Abstract
Background
The Cape horseshoe bat, Rhinolophus capensis, is endemic to the Cape region of South Africa. Coalescent analysis of mitochondrial DNA sequence data suggests extensive historical gene flow between populations despite strong geographic variation of their echolocation call phenotype. Nevertheless the fine-scale genetic structure and evolutionary ecology of R. capensis remains poorly understood. Here we describe the development of 10 novel polymorphic microsatellite loci to investigate of the dispersal ecology of R. capensis and to facilitate taxonomic studies of Rhinolophus species in southern Africa.
Findings
We report 10 microsatellite primer pairs that consistently amplify scorable and polymorphic loci across 12 African rhinolophid species. Initial analysis of two populations of R. capensis from South Africa revealed moderate to high levels of allelic variation with 4–14 alleles per locus and observed heterozygosities of 0.450–0.900. No evidence of linkage disequilibrium was observed and eight of the loci showed no departure from Hardy–Weinberg equilibrium. Cross-species utility of these markers revealed consistently amplifiable polymorphic loci in eleven additional rhinolophid species.
Conclusions
The cross-amplification success of the microsatellites developed here provides a cost-effective set of population genetic marker for the study of rhinolophid evolutionary ecology and conservation in southern Africa.
Similar content being viewed by others
Findings
The Cape horseshoe bat (Rhinolophus capensis) has a predominantly coastal distribution encompassing several major biomes in South Africa [1]. Despite evidence for extensive historical gene flow the species is characterised by geographically structured variation in echolocation frequency which appears to be closely coupled to local environmental conditions [2]. Such phenotypic variation is evident in a number of rhinolophids (1; DSJ, unpublished data). Here we develop and test a suite of microsatellite markers (1) to test hypotheses of adaptive evolution in R. capensis and (2) to better understand population genetic structuring and the recent evolutionary history of African rhinolophids.
Twenty-six species of horseshoe bats have been described in sub-Saharan Africa with 11 occurring in the southern African countries of Botswana, Lesotho, Mozambique, Namibia, South Africa, Swaziland, and Zimbabwe [1]. The taxonomy and true number of species of African Rhinolophus nevertheless remains unresolved because of the highly convergent morphology observed across taxa. Currently species delimitation is based on slight variations in morphological measurements and echolocation call frequency [1, 3] but there is much ambiguity in these characters as a result of extensive intraspecific phenotypic variation and the true diversity of species is likely to be underestimated when using these methods alone. The recent inclusion of genetic data has uncovered a number of species complexes in the African rhinolophids (e.g. R. hildebrandtii and R. darlingi) [4, 5]. The use of an integrative approach combining molecular techniques and morphological analyses has enabled the recognition and description of several new species and will undoubtedly contribute to the improved taxonomic resolution of African rhinolophid diversity.
Microsatellite loci are characterised by high polymorphism and co-dominance which make them ideal genetic markers for use in population genetic studies. These molecular markers have also been widely used to study demographic and ecological processes within species and to resolve taxonomic problems among recently diverged lineages [6]. Their value in such studies is further enhanced by the fact that microsatellite markers primarily developed for a specific taxon can also be used on closely related species, making them a cost effective tool for taxonomic, population and conservation genetic studies.
In this study microsatellite markers were isolated following an enrichment protocol developed by Glenn and Schable [7]. To construct the library we used genomic DNA (gDNA) from both R. capensis and a closely related taxa Dent’s horseshoe bat, R. denti. Total DNA was extracted using a DNeasy Blood and Tissue Kit (Qiagen) from biopsy wing punch samples (3 mm) collected from the wing or tail membrane. To increase the amount of enriched fragments, a ‘recovery’ PCR was performed after the initial round of enrichment. Reactions were performed in a final volume of 25 μL containing 1X PCR buffer (10 mM Tris–HCl, 50 mM KCl, pH 8.3), 1.5 mM MgCl2, 0.16 mM of each dNTP, 10 × BSA, 0.5 μM of the Super SNX24 forward primer, 1U Taq DNA polymerase, and approximately 25 ng enriched gDNA fragments. Thermal cycling, performed in an MJ Research DYAD, was performed as follows: 95 °C for 2 min followed by 25 cycles of 95 °C for 20 s, 60 °C for 20 s, and 72 °C for 90 s, and a final elongation step of 72 °C for 30 min. Subsequent PCR fragments were cloned using the TOPO-TA Cloning kit (Invitrogen) following the manufacturer’s protocol. DNA sequencing was performed using the BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) and run on an ABI 3730 DNA Analyzer. From the resulting libraries we identified 50 R. capensis and 55 R. denti clones that contained repetitive elements with sufficient flanking regions for primer development. A total of 55 primer pairs were designed from the positive clones both libraries using the web-based program Primer3Plus [8].
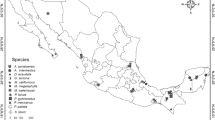
For the initial PCR trials, amplification of 21 and 15 microsatellite loci from the R. capensis and R. denti libraries, respectively, were tested using eight individuals of each species. PCR was carried out in a final volume of 20 µL containing ~25 ng of DNA template, 0.5 µM of forward and reverse primers, 1X PCR buffer, 1.5 mM MgCl2, 0.2 mM of each dNTP and 5 U/µL Kapa Taq DNA polymerase (Kapa Biosystems). PCR amplification was performed under the following conditions: initial denaturation at 95 °C for 5 min, 35 cycles of 95 °C for 45 s, annealing for 45 s at 48–60 °C (gradient tests) and 72 °C for 45 s, followed by a final extension of 72 °C for 10 min. To confirm amplification and identify unambiguous loci PCR products were first visualised on a 3 % agarose gel under UV. Of the initial 36 loci tested, seven loci isolated from R. capensis and three loci from R. denti consistently amplified scorable loci in our target study species. These loci were then used to determine levels of polymorphism using a sample of 40 R. capensis individuals. All loci were then tested for cross-species amplification and polymorphism in 11 additional rhinolophids distributed in southern and central Africa using 2–5 individuals from across their respective ranges (geographic localities indicated in the Additional file 1). The 5′ end of each forward primer was labelled with a fluorescent dye (6-FAM, NED, PET and VIC) for visualisation of the PCR products. For the initial characterisation of loci amplification for each locus was performed individually using 40 R. capensis individuals from two populations (see Table 1 for PCR conditions). PCR products were then pooled for each individual in two different multiplex panels (Table 1).
For cross-species testing of the loci PCR amplification was carried out using a QIAGEN Multiplex PCR Kit on two multiplex panels containing primers labelled with different fluorescent dyes (seven and two loci); one locus (RCA3, T 50 °C) was amplified alone using the protocol described earlier. Multiplex PCR amplifications were performed in a final volume of 20 µL containing 10 µL of QIAGEN Multiplex PCR Master Mix, 2 µL of the 10× primer mix with each primer at a final concentration of 2 µM and 1 µL of DNA template. Thermal cycling, performed in an Applied Biosystems Veriti 96-Well Thermal Cycler, was performed as follows: 95 °C for 15 min; 35 cycles of 94 °C for 30 s, 57 °C for 90 s, 72 °C for 60 s and a final step of 60 °C for 30 min. PCR products separated by capillary electrophoresis on an AB3730 DNA analyser (Central Analytical Facilities, Stellenbosch, South Africa). Alleles were sized using an internal size standard (GeneScan600LIZ) and scored using the software GeneMarker 2.6.3 (SoftGenetics).
To characterise variation at these loci individuals of R. capensis were genotyped from two colonies representing the full geographic extent of the species (Table Farm: −33.28, 26.42; Heidehof: −34.62, 19.50). For each locus a number of parameters were calculated in GenAlEx 6.5 [9], including the number of alleles observed (NA), effective number of alleles (NE), observed and expected heterozygosities (HO and HE). Tests of Hardy–Weinberg equilibrium (PHW) and tests of linkage of disequilibrium (LD) between loci and populations were calculated in Genepop 4.3 [10]. MicroChecker 2.2.3 [11] was used to detect typing errors and null alleles. The polymorphism information content (PIC) for each locus was assessed with Cervus 3.0.7 [12].
Across the ten loci the number of alleles per locus, NA, ranged from 4 to 14, while the effective number of alleles per locus, NE, ranged from 2.48 to 9.63 in R. capensis (Table 1). The observed (HO) and expected (HE) heterozygosities ranged from 0.450 to 0.900 and 0.597 to 0.896, respectively. Deviation from Hardy–Weinberg equilibrium (HWE) was not found evident except for locus RCA2 (P < 0.05 after Bonferroni adjustment). Null alleles was found for two loci (Table Farm, RDE2 r = 0.122; Heidehof, RDE2, r = 0.172, RCA2, r = 0.201). All of the loci revealed moderate to high PIC values (0.631–0.904).
Cross-amplification tests of the 10 loci were successful on all the 11 species selected, and 98 % of loci were polymorphic (Table 2). The number of Rhinolophus species that showed polymorphism across the 10 loci ranged from 11 to 12 and the number of polymorphic loci within taxa ranged from 9 to 10.
The 10 microsatellite markers reported here will be highly useful for a wide range of population and evolutionary genetic studies in the African Rhinolophidae. Microsatellite markers presented here, together with mitochondrial and additional nuclear markers, will provide valuable tools to resolve the systematic and taxonomic relationships of several species complexes of Rhinolophus.
Availability of the supporting data
The microsatellite sequences are available through the National Centre for Biotechnology Information (http://www.ncbi.nlm.nih.gov); GenBank accession numbers KP842262–KP842271.
References
Monadjem A, Taylor PJ, Cotterill FPDW, Schoeman MC. Bats of Southern and Central Africa a biogeographic and taxonomic synthesis. Johannesburg: Wits University Press; 2010. p. 534.
Odendaal LJ, Jacobs DS, Bishop JM. Sensory trait variation in an echolocating bat suggests roles for both selection and plasticity. BMC Evol Biol. 2014;14:60.
Csorba G, Ujhelyi P, Thomas N. Horseshoe bats of the world (Chiroptera: Rhinolophidae). Alana Books; 2003. p. 160i.
Taylor PJ, Stoffberg S, Monadjem A, Schoeman MC, Bayliss J, Cotterill FPD. Four new bat species (Rhinolophus hildebrandtii Complex) reflect Plio-Pleistocene divergence of dwarfs and giants across an afromontane archipelago. PLoS One. 2012;7:e41744.
Jacobs DS, Babiker H, Bastian A, Kearney T, van Eeden R, Bishop JM. Phenotypic convergence in genetically distinct lineages of a Rhinolophus species complex (mammalia, chiroptera). PLoS One. 2013;8:e82614.
Selkoe KA, Toonen RJ. Microsatellites for ecologists: a practical guide to using and evaluating microsatellite markers. Ecol Lett. 2006;9:615–29.
Glenn TC, Schable NA. Isolating microsatellite DNA loci. Methods Enzymol. 2005;395:202–22.
Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JAM. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007; 35 (Web Server issue):W71–4. http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi/.
Peakall R, Smouse PE. GenALEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics. 2012;28:2537–9.
Rousset F. GENEPOP’007: a complete re-implementation of the GENEPOP software for Windows and Linux. Mol Ecol Resour. 2008;8:103–6.
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. Micro-Checker: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes. 2004;4:535–8.
Kalinowski ST, Taper ML, Marshall TC. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol Ecol. 2007;16:1099–106.
Authors’ contributions
JMB and NN were responsible for the design of the study and the interpretation of results. KF developed the clone libraries. NN performed the lab work and data analysis. NN and JMB wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgements
We thank L. Odendaal for R. capensis and R. denti tissues samples. Microsatellite development was carried out in the Field Museum’s Pritzker Laboratory for Molecular Systematics and Evolution operated with support from the Pritzker Foundation. This research was funded by grants to DSJ from the South African Research Chair Initiative of the Department of Science and Technology and administered by the National Research Foundation of South Africa. NN is funded by aSouth African Research Chair Initiative Postdoctoral fellowship.
Compliance with ethical guidelines
Competing interests The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Nesi, N., Jacobs, D.S., Feldheim, K. et al. Development and characterization of 10 microsatellite markers in the Cape horseshoe bat, Rhinolophus capensis (Chiroptera, Rhinolophidae) and cross-amplification in southern African Rhinolophus species. BMC Res Notes 8, 477 (2015). https://doi.org/10.1186/s13104-015-1465-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13104-015-1465-5