijpkema SG, Schot CS. Detection and identification of Ehrlichia, Borrelia burgdorferi sensu lato, and Bartonella species in Dutch Ixodes ricinus ticks. J Clin Microbiol. 1999;37:2215–22.
Chang CC, Chomel BB, Kasten RW, Romano V, Tietze N. Molecular evidence of Bartonella spp. in questing adult Ixodes pacificus ticks in California. J Clin Microbiol. 2001;39:1221–6.
Chang CC, Hayashidani H, Pusterla N, Kasten RW, Madigan JE, Chomel BB. Investigation of Bartonella infection in ixodid ticks from California. Comp Immunol Microbiol Infect Dis. 2002;25:229–36.
Adelson ME, Rao RVS, Tilton RC, Cabets K, Eskow E, Fein L, et al. Prevalence of Borrelia burgdorferi, Bartonella spp., Babesia microti, and Anaplasma phagocytophila in Ixodes scapularis ticks collected in Northern New Jersey. J Clin Microbiol. 2004;42:2799–801.
Morozova OV, Cabello FC, Dobrotvorsky AK. Semi-nested PCR detection of Bartonella henselae in Ixodes persulcatus ticks from Western Siberia, Russia. Vector Borne Zoonotic Dis. 2004;4:306–9.
Halos L, Jamal T, Maillard R, Beugnet F, Le Menach A, Boulouis HJ, et al. Evidence of Bartonella sp. in questing adult and nymphal Ixodes ricinus ticks from France and co-infection with Borrelia burgdorferi sensu lato and Babesia sp. Vet Res. 2005;36:79–87.
Kim CM, Kim JY, Yi YH, Lee MJ, Cho MR, Shah DH, et al. Detection of Bartonella species from ticks, mites and small mammals in Korea. J Vet Sci. 2005;6:327–34.
Sréter-Lancz Z, Tornyai K, Széll Z, Sréter T, Márialigeti K. Bartonella infections in fleas (Siphonaptera: Pulicidae) and lack of bartonellae in ticks (Acari: Ixodidae) from Hungary. Folia Parasitol (Praha). 2006;53:313–6.
Holden K, Boothby JT, Kasten RW, Chomel BB. Co-detection of Bartonella henselae, Borrelia burgdorferi, and Anaplasma phagocytophilum in Ixodes pacificus ticks from California, USA. Vector Borne Zoonotic Dis. 2006;6:99–102.
Swanson KI, Norris DE. Co-circulating microorganisms in questing Ixodes scapularis nymphs in Maryland. J Vector Ecol. 2007;32:243–51.
Chae JS, Yu DH, Shringi S, Klein TA, Kim HC, Chong ST, et al. Microbial pathogens in ticks, rodents and a shrew in northern Gyeonggi-do near the DMZ. Korea J Vet Sci. 2008;9:285–93.
Cotté V, Bonnet S, Cote M, Vayssier-Taussat M. Prevalence of five pathogenic agents in questing Ixodes ricinus ticks from western France. Vector Borne Zoonotic Dis. 2010;10:723–30.
Dietrich F, Schmidgen T, Maggi RG, Richter D, Matuschka FR, Vonthein R, et al. Prevalence of Bartonella henselae and Borrelia burgdorferi sensu lato DNA in Ixodes ricinus ticks in Europe. Appl Environ Microbiol. 2010;76:1395–8.
Tijsse-Klasen E, Fonville M, Gassner F, Nijhof AM, Hovius EK, Jongejan F, et al. Absence of zoonotic Bartonella species in questing ticks: first detection of Bartonella clarridgeiae and Rickettsia felis in cat fleas in the Netherlands. Parasit Vectors. 2011;4:61.
Corrain R, Drigo M, Fenati M, Menandro ML, Mondin A, Pasotto D, et al. Study on ticks and tick-borne zoonoses in public parks in Italy. Zoonoses Public Health. 2012;59:468–76.
Janecek E, Mietze A, Goethe R, Schnieder T, Strube C. Bartonella spp. infection rate and B. grahamii in ticks. Emerg Infect Dis. 2012;18:1689–90.
Sytykiewicz H, Karbowiak G, Werszko J, Czerniewicz P, Sprawka I, Mitrus J. Molecular screening for Bartonella henselae and Borrelia burgdorferi sensu lato co-existence within Ixodes ricinus populations in central and eastern parts of Poland. Ann Agric Environ Med. 2012;19:451–6.
Vayssier-Taussat M, Moutailler S, Féménia F, Raymond P, Croce O, La Scola B, et al. Identification of Novel Zoonotic Activity of Bartonella spp. France Emerg Infect Dis. 2016;22:457–62.
Müller A, Reiter M, Schötta AM, Stockinger H, Stanek G. Detection of Bartonella spp. in Ixodes ricinus ticks and Bartonella seroprevalence in human populations. Ticks Tick Borne Dis. 2016;7:763–7.
Bonnet SI, Paul RE, Bischoff E, Cote M, Le Naour E. First identification of Rickettsia helvetica in questing ticks from a French Northern Brittany Forest. PLoS Negl Trop Dis. 2017;11:e0005416.
Maggi RG, Toliver M, Richardson T, Mather T, Breitschwerdt EB. Regional prevalences of Borrelia burgdorferi, Borrelia bissettiae, and Bartonella henselae in Ixodes affinis, Ixodes pacificus and Ixodes scapularis in the USA. Ticks Tick Borne Dis. 2019;10:360–4.
Livengood J, Hutchinson ML, Thirumalapura N, Tewari D. Detection of Babesia, Borrelia, Anaplasma, and Rickettsia spp. in adult black-legged ticks (Ixodes scapularis) from Pennsylvania, United States, with a Luminex Multiplex Bead Assay. Vector Borne Zoonotic Dis. 2020;20:406–11.
Eisen L. Vector competence studies with hard ticks and Borrelia burgdorferi sensu lato spirochetes: a review. Ticks Tick Borne Dis. 2020;11:101359.
Sormunen JJ, Penttinen R, Klemola T, Hanninen J, Vuorinen I, Laaksonen M, et al. Tick-borne bacterial pathogens in southwestern Finland. Parasit Vectors. 2016;9:168.
Eisen L. Tick species infesting humans in the United States. Ticks Tick Borne Dis. 2022;13:102025.
Piesman J, Eisen L. Prevention of tick-borne diseases. Annu Rev Entomol. 2008;53:323–43.
Krause PJ, Fish D, Narasimhan S, Barbour AG. Borrelia miyamotoi infection in nature and in humans. Clin Microbiol Infect. 2015;21:631–9.
Eisen RJ, Eisen L. The blacklegged tick, Ixodes scapularis: an increasing public health concern. Trends Parasitol. 2018;34:295–309.
Bai Y, Osikowicz LM, Hojgaard A, Eisen RJ. Development of a quadruplex PCR amplicon next generation sequencing assay for detection and differentiation of Bartonella spp. Front Microbiol. 2023;14:1243471.
Eisen RJ, Paddock CD. Tick and Tickborne Pathogen Surveillance as a Public Health Tool in the United States. J Med Entomol. 2021;58:1490–502.
Lehane A, Maes SE, Graham CB, Jones E, Delorey M, Eisen RJ. Prevalence of single and coinfections of human pathogens in Ixodes ticks from five geographical regions in the United States, 2013–2019. Ticks Tick Borne Dis. 2021;12:101637.
Eisen L, Eisen RJ, Lane RS. Geographical distribution patterns and habitat suitability models for presence of host-seeking ixodid ticks in dense woodlands of Mendocino County, California. J Med Entomol. 2006;43:415–27.
Eisen RJ, Eisen L, Girard YA, Fedorova N, Mun J, Slikas B, et al. A spatially-explicit model of acarological risk of exposure to Borrelia burgdorferi-infected Ixodes pacificus nymphs in northwestern California based on woodland type, temperature, and water vapor. Ticks Tick Borne Dis. 2010;1:35–43.
Norris DE, Klompen JS, Keirans JE, Black WC 4th. Population genetics of Ixodes scapularis (Acari: Ixodidae) based on mitochondrial 16S and 12S genes. J Med Entomol. 1996;33:78–89.
Schrader C, Schielke A, Ellerbroek L, Johne R. PCR inhibitors—occurrence, properties and removal. J Appl Microbiol. 2012;113:1014–26.
Osikowicz LM, Hojgaard A, Maes S, Eisen RJ, Stenglein MD. A bioinformatics pipeline for a tick pathogen surveillance multiplex amplicon sequencing assay. Ticks Tick Borne Dis. 2023;14:102207.
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. https://doi.org/10.1016/S0022-2836(05)80360-2.
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: Architecture and applications. BMC Bioinformatics. 2009;10:421. https://doi.org/10.1186/1471-2105-10-421.
Bogumiła S, Adamska M. Capreolus capreolus and Ixodes ricinus as a reservoir of Bartonella in north-western Poland. Wiad Parazytol. 2005;51:139–43.
Duplan F, Davies S, Filler S, Abdullah S, Keyte S, Newbury H, et al. Anaplasma phagocytophilum, Bartonella spp., haemoplasma species and Hepatozoon spp. in ticks infesting cats: a large-scale survey. Parasit Vectors. 2018;11:201.
Bai Y, Calisher CH, Kosoy MY, Root JJ, Doty JB. Persistent infection or successive reinfection of deer mice with Bartonella vinsonii subsp. arupensis. Appl Environ Microbio. 2011;77:1728–31.
Ziedins AC, Chomel BB, Kasten RW, Kjemtrup AM, Chang CC. Molecular epidemiology of Bartonella species isolated from ground squirrels and other rodents in northern California. Epidemiol Infect. 2016;144:1837–44.
Kosoy M, Mandel E, Green D, Marston E, Jones D, Childs J. Prospective studies of Bartonella of rodents. Part II. Diverse infections in a single rodent community. Vector Borne Zoonotic Dis. 2004;4:296–305.
Telford SR 3rd, Wormser GP. Bartonella spp. transmission by ticks not established. Emerg Infect Dis. 2010;16:379–84.
Tokarz R, Tagliafierro T, Sameroff S, Cucura DM, Oleynik A, Che X, et al. Microbiome analysis of Ixodes scapularis ticks from New York and Connecticut. Ticks Tick Borne Dis. 2019;10:894–900.
Cotté V, Bonnet S, Le Rhun D, Le Naour E, Chauvin A, Boulouis HJ, et al. Transmission of Bartonella henselae by Ixodes ricinus. Emerg Infect Dis. 2008;14:1074–80.
Reis C, Cote M, Le Rhun D, Lecuelle B, Levin ML, Vayssier-Taussat M, et al. Vector competence of the tick Ixodes ricinus for transmission of Bartonella birtlesii. PLoS Negl Trop Dis. 2011;5:e1186.
Acknowledgements
This project was supported in part by an appointment to the Research Participation Program at the Centers for Disease Control and Prevention administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and the Centers for Disease Control and Prevention.
Funding
This research was supported by intramural funding within the Centers for Disease Control and Prevention.
Author information
Authors and Affiliations
Contributions
YB and RJB conceived and designed the study. YB and KM performed the lab testing. LMO, YB, and KM conducted data analysis. SM prepared and organized samples. YB prepared the manuscript draft. All authors reviewed and edited the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Bai, Y., McClung, K.L., Osikowicz, L.M. et al. No evidence of Bartonella infections in host-seeking Ixodes scapularis and Ixodes pacificus ticks in the United States. Parasites Vectors 17, 345 (2024). https://doi.org/10.1186/s13071-024-06386-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-024-06386-3