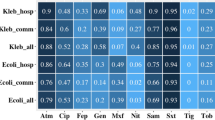
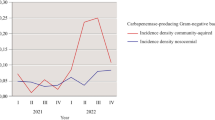
Abstract
Background
The burden of antibiotic resistant infection is mainly felt in low-to-middle income countries, where the rate of antimicrobial resistance is largely under-surveyed and under huge pressure from unregulated, disparate and often self-guided access to antimicrobials. Nosocomial infections from hospital environments have been shown to be a particularly prevalent source of multi-drug resistant strains, yet surveillance of hospital environmental contamination is often not investigated.
Methods
The study was prospective, observational and cross-sectional, sampling 231 high and low touch surfaces from 15th March to 13th April 2021, from five wards in the Cape Coast Teaching Hospital, Ghana. Microbial growth in the presence of vancomycin and either meropenem or cefotaxime was examined and bacterial species were identified by MALDI-TOF. The presence of common extended-spectrum β-lactamases (ESBL) and carbapenemase antimicrobial resistance genes (ARG) were identified through PCR screening, which were confirmed by phenotypic antimicrobial susceptibility determination. Isolates positive for carbapenem resistance genes were sequenced using a multi-platform approach.
Results
We recovered microbial growth from 99% of swabs (n = 229/231) plated on agar in the absence of antimicrobials. Multiple sites were found to be colonised with resistant bacteria throughout the hospital setting. Bacteria with multi-drug resistance and ARG of concern were isolated from high and low touch points with evidence of strain dissemination throughout the environment. A total of 21 differing species of bacteria carrying ARG were isolated. The high prevalence of Acinetobacter baumannii carrying blaNDM-1 observed was further characterised by whole genome sequencing and phylogenetic analysis to determine the relationship between resistant strains found in different wards.
Conclusion
Evidence of multiple clonal incursions of MDR bacteria of high sepsis risk were found in two separate wards for a regional hospital in Ghana. The prevalence of multiple blaNDM carrying species in combination with combinations of ESBLs was particularly concerning and unexpected in Africa. We also identify strains carrying tet(X3), blaVIM-5 or blaDIM-1 showing a high diversity of carbapenamases present as a reservoir in a hospital setting. Findings of multi-drug resistant bacteria from multiple environmental sites throughout the hospital will inform future IPC practices and aid research prioritisation for AMR in Ghana.
Similar content being viewed by others
Background
Antimicrobial resistance (AMR) is a recognized global health crisis with annual projections estimating up to 10 million lives being lost and costing $100 trillion globally by the year 2050 [1]. Of particular concern is antibiotic resistance among Gram-negative bacterial species which presents an ever-increasing threat due to the diverse pathologies they are capable of causing, as well as their acquired and intrinsic resistance mechanisms which render many existing antibiotics ineffective. This leads to increased morbidity, mortality, and adverse patient outcomes [2]. In light of this problem, the World Health Organization (WHO) published the global Pathogen Priority List which contains selected bacteria pathogens for which new treatments are urgently needed. The ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp.) pathogens, out of which four are Gram-negative organisms designated as “critical priority status” [3] further highlights their importance in the spread and propagation of AMR and disease burden. These pathogens have been well documented for their significant contribution to mortality in a wide range of clinical presentations such as ventilator-acquired pneumonia, central line-associated bacteremia [4] nosocomial meningitis [5], neonatal sepsis [6], and bloodstream infections [7, 8], with most of these being hospital-associated infections [9]. Additionally, several strains of these bacterial species have propagated worldwide with the concomitant spread of the mobile resistance genes they harbour, especially those conferring resistance to key classes of antibiotics namely the β-lactam class (including carbapenems) of antibiotics [10,11,12,13,14,15,16]. Carbapenem antibiotics within this class are more expensive and mostly reserved as second or last-line options for hard-to-treat infections or when first-line therapy proves to be ineffective in Ghana [17]. The WHO AWaRe (Access, Watch, and Reserve) classification categorizes most third-generation cephalosporins and carbapenems as “Watch” drugs based on their likely potential to induce antibiotic resistance which could have negative consequences for wider populations [18]. Extended-Spectrum β-Lactamase (ESBL) and carbapenemase genes are implicit in this occurrence and are known to spread relatively rapidly within and between countries and continents [6, 19].
In Africa, data on the prevalence and characterization of healthcare-associated infections (HAI’s) caused by these organisms is available though not comprehensive [20,21,22]. Additionally, where infectious diseases have a high burden coupled with sometimes inadequate laboratory capacity, the effects of AMR may be higher especially in low- and low-middle-income countries in the region [23]. Other factors such as high poverty rates, burdened healthcare systems, and poor infection prevention and control practices (IPC) may further exacerbate the issue [24]. In Ghana specifically, several studies exist on phenotypic and molecular characterization of these priority pathogens mainly with a focus on clinical isolates. Intermediate to high rates of resistance to cephalosporins and carbapenems similar to estimates in the continent and globally were reported in these studies despite limitations of small sample sizes in some cases [25,26,27,28,29,30,31,32,33]. With regards to surveillance of AMR from hospital environments, research studies exist [34,35,36,37] but are inadequate especially for Gram-negative pathogens considering the important role hospital environments can play in transmission. The presence of resistant bacteria in hospital environments are a critical component of nosocomial infection. Contaminated intermediate objects represent a common segue of transmission between patients, from visitors to patients, or from healthcare workers to patients [38, 39], which also impact on the choice of antibiotic prophylaxis for surgeries [40]. The Ghanaian National policy on IPC overseen by the Ministry of Health delineates roles and good practices to be followed among all levels of healthcare [38] and emphasizes the importance of environmental cleaning and disinfection and generation of robust data on monitoring and evaluating these practices to advise the MOH research agenda on priority areas of IPC at either national, regional, and district levels.
This study investigated the presence and prevalence of ESBL- and carbapenemase-carrying bacterial species by swabbing "high touch" and “low touch” areas in five wards in the Cape Coast Teaching Hospital, Ghana. We performed microbial culture and PCR on the total growth to detect the presence of ESBL genes (blaCTX-M-15, blaOXA-1, blaTEM, blaSHV) and carbapenemase genes (blaNDM, blaOXA-48-like, blaKPC). Positive samples were characterised by MALDI-TOF to identify the bacterial isolate containing the antimicrobial resistance genes (ARG), and isolates containing carbapenemase genes were further analysed by whole- genome sequencing (WGS) and antibiotic susceptibility testing (AST) with a range of antibiotics.
Methods
Study design
This study aimed to establish environmental antimicrobial resistance surveillance in the Cape Coast Teaching Hospital (CCTH) in Ghana to enable adequate identification of resistance patterns and genetic mechanisms of bacterial isolates recovered from fomites classified as “high touch” and “low touch” areas in inpatient wards and to determine their potential as sources of these pathogens and implications in hospital-acquired infections. The study was a prospective, observational, cross-sectional study that employed sampling from 15 March to 13 April 2021. A total of 231 swabs were collected from five wards in CCTH namely, Obstetrics and gynaecology (OG), Neonatal Intensive Care Unit (NICU), Female Medical Ward (FMW), Accident and Emergency Ward (AE), and the Paediatric Ward (PW). These were selected purposively based on local clinical susceptibility data (based on CLSI M-100 30ed. Guidelines) [41] indicating a high occurrence of potential ESBLs i.e., resistance to third-generation cephalosporins (cefotaxime, ceftriaxone, or cefuroxime) for the year 2020 via the WHONET software (unpublished data).
CCTH is a major tertiary hospital located in the Central Region of Ghana, West Africa. It is a 400-bed capacity institution that offers specialist services and serves a population of 170,000 in the Cape Coast Metropolitan area [42]. It also receives referrals from within the Central, Western and other regions in the country.
Swab sites were selected based on guidance from the National IPC policy and published literature. Items were further classified as “high touch” and “low touch” areas based on how likely healthcare workers and patients would encounter them based on the investigators’ judgments and observations, including:
-
1.
Low touch areas IV stands, bed wheels, bedpans or trays, sphygmomanometer cuffs, desk surfaces, waiting area chair handles, thermometers and stethoscopes.
-
2.
High touch areas Bed handles, mattress, pillows, bedside carts, bedside cabinet tops and drawers, computer keyboards, window levers, light switches, wall socket switches, tap handles, and washroom door handles.
Items were swabbed between 9:00 am and 2:00 pm on weekdays. Ward staff were not informed of the swabbing date to capture the true prevalence of gram-negative bacteria (GNB) after routine ward cleaning. Swabs were stored in Amies charcoal transport medium and were refrigerated at 4 °C, shipped via a courier service per UN3373 regulations to Cardiff University and stored at 4 °C prior to testing.
Microbial culture and PCR of target antibiotic resistance genes
Swabs were streaked onto three chromogenic agars; no antibiotics, vancomycin and cefotaxime (VC, 10 mg/L and 1 mg/L) to select for cefotaxime resistance an indicator of ESBLs, and vancomycin and meropenem (VM, 10 mg/L and 1 mg/L) to screen for the presence of carbapenemases. Agar plates were incubated aerobically for 48 h at 37 °C.
VC and VM plates recording growth were streaked and 10 µl from the first quadrant (mixed microbial community) was suspended into 150 µl of sterile distilled molecular grade water. Three PCR Master Mix preparations were made: ESBL Multiplex Master mix was used in detecting blaTEM blaSHV, blaOXA-1 [43], Carbapenemase Multiplex Master mix for detecting blaNDM, blaOXA-48-like variants, and blaKPC and CTX-M Master mix for detection of blaCTX-M-15 (see Additional file 1: Table S1 for primer sequences and Additional file 1: Table S2 for conditions).
One microlitre of the bacterial suspension was added to 24 µl of each of the three PCR Mastermixes, PCR and gel electrophoresis (300 V for 40 min) were performed on isolates. Those yielding positive for any of the tested resistance genes were further sub-cultured to obtain pure single colonies which were then re-screened to confirm the presence of the gene of interest.
Positive isolates were identified using a Microflex LT MALDI-TOF MS (Bruker Daltonik) with α-cyano-4-hydroxycinnamic acid matrix (Sigma–Aldrich). Bacterial isolates were stored in TS/72 beads (Technical Service Consultants) at − 80 °C and the original swabs were stored at 4 °C.
Descriptive statistics were used in determining the prevalence of ESBL- and carbapenemase-producing GNB recovered from the ward environments as well the number and types of resistance genes they harboured. Microbial growth and PCR data were recorded and analysed using Microsoft Excel.
Antimicrobial susceptibility testing (AST)
Minimum inhibitory concentrations (MIC) were determined by agar dilution with a multipoint inoculator (MAST Uri-dot) using cation adjusted Mueller–Hinton agar for meropenem, ceftazidime, tigecycline, gentamicin and colistin. Escherichia coli ATCC 25922, Klebsiella pneumoniae ATCC 700603, Pseudomonas aeruginosa ATCC 2785, and Escherichia coli ATCC 13846 (mcr1 positive) were used for quality control and the results were interpreted according to the EUCAST v11 guidelines and ECOFF values where applicable [44]. Agar dilution was used for colistin MIC as concordance between agar and microbroth dilution methods has previously been published [45].
Whole-genome sequencing (WGS)
Isolates with a carbapenemase gene were selected for WGS using an Illumina MiSeq as described in Sands et al. [6]. Isolates were selected for additional long read sequencing guided by the initial short read bioinformatics analysis. gDNA was prepared as previously described [6] using the Qiagen QIAamp DNA kit and extracted on the QIAcube platform (Qiagen). gDNA was quantified using the dsDNA HS assay kit and the Qubit 4.0 fluorometer (Thermofisher), purified and concentrated using SPRI beads (Mag-Bind TotalPure, Omega) at a 1:1 ratio with a final elution volume of 15 µl to achieve an optimal range between 40 and 60 ng/µl. gDNA was re-quantified with the dsDNA BR assay kit. Genomic libraries were prepared using the Rapid Barcoding Kit (SQK-RBK004; Oxford Nanopore), sequenced on a R9.4 flow cell using a MinION (Oxford Nanopore Technologies) and basecalling was performed using Guppy v4.0.9 within MinKNOW v20.06.4.
Short read bioinformatics analysis
Fastq reads were subject to quality control (QC) analysis and trimming using fastqc (v0.8.11) [46] and trimgalore (v0.5.0) [47] respectively. Reads were assembled into contigs using shovill (v0.9.0) [48] and assembly metrics were assessed using quast (v5.0.2) [49]. ABRicate (v0.9.7) [50] (> 98% coverage and identity) was used to detect ARG and virulence factors (VF) with accompanying databases Resfinder and VFDB. In silico MLST was determined using mlst (v2.17.6) [51] and the Pasteur scheme was used [52]. Genomes (contigs) were annotated using Prokka (v1.14.0) [53] and the resulting.gff files were used to create a core genome alignment of Acinetobacter spp. using Roary (v3.12.0) [54], and IQ-tree (v2.0) [55] was used to create a maximum likelihood phylogeny. ModelFinder within IQ-tree utilised the GTR+F+I+G4 model and 1000 bootstrap replicates were performed. For SNP analysis a local reference genome was selected for additional long read sequencing using Oxford Nanopore MinION (details described below) to produce a high-quality genome. Snippy (v4.4.5) [56] was used to map the fastq reads against the reference. The SNPs were aligned using snippy-core and the SNP sites were extracted using snp-sites (v2.5.1) [57]. Recombination events were removed with Gubbins (v.2.3.4) [58] and IQ-tree (v2.0) [55] was used to create a maximum likelihood phylogeny. Pairwise SNP distances were generated using snp-dists (v0.7) [59]. Phylogenetic trees were mid-rooted, visualised and annotated using iTOL (v5.7) [60].
Long read bioinformatics analysis
Long reads were demultiplexed using porechop (v0.2.4) [61] and assembled against corresponding short reads generated from the Illumina MiSeq using Unicycler (v0.4.7) [62] with default parameters. The hybrid assembly was assessed using quast (v5.0.2) and ABRicate (v0.9.7) [50] (> 98% coverage and identity) was used to detect antimicrobial resistance genes (ARG). The contig containing the ARG of interest was exported to a separate fasta file for comparative analysis using Bandage (v0.8.1) [63]. Plasmid sequences were uploaded to PLSDB [64] and mash dist search strategy was used to assess for similar plasmids in the database. The MobileElementFinder database (v1.0.2) was downloaded and ABRicate [50] was used to search for the mobile genetic elements (MGE) genomic context of ARG.
Data for patient isolates
Comparison for data collected between 1st January and 30th June 2021, for bacterial isolates submitted to the local microbiological diagnostic laboratory at CCTH were examined for comparison to the in-depth swab analysis performed at Cardiff University. Ethical approval to include patient isolate data in the analysis was granted (CCTHERC/EC/2021/027) by the local ethical committee at CCTH. Data was restricted to ward origin of isolates, bacterial species and AMR investigation. All patient isolates included were inoculated on MacConkey agar with urine isolates inoculated exclusively on Cysteine Lactose Electrolyte Deficient (CLED) agar (Himedia, India) plates for 24 h at 35 ± 2 °C. Sub-culturing was employed to obtain pure colonies which were subjected to gram-staining and microscopy. Biochemical analysis was performed under aerobic conditions and these plates were incubated for 24 h at 35 ± 2 °C and included metabolism of indole, citrate, urea, glucose, lactose, hydrogen sulphide production, gas, oxidase presence and motility (differential determination given in Additional file 1: Table S3). Antibiotic susceptibility testing (AST) via the disk diffusion method using a 0.5 McFarland standard bacterial suspension of pure isolates on Muller Hinton agar (Himedia, India) incubated for 24 h aerobically at 35 ± 2 °C. AST data were collated by WHONET software (version 21.11.24) utilising CLSI 2021 breakpoints, and the following parameters: 1. analysis type – study = RIS and test measurements, antibiotics = CRO, CTX, SAM, TZP; 2. Organisms – ALL = All organisms; 3.All other parameters were used in their default.
Results
Microbial culture and PCR of environmental swabs
The largest number of swabs collected was in the PW (n = 71) followed by the NICU (n = 53). For the FMW, OG and AE n = 36, 38, and 33 swabs were collected respectively (Table 1). We recovered bacterial growth from all but 2 swabs (n = 229/231) collected during this study when plated onto non-selective agar, in the absence of antibiotics. Surfaces from OG produced the highest level of GNB growth on both VC and VM agar (Table 1). Swabs taken from the FMW produced the least GNB growth on VC agar (n = 24/36, 67%) and second to least growth on VM (n = 17/36, 47%). Surfaces from NICU produced the least growth on VM agar (n = 23/53, 43%) (Table 1).
Swabs (n = 231) were collected from a total of 28 different surfaces (Additional file 1: Table S4) with one-third (n = 77/231, 33%) collected from the immediate surroundings of patient beds (sheets, pillows, bed wheels, bed pans, etc., Additional file 1: Table S4). The majority of swabs collected around patient beds produced growth on both agars supplemented with antibiotics (VC n = 71/77, 92% and VM n = 61/77, 86%). Computer keyboards and desk surfaces produced very high GNB growth rates on VC agar (n = 12/13, 92% and n = 11/13, 85% respectively), similar to tap handles and thermometers (n = 14/17, 82% and n = 9/10, 90%). Surface swabs across computer keyboards, desks, tap handles and thermometers produced a similar GNB growth recovery on VM (between 60 and 71%, Additional file 1: Table 4).
In total, 26 surfaces (n = 26/231, 11%) were positive for blaNDM (n = 13/26 50% PW, n = 11/26 42% OG and n = 2/26 8% NICU), 12/26 were collected from the immediate surroundings of patient beds, two from bed wheels (NICU and PW), three from computer keyboards and desks from different wards (OG and PW), three from tap handles or washroom doors from different wards (OG and PW), as well as assorted other sites including a thermometer (PW), sphygmomanometer (OG), medicine trolley (NICU) IV stand (PW) and a wall socket (OG) (Fig. 1). One surface (PW-0012) was positive for multiple ARG including blaNDM, blaOXA-48-like (the only surface positive for blaOXA-48-like), blaSHV and blaCTX-M-15. There were 28 surfaces (n = 28/231, 12%) positive for at least one ESBL gene (Fig. 1) and five surfaces (n = 3 OG, n = 2 PW) contained both carbapenemase and ESBL ARG genes.
Gram-negative bacterial (GNB) isolates positive for ESBL and/or carbapenemase ARG
A total of 71 bacterial isolates from 21 bacterial species (including three isolates into the ‘unidentified’ category where the MALDI-TOF failed to accurately speciate the colony) containing at least one target ESBL (n = 43, 61%) and/or carbapenemase (n = 28, 39%) gene (Table 2) were recovered. Ten isolates from six GNB species were recovered from the AE, all of which contained at least one ESBL gene (Fig. 2a), as were 13 GNB isolates from NICU, 10 containing ESBL genes, and three with blaNDM genes (Fig. 2a). In OG 8/11 ESBL containing isolates were K. pneumoniae whereas A. baumannii (n = 11/11, 100%) isolates carried blaNDM (Fig. 2a). The highest frequency of isolates recovered was from PW (n = 24), with a similar split between GNB isolates carrying ESBL genes and carbapenemase genes. Eight different species were identified harbouring ESBL in PW and similarly to OG the most frequent species was K. pneumoniae (n = 3/11, 27%) (Fig. 2a).
a Sunburst diagram summarising bacterial species identified (outer ring); ACB, Acinetobacter baumannii complex; ACS, Acinetobacter spp.; ECO, E. coli; EBU, Enterobacter bugandensis; ECL, Enterobacter cloacae; EHE, Escherichia hermannii; KPN, Klebsiella pneumoniae; LEA, Leclercia adecarboxylata; MIX, Mixta calida; PAA, Pantoea agglomerans; PSF, Pseudomonas fulva; PSS, Pseudomonas stutzeri; UNI, Unidentified. The carriage of ESBL and/or carbapenemase genes (middle ring); ESBL (blaCTX-M-15 and/or blaOXA-1 and/or blaSHV and/or blaTEM), All (all ESBL genes identified and blaNDM and blaOXA-48-like). The ward the sample was collected from (inner ring); AE, accident and emergency; OG, obstetrics and gynaecology; NICU, Neonatal Intensive Care Unit; PW, paediatric ward
When delineating the occurrence of ESBL genes, six isolates from five different species carried blaCTX-M-15 only (Fig. 2b). Three different groups of isolates were detected containing dual ESBL genes (blaTEM + blaSHV, blaOXA-1 + blaCTX-M-15, blaTEM + blaOXA-1) from AE, NICU, and OG, however the majority of isolates (n = 30/43, 70%) contained ≥ 3 ESBL genes, and n = 19 isolates carried all four target ESBL ARG (Fig. 2b), including all 14 K. pneumoniae (10 from OG, 2 from PW, 2 from NICU), three Enterobacter cloacae (2 from AE, 1 from OG), and two A. baumannii complex (both from AE: one A. baumannii and one Acinetobacter pittii). Most carbapenemase genes encountered in the study were detected from Acinetobacter isolates (n = 25/28), the remaining belonging to Pseudomonas stutzeri (n = 2/28) and Klebsiella pneumoniae (n = 1/28). The carbapenemase-carrying K. pneumoniae was also the only isolate positive for blaOXA-48-like, and the only isolate carrying all five target ARG selected for the study. Due to the multi-drug resistance (MDR) focus of this study the carbapenemase isolates were selected for further characterisation by AST and WGS.
Species diversity, sequence type (ST) analysis, and genomic isolate relatedness
There were 28 isolates with WGS data, and n = 25 isolates were Acinetobacter, from eight different species. Raw sequencing reads from NICU-0042 (unidentified on the MALDI-TOF) were excluded from analysis after failing QC. The most frequent species identified was A. baumannii (n = 14) (Table 3, Fig. 3), three isolates were identified as Acinetobacter indicus and three were identified as Acinetobacter nosocomialis. A single isolate each represented five other Acinetobacter species (Table 3). Two isolates were identified as Pseudomonas stutzeri, and although both were negative for blaNDM-1 by WGS analysis, one was found to contain blaVIM-5 and the other was found to carry blaDIM-1 and were therefore kept within the dataset for analysis. Additionally, one carbapenemase positive isolate was identified as K. pneumoniae (ST152) and contained both blaNDM-1 and blaOXA-48-like.
A mid-point rooted core genome phylogenetic tree of blaNDM positive Acinetobacter isolates. Leaf labels are isolate codes with the ward abbreviation (PW, paediatric ward; OG, obstetrics and gynaecology; NICU, neonatal intensive care) and swab number. The sequence type (ST) follows the leaf node where applicable. The presence/absence of antibiotic resistance genes is colour coded per class; aminoglycoside (green), β-lactam (purple), trimethroprim (olive), phenicol (lilac), macrolide (pink), sulphonamide (blue), tetracycline (orange)
The most frequent ST for A. baumannii was ST109 (ST542 in the Oxford scheme) with n = 10 isolates (Fig. 3). All ten ST109 A. baumannii isolates were isolated from hospital swabs collected on the same day from the same ward, OG (Fig. 4a). Of the eight isolates of sufficient genome coverage for SNP analysis, the pairwise SNP distance was ≤ 6 SNPs. The most genetically distant ST109 isolate, OG-0027, was isolated from a tap handle in a patient washroom and was between 3 and 6 pairwise SNPs distant from the remaining ST109 isolates. Four isolates had 0 pairwise SNPs (OG-0001, OG-0004, OG-0018, and OG-0020) and these were isolated from samples collected from both low touch and high touch surfaces including patient bed materials across the ward from west wing B to west wing C, a sphygmomanometer and a desk surface used by staff at the general waiting area suggesting a transmission cluster and spread of carbapenem resistant A. baumannii (CRAB) (Fig. 4ab). Each ST109 isolate was resistant to β-lactam antibiotics tested (meropenem and cefotaxime) and gentamicin, whereas all were sensitive to colistin and tigecycline (Table 3, Fig. 4c).
a An outline of the obstetrics and gynaecology ward with red dots indicating the locations of the swabs containing the cluster of ST109 A. baumannii. b A SNP heatmap revealing the pairwise SNPs between the isolates with sufficient genome coverage for analysis. c A phenotype to genotype linkage for the antibiotics tested, where β-lactam resistance includes antibiotics meropenem and cefotaxime
In silico virulence factors (VF) were screened for in A. baumannii isolates and we were specifically screening for the presence of VF associated with biofilm production including biofilm-associated protein (bap) gene, ompA, csuE, fimH, epsA, bfmS, ptk, pgaB, csgA, kpsMII and blaPER-1 (a β-lactamase shown to enhance biofilm formation and attachment, especially to epithelial cells). All ST109 A. baumannii contained the greatest number of biofilm-related genes (bap, bfmS, csuE, ompA and pgaB), whereas the four single ST A. baumannii isolates (ST1, ST78, ST132 and ST1136) did not carry bap. The only other notable identification of VFs from in silico screening of Acinetobacter spp, was the identification of both bfmS and ompA in all three A. nosocomialis strains isolated from PW (Table 3).
Other in silico STs detected for Acinetobacter included four A. baumannii isolates each of a different ST; ST1, a globally disseminated strain clonal complex (CC) 1 from OG, ST78 and ST1136 also from OG, and ST132 from PW. All isolates (n = 3) identified as Acinetobacter nosocomialis were typed as ST1326 and were from the PW. There were two sampling time points whereby the first A. nosocomialis isolate was recovered from swabbing the insides surfaces of the finger clip of a pulse oximeter in the west wing of the ward. One week later two additional A. nosocomialis isolates were recovered from a thermometer tip and handle in the west wing and a keyboard and mouse at the general nursing station. Although a small sample size, pairwise SNP distances suggest that these isolates are within 10 SNPs and are therefore likely to be related in a transmission cluster given the locality within the same ward and time frame. For this analysis the internal short read reference genome was utilised (PW0008) as whilst scanning the NCBI database and the PubMLST database [72] for an appropriate reference genome for SNP calling, we found only one ST1326 A. nosocomialis genome from a neonatal blood culture collected in 2017 in Nigeria [6] however this was approximately 10,000 SNPs distant from the PW ST1326 A. nosocomialis.
Mechanisms of antimicrobial resistance in bla NDM-1 positive isolates
We performed AST via determination of the minimum inhibitory concentrations (MIC) for all isolates subject to WGS analysis. Due to the selective screening of ARG during this study and the focus for the detection of MDR bacteria, we tested meropenem, gentamicin, cefotaxime, colistin and tigecycline. All isolates were resistant to meropenem with the MIC range 16–256 mg/L (Table 3) (n = 27, 100%, it was not possible to test one A. nosocomialis and one P. stuzteri isolates due to recovery issues from −80 °C).
The K. pneumoniae isolate characterised by WGS and MIC (PW0012.2) contained blaCTX-M-15, blaSHV-1, blaOXA-1 in addition to blaNDM-1 and blaOXA-48-like, was resistant to meropenem (MIC = 128 mg/L), cefotaxime (MIC = > 256 mg/L), gentamicin (MIC = 64 mg/L) and colistin (MIC = 32 mg/L) although mcr genes were not detected nor were mutations in the mgrB gene often conferring resistance to colistin in K. pneumoniae, this isolate was susceptible to tigecycline (MIC = 2 mg/L) (Table 3). Hybrid de novo genome assembly (short read and long read sequences) for PW0012.2 revealed the blaNDM-1 and blaOXA-48 were located on the same 273,549 bp IncHI1B/IncFIB plasmid along with six other ARG from four antibiotic classes (Table 4).
All 25 Acinetobacter isolates were blaNDM-1 positive and the hybrid assembly of three different Acinetobacter isolates reveals different size plasmids circulating in the hospital environment all containing blaNDM (Table 4). The blaNDM-1 gene in a 309,568 bp plasmid in an A. baumannii, ST132 isolate (PW0012-1) was carried by the transposon Tn125. The isolate identified to Acinetobacter sp. by WGS carried a 53,570 bp plasmid with blaNDM-1 and blaOXA-58, whereas a 268,855 bp plasmid carried by ST109 A. baumannii co-harboured blaNDM-1 and blaOXA-23 (Table 4). Regardless of the plasmid genomic context, bleMBL (a gene conferring resistance to bleomycin and bleomycin-like antibiotics) was identified downstream of all four isolates.
There were 14 different blaOXA variants (both ESBL and carbapenemase variants) detected with n = 24/25 (96%) of Acinetobacter genomes containing carbapenem-hydrolysing oxacillinase (CHO) ARG including blaOXA-23, blaOXA-58, and blaOXA-67 (blaOXA-51-like), although blaOXA-67 were only detected in the ST109 A. baumannii cluster (Table 3, Fig. 3). Six Acinetobacter from six different species carried blaOXA-58 however blaOXA-58 was not detected in A. baumannii isolates whereas blaADC was detected in all A. baumannii isolates only. Whilst there is no Acinetobacter EUCAST breakpoint or ECOFF value for cefotaxime, the MIC values obtained were very high, ranging between 128 and > 256 mg/L (Table 3).
Resistance to gentamicin was high (n = 25/27, 93% [n = 27 for MIC, n = 28 with WGS/ARG data) with 56 aminoglycoside ARG from 13 variants found (Table 3, Fig. 3). The most frequent were aac(3)-IId (n = 19/28, 68%), aph(6′)-Id (n = 8/28, 29%) and aph(3′)-Id (n = 7/28, 25%). On the other hand, resistance to colistin was low overall (n = 2/27, 7%), colistin resistance was observed within the K. pneumoniae isolate (PW0012.2; MIC 32 mg/L) and an Acinetobacter junii isolate (PW0056; 8 mg/L). Only one isolate (n = 1/27, 4%) had a tigecycline MIC of 8 mg/L, an A. baumannii harbouring a tet(X3) gene (PW0012.1), however (n = 9/26, 35%) Acinetobacter isolates (inclusive of PW0012.1) had resistant MICs ranging between 1 and 8 mg/L. Furthermore, we detected tet(X3) in an additional isolate, identified as A. johnsonii, although this was sensitive to tigecycline (MIC 0.5 mg/L). The two Acinetobacter carried tet(X3) at 100% coverage and 100% identity to the reference, a mobile resistance gene that confers resistance to tigecycline. The A. baumannii isolate with tet(X3) (PW-0012.1) was typed as ST132 (ST1181 Oxford scheme). Complementary long read sequencing and annotation of the complete plasmid with tet(X3) reveals the tet(X3) gene is carried on the same plasmid as blaNDM-1. Only short read WGS was available for the Acinetobacter johnsonii isolate positive for tet(X3), genetic context could not be extrapolated as the contig containing the tet(X3) was 4304 bp.
In terms of additional ARGs detected in the WGS data, trimethoprim (dfrA1, dfrA12, dfrA14, and dfrA20) ARGs were detected in four species including A. baumannii, A. indicus and K. pneumoniae whereas fluoroquinolone ARG (OqxA/B, qnrB and qnrVC1) were detected in two isolates, K. pneumoniae and one P. stutzeri. Macrolide ARG (mph(E) and msr(E)) were detected in (n = 26/28, 93%) and sulphonamide ARG (sul1 and sul2) were detected in all isolates except the P. stutzeri isolates.
Antimicrobial resistance data for patient isolates collected locally at CCTH
Ninety-five patient isolates were collected between 1st January to 30th June 2021 (pre- and post-swab collection) and these consisted of 48. Escherichia coli, 26 Klebsiella spp., 1 K. oxytoca, 2 K. pneumoniae, 5 Enterobacter spp., 12 Pseudomonas spp. and 1 P. aeruginosa isolates. MICs were determined against ampicillin-sulbactam, carbapenems (ceftriaxone or cefotaxime), and piperacillin/tazobactam. All isolates tested were resistant to both ceftriaxone and cefotaxime, with the exception of 7/45 E. coli isolates examined, indicative of prevalent ESBLs and carbapenemases consistent with our environmental swab findings. A smaller sub-set (N = 27, due to restricted availability of antimicrobial disks) was examined for susceptibility to β-lactam + β-lactamase inhibitor combinations, and also showed 100% resistance except for 1/12 of the E. coli isolates. However, no data was recorded for Acinetobacter spp. during this period because the local microbiology laboratory is unable to identify this species through the limited available biochemical tests available locally. Acinetobacter spp. could have been recorded as Enterobacteriaceae, Enterobacter spp. or Citrobacter spp. based on the local SOP as Acinetobacter spp. was not one of the potential determinants for the biochemical outcome flowchart. Therefore, no AST values can be specifically assigned to Acinetobacter for comparison.
Discussion
This article reports data from a pilot study evidencing hospital surface colonisation with MDR GNB across five different wards; AE, FMW NICU, OG, and PW. Importantly, local scientists and members of the hospital staff collected the clinical environmental swabs without notice, limiting the use of disinfectant immediately before sampling or other behavioural bias among staff responsible for disinfection (e.g., the Hawthorne effect). PCR-confirmed ESBL and carbapenemase-producing isolates were recovered from all wards except FMW where all screened isolates produced negative results for AMR genes of interest despite high growth rates of 67% VE and 47% plates from swabs taken from the ward’s sampled environments during screening (Table 1, Fig. 1). Although the FMW ward is proximal to the other sampled wards the difference in terms of ARG detection indicates variation among environmental contaminating bacteria within a single institution. One of the most striking findings of this study was the high growth of bacteria in the presence of cefotaxime and meropenem and the diversity of Acinetobacter species (Fig. 3) harbouring multiple carbapenemase antibiotic resistance genes, noticeably blaNDM-1, blaOXA-23, blaOXA-58 and blaOXA-67. CRAB have often been linked to hospital-acquired infections (HAI) [9, 20, 65, 66] and have regularly been detected colonising abiotic hospital surfaces [67]. We also detected A. nosocomialis within the PW on thermometers and computer keyboards suggesting multiple carbapenem-resistant Acinetobacter species (CRAS) are present with potential to transmit within the wards. Of the 10 different Acinetobacter species identified in this study in total, eight harboured blaNDM-1 in addition to CHO. Similarly, Bhatta et al. performed a study in a tertiary care hospital in Nepal whereby 232 samples were collected from various sites and Acinetobacter species were the most prevalent GNB [68], and Aliramezani et al. detected clonal expansion of CRAB in hospital environments in Iran [69]. There are limited published data available from Africa, however high emerging prevalence of carbapenemase A. baumannii has been documented in wound infections in Ghana [32, 33].
Collectively CRAS were recovered from a wide variety of high-touch hospital surfaces including wall sockets, cots, mattresses, sheets, pillows, computer keyboards, medical equipment, bed handles and tap handles. This finding is in accordance with previous studies suggesting Acinetobacter successfully colonise abiotic surfaces [70]. The genetic relationship between the cluster of ST109 blaNDM-1 positive A. baumannii isolates identified in OG was investigated with a mapping-based SNP analysis using a local complete reference genome to improve coverage and SNP calling across the whole genome [71]. All ST109 A. baumannii were within 6 pairwise SNPs, with four identical isolates (0 SNPs), suggesting the spread of this strain throughout the OG ward. Scanning the PubMLST database [72] revealed that only 5/6640 (on 23 July 2021) isolates submitted were ST109 (Pasteur scheme) suggesting this is a relatively uncommon ST globally. Acinetobacter baumannii can persist in the healthcare environment by tolerating desiccation and producing biofilms on abiotic surfaces [70]. CRAB isolates recovered in this study were screened for biofilm-associated VF and detection of bap genes indicates biofilm production was possible. Future studies combining biofilm production assays and WGS could be beneficial to determine whether specific STs have the potential to persist longer in the environment. Studies combining antibiotic susceptibility testing, PFGE and biofilm assays have shown distinct clusters of CRAB persisting in the environment, including in high-risk wards like NICU [69]. During this study we identified three carbapenem resistant isolates, including Acinetobacter and Pseudomonas from cots, a medical trolly and bed wheels. The detection of a smaller cluster (three) of A. noscomialis isolates from a different ward, PW indicates multiple introductions of MDR Acinetobacter, and all these isolates originated from swabs from pieces of equipment regularly used by hand (Table 3). Unfortunately, the limited diagnostic capacity for the accurate speciation of certain isolates which include Acinetobacter baumannii, Pseudomonas spp. (which do not produce pigments), Klebsiella spp. (aside from that of K. pneumoniae and Enterobacter spp.) in the CCTH microbiology lab would likely result in misidentification. Four biochemical tests namely; indole, urease, citrate, and triple sugar iron test are utilised in this regard. This limited panel could lead to the misidentification of species especially within gram-negative non-fermenters due to similarities in the test outcomes especially for Pseudomonas spp. and Acinetobacter spp. Furthermore, the lack of PCR and visualisation equipment currently make it impossible to screen for ARGs. Rectifying these short-falls, especially in light of the findings for this study, are currently being addressed.
In 2019 He et al. first reported and characterised two mobile resistance genes named tet(X3) and tet(X4) which were isolated from (food-producing) animals in China [73]. When they performed genomic data mining, they found the presence of both tet(X3) in Acinetobacter and tet(X4) in Enterobacterales isolated from several countries including Cote d’Ivoire predating 2014 suggesting the dissemination into human pathogens of these genes. Here we report two Acinetobacter isolates carrying tet(X3) in addition to blaNDM. We often cultured multiple bacterial isolates from the same sample, containing a mixture of ESBL and/or carbapenemase genes (Fig. 2); most notably in this study a particular sample PW0012, a swab of the top surface and drawer handles of a bedside cabinet in the east wing of PW. It is concerning that bacterial isolates from this sample alone were resistant to aminoglycosides, cephalosporins, β-lactams, carbapenems, tigecycline and colistin. To the best of our knowledge this is the first report of multidrug-resistant Acinetobacter also harbouring tet(X3) in Ghana.
CCTH in Ghana has around 16 inpatient wards and this study collected 231 swabs across five wards over one month from March 2021. Surveillance across all wards is vital to determine which wards are at higher risk of greater MDR colonisation rates and such data is invaluable to focus on interventional infection control policies. Currently, the hospital performs infection prevention and control practices according to the national policy’s technical guidelines encompassing hand hygiene, environmental management and controls, handling linen, and environmental and engineering considerations which state the appropriate disinfectants/antiseptic agents to use and the procedures for using them. Some wards uphold more rigorous cleaning schedules and implement additional cleaning requirements, depending on the presence of certain equipment and other demands. However, there is limited infection control surveillance in the laboratory and inadequate monitoring and evaluation mechanisms in place to stringently enforce these IPC policies and practices. Data generated from such a study places emphasis on the need to implement increased measures to limit the spread of AMR. Limitations of this study include a short sampling period, small sample sizes for both swab sites and wards, and selective WGS, however the detection of clusters of closely related CRAS, mobile tigecycline resistance genes, ESBL and carbapenemase gene within K. pneumoniae, E. cloacae, and E. coli warrants larger and continued surveillance to reduce possible transmission events between hospital surfaces and patients. It would be beneficial for further studies to include sampling of health care workers’ hands, especially those working across multiple wards in a given time period, and also to link to potential HAI and AMR cases within the hospital. Future surveillance studies adopting a ‘One-Health’ approach including local community AMR surveillance are essential to provide context and linkage across all sectors [74] to provide data to inform intervention and policy change.
Conclusion
A high level of AMR-containing species and contaminated surfaces were detected within the hospital environment, noticeably in the OG and PW wards. A diversity of resistance mechanisms and species were detected within the hospital environment, including isolates with multi-drug resistance and evidence multiple of localised clonal transmission. Many of the surfaces swabbed harboured bacteria of multiple species carrying different ARG conferring resistance to several classes of antibiotics. This indicates surface colonisation could be acting as a potential reservoir for the transmission of MDR pathogens within the hospital environment. Noting high rates of healthcare-associated infections in sub-Saharan Africa and the role hospital environments play in these as evidenced in studies globally, environmental surveillance is an invaluable tool in reducing its incidence thus reducing AMR rates in the region. In Ghana, the National Action Plan for Antimicrobial Use and Resistance policy in strategic objective 7.1.1.1 focuses on the implementation of the National IPC policy. This study builds the needed foundation for the establishment of routine environmental surveillance as a key indicator for evaluating IPC practices within healthcare institutions in the country.
Availability of data and materials
All fastq reads (Illumina) were submitted to the ENA repository under the project accession PRJEB46496. Hybrid genomes were upload to the NCBI under the project accession PRJNA750808.
References
O’Neill J. Infection prevention, control and surveillance: limiting the development and spread of drug resistance. Rev Antimicrob Resist. 2016.
Founou RC, Founou LL, Essack SY. Clinical and economic impact of antibiotic resistance in developing countries: a systematic review and meta-analysis. PLoS ONE. 2017;12(12):e0189621.
World Health Organization. WHO publishes list of bacteria for which new antibiotics are urgently needed. 2017.
Weiner LM, Webb AK, Limbago B, Dudeck MA, Patel J, Kallen AJ, et al. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2011–2014. Infect Control Hosp Epidemiol. 2016;37(11):1288–301.
Kim BN, Peleg AY, Lodise TP, Lipman J, Li J, Nation R, et al. Management of meningitis due to antibiotic-resistant Acinetobacter species. Lancet Infect Dis. 2009;9(4):245–55.
Sands K, Carvalho MJ, Portal E, Thomson K, Dyer C, Akpulu C, et al. Characterization of antimicrobial-resistant Gram-negative bacteria that cause neonatal sepsis in seven low- and middle-income countries. Nat Microbiol. 2021;6(4):512–23.
Tumbarello M, Maria Trecarichi E, Giuseppe De Rosa F, Giannella M, Roberto Giacobbe D, Bassetti M, et al. Infections caused by KPC-producing Klebsiella pneumoniae: differences in therapy and mortality in a multicentre study. JAC Antimicrob Resist. 2015;70:2133–43.
Tzouvelekis LS, Markogiannakis A, Piperaki E, Souli M, Daikos GL. Treating infections caused by carbapenemase-producing Enterobacteriaceae. Clin Microbiol Infect. 2014;20(9):862–72.
De Oliveira DMP, Forde BM, Kidd TJ, Harris PNA, Schembri MA, Beatson SA, et al. Antimicrobial resistance in ESKAPE pathogens. Clin Microbiol Rev. 2020;33(3):e00181.
Farzana R, Jones LS, Rahman MA, Andrey DO, Sands K, Portal E, et al. Outbreak of hypervirulent multidrug-resistant klebsiella variicola causing high mortality in neonates in Bangladesh. Clin Infect Dis. 2018;68:1225–7.
Nordmann P, Naas T, Poirel L. Global Spread of Carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis. 2011;17(10):1791.
Nordmann P, Cuzon G, Naas T. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect Dis. 2009;9(4):228–36.
Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, et al. Characterization of a new metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother. 2009;53(12):5046.
Walsh TR, Toleman MA, Poirel L, Nordmann P. Metallo-β-lactamases: the quiet before the storm? Clin Microbiol Rev. 2005;18(2):306–25.
Giske CG, Sundsfjord AS, Kahlmeter G, Woodford N, Nordmann P, Paterson DL, et al. Redefining extended-spectrum β-lactamases: Balancing science and clinical need. J Antimicrob Chemother. 2009;63(1):1–4.
Walsh T, Weeks J, Livermore D, Toleman M. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect Dis. 2011;11(5):355–62.
Ministry of Health Ghana. Standard treatment guidelines. Seventh. Accra: Ghana National Drugs Programme; 2017.
World Health Organization. 2019 WHO AWaRe classification database of antibiotics for evaluation and monitoring of use. 2019.
Johnson AP, Woodford N. Global spread of antibiotic resistance: the example of New Delhi metallo-β-lactamase (NDM)-mediated carbapenem resistance. J Med Microbiol. 2013;62:499–513.
Nejad SB, Allegranzi B, Syed SB, Ellis B, Pittet D. Health-care-associated infection in Africa: a systematic review. Bull World Health Organ. 2011;89:757–65.
Saleem Z, Godman B, Hassali MA, Hashmi FK, Azhar F, Rehman IU. Point prevalence surveys of health-care-associated infections: a systematic review. Pathog Glob Health. 2019;113(4):191–205.
World Health Organization. Antimicrobial resistance: global report on surveillance. Geneva; 2014.
Seale AC, Hutchison C, Fernandes S, Stoesser N, Kelly H, Lowe B, et al. Supporting surveillance capacity for antimicrobial resistance: laboratory capacity strengthening for drug resistant infections in low and middle income countries. Wellcome Open Res. 2017;2:91.
Ataiyero Y, Dyson J, Graham M. Barriers to hand hygiene practices among health care workers in sub-Saharan African countries: a narrative review. Am J Infect Control. 2019;47(5):565–73.
Ayibieke A, Kobayashi A, Suzuki M, Sato W, Mahazu S, Prah I, et al. Prevalence and characterization of carbapenem-hydrolyzing class D β-lactamase-producing acinetobacter isolates from Ghana. Front Microbiol. 2020;11:1–13.
Ayibieke A, Sato W, Mahazu S, Prah I, Addow-Thompson J, Ohashi M, et al. Molecular characterisation of the NDM-1encoding plasmid p2189-NDM in an Escherichia coli ST410 clinical isolate from Ghana. PLoS ONE. 2018;13(12):1–10.
Labi AK, Nielsen KL, Marvig RL, Bjerrum S, Enweronu-Laryea C, Bennedb KM, et al. Oxacillinase-181 carbapenemase-producing Klebsiella pneumoniae in Neonatal Intensive Care Unit, Ghana, 2017–2019. Emerg Infect Dis. 2020;26(9):2235–8.
Forson OA, Ayanka E, Olu-Taiwo M, Pappoe-Ashong PJ, Ayeh-Kumi PJ. Bacterial infections in burn wound patients at a tertiary teaching hospital in Accra, Ghana. Ann Burns Fire Disasters. 2017;30(2):116.
Obeng-nkrumah N, Twum-danso K, Krogfelt KA, Newman MJ. High levels of extended-spectrum beta-lactamases in a major teaching hospital in Ghana: the need for regular monitoring and evaluation of antibiotic resistance. Am J Trop Med Hyg. 2013;89(5):960–4.
Agyepong N, Govinden U, Owusu-ofori A. Multidrug-resistant gram-negative bacterial infections in a teaching hospital in Ghana. Antimicrob Resist Infect Control. 2018;7:1–8.
Alexander B, Roland A, Anthony A, Matthew GA. Antibiotic resistance and genotypic detection of extended spectrum beta-lactamase producing pathogens in catheter associated urinary tract infection at a teaching facility in Kumasi. Ghana Afr J Microbiol Res. 2020;14(8):395–401.
Codjoe FS, Brown CA, Smith TJ, Miller K, Donkor ES. Genetic relatedness in carbapenem-resistant isolates from clinical specimens in Ghana using ERIC-PCR technique. PLoS ONE. 2019;14(9):1–11.
Monnheimer M, Cooper P, Amegbletor HK, Pellio T, Groß U, Pfeifer Y, et al. High prevalence of carbapenemase-producing acinetobacter baumannii in wound infections, Ghana, 2017/2018. Microorganisms. 2021;9(3):1–10.
George DF, Gbedema SY, Agyare C, Adu F, Boamah VE, Tawiah AA, et al. Antibiotic resistance patterns of Escherichia coli isolates from hospitals in Kumasi, Ghana. ISRN Microbiol. 2012;2012:1–5.
Saba CKS, Amenyona JK, Kpordze SW. Prevalence and pattern of antibiotic resistance of Staphylococcus aureus isolated from door handles and other points of contact in public hospitals in Ghana. Antimicrob Resist Infect Control. 2017;6(1):1–6.
Donkor ES, Jamrozy D, Mills RO, Dankwah T, Amoo P, Egyir B, et al. A genomic infection control study for Staphylococcus aureus in two Ghanaian hospitals. Infect Drug Resist. 2018;11:1757–65.
Duedu KO, Offei G, Codjoe FS, Donkor ES. Multidrug resistant enteric bacterial pathogens in a psychiatric hospital in Ghana: implications for control of nosocomial infections. Int J Microbiol. 2017. https://doi.org/10.1155/2017/9509087.
Ministry of Health Ghana. National policy and guidelines for infection prevention and control in health care settings. Accra; 2015. 160 p. https://www.ghanahealthservice.org/downloads/National_Policy_and_Guidelines_for_Infection_Prevention_and_Control_in_Health_Care_Settings_2015.pdf.
World Health Organization. Core competencies for infection prevention and control professionals. Geneva: World Health Organization; 2020.
Mwita JC, Ogunleye OO, Olalekan A, Kalungia AC, Kurdi A, Saleem Z, et al. Key issues surrounding appropriate antibiotic use for prevention of surgical site infections in low- and middle-income countries: a narrative review and the implications. Int J Gen Med. 2021;14:515.
CLSI. M100 performance standards for antimicrobial susceptibility testing. 30th ed, vol 8. Pennsylvania: Clinical and Laboratory Standards Institute; 2020.
Ghana Statistical Service. 2010 Population and housing census final results Ghana statistical service. 2012; pp. 1–11.
Dallenne C, da Costa A, Decré D, Favier C, Arlet G. Development of a set of multiplex PCR assays for the detection of genes encoding important β-lactamases in Enterobacteriaceae. J Antimicrob Chemother. 2010;65(3):490–5.
EUCAST. EUCAST V11: The European Committee on antimicrobial susceptibility testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 11.0, 2021. 2021.
Turlej-Rogacka A, Xavier BB, Janssens L, Lammens C, Zarkotou O, Pournaras S, et al. Evaluation of colistin stability in agar and comparison of four methods for MIC testing of colistin. Eur J Clin Microbiol Infect Dis. 2018;37(2):345–53.
Andrews S. FastQC: a quality control tool for high throughput sequence data. 2009.
Krueger F. Trimgalore. 2018.
Seemann T. Shovill. 2018.
Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29(8):1072–5.
Seemann T. ABRicate. 2019.
Seemann T. mlst, Github. 2019.
Diancourt L, Passet V, Nemec A, Dijkshoorn L, Brisse S. The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS ONE. 2010;5(4):e10034.
Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–9.
Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31(22):3691–3.
Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, Von Haeseler A, et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020;37(5):1530–4.
Seemann T. Snippy: fast bacterial variant calling from NGS reads. 2015.
Page AJ, Taylor B, Delaney AJ, Soares J, Seemann T, Keane JA, et al. SNP-sites: rapid efficient extraction of SNPs from multi-FASTA alignments. Microb Genomics. 2016;2(4):e000056.
Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43(3):e15.
Seemann T. snp-dists.
Letunic I, Bork P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019;47(W1):256–9.
Wick RR. Porechop v0.2.4.
Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13(6):e1005595.
Wick RR, Schultz MB, Zobel J, Holt KE. Bandage: Interactive visualization of de novo genome assemblies. Bioinformatics. 2015;31(20):3350–2.
Galata V, Fehlmann T, Backes C, Keller A. PLSDB: a resource of complete bacterial plasmids. Nucleic Acids Res. 2019;47(D1):D195-202.
Ayobami O, Willrich N, Harder T, Okeke IN, Eckmanns T, Markwart R. The incidence and prevalence of hospital-acquired (carbapenem-resistant) Acinetobacter baumannii in Europe, Eastern Mediterranean and Africa: a systematic review and meta-analysis. Emerg Microbes Infect. 2019;8(1):1747–59.
Nguyen M, Joshi SG. Carbapenem resistance in Acinetobacter baumannii, and their importance in hospital-acquired infections: a scientific review. J Appl Microbiol. 2021;131:2715–38.
Nutman A, Lerner A, Schwartz D, Carmeli Y. Evaluation of carriage and environmental contamination by carbapenem-resistant Acinetobacter baumannii. Clin Microbiol Infect. 2016;22(11):949.e5-949.e7.
Bhatta DR, Hamal D, Shrestha R, Hosuru Subramanya S, Baral N, Singh RK, et al. Bacterial contamination of frequently touched objects in a tertiary care hospital of Pokhara, Nepal: how safe are our hands? Antimicrob Resist Infect Control. 2018;7(1):4–9.
Aliramezani A, Douraghi M, Hajihasani A, Mohammadzadeh M, Rahbar M. Clonal relatedness and biofilm formation of OXA-23-producing carbapenem resistant Acinetobacter baumannii isolates from hospital environment. Microb Pathog. 2016;99:204–8.
Weinberg SE, Villedieu A, Bagdasarian N, Karah N, Teare L, Elamin WF. Control and management of multidrug resistant Acinetobacter baumannii: a review of the evidence and proposal of novel approaches. Infect Prev Pract. 2020;2(3):100077.
Bush SJ, Foster D, Eyre DW, Clark EL, de Maio N, Shaw LP, et al. Genomic diversity affects the accuracy of bacterial single-nucleotide polymorphism-calling pipelines. Gigascience. 2020;9(2):1–21.
Jolley KA, Bray JE, Maiden MCJ. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications [version 1; referees: 2 approved]. Wellcome Open Res. 2018;3:1–20.
He T, Wang R, Liu D, Walsh TR, Zhang R, Lv Y, et al. Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat Microbiol. 2019;4(9):1450–6.
Walsh TR. A one-health approach to antimicrobial resistance. Nat Microbiol. 2018;3(8):854–5.
Acknowledgements
We are grateful to the Chief Executive, the Laboratory manager, and all the nurse-in-charges of the various wards sampled in the Cape Coast Teaching Hospital for their support during this study. We are thankful to Dr Maria Mendes De Carvalho for the development of the CTX-M-15 PCR and to Dr Brekhna Hassan for developing the carbapenemase multiplex PCR (data unpublished). We would like to thank the NHS staff in the Specialist Antimicrobial Chemotherapy Unit (SACU) Public Health Wales for supporting access to the MALDI-TOF MS instrument. Bioinformatics analysis was undertaken using the supercomputing facilities at Cardiff University operated by Advanced Research Computing at Cardiff (ARCCA) on behalf of the Cardiff Supercomputing Facility and the HPC Wales and Supercomputing Wales (SCW) projects. We acknowledge the support of the latter, which is part-funded by the European Regional Development Fund (ERDF) via the Welsh Government. Joseph Elikem Efui Acolatse has received support and guidance for the study/project through ESCMID's mentorship programme by Victoria J. Chalker and Owen Brad Spiller.
Funding
This study was partly funded by the Microbiology Society International Development Fund Grant (GA002502). The funding body had no role in the design of the study, collection of data, analysis, interpretation of data, and writing the manuscript.
Author information
Authors and Affiliations
Contributions
JA, KS, EP, VC and OS contributed to study design. JA, GA and MD collected the samples. JA, KS and EP contributed to manuscript writing and figure production. JA, KS, EP, IB and OS performed the microbiology experiments. KS and EP performed the WGS and bioinformatics analysis. All authors reviewed and read the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
All authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1
. Additional data for antimicrobial resistance testing of patient isolates of Gram-negative bacteria isolated before, during and after the environmental swabs were collected, collated by corresponding wards sampled.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Acolatse, J.E.E., Portal, E.A.R., Boostrom, I. et al. Environmental surveillance of ESBL and carbapenemase-producing gram-negative bacteria in a Ghanaian Tertiary Hospital. Antimicrob Resist Infect Control 11, 49 (2022). https://doi.org/10.1186/s13756-022-01090-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13756-022-01090-2