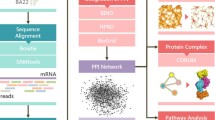
Abstract
Background
Psychiatric disorders are a major burden on global health. These illnesses manifest as co-morbid conditions, further complicating the treatment. There is a limited understanding of the molecular and regulatory basis of psychiatric co-morbidities. The existing research in this regard has largely focused on epigenetic modulators, non-coding RNAs, and transcription factors. RNA-binding proteins (RBPs) functioning as multi-protein complexes are now known to be predominant controllers of multiple gene regulatory processes. However, their involvement in gene expression dysregulation in psychiatric co-morbidities is yet to be understood.
Results
Ten RBPs (QKI, ELAVL2, EIF2S1, SRSF3, IGF2BP2, EIF4B, SNRNP70, FMR1, DAZAP1, and MBNL1) were identified to be associated with psychiatric disorders such as schizophrenia, major depression, and bipolar disorders. Analysis of transcriptomic changes in response to individual depletion of these RBPs showed the potential influence of a large number of RBPs driving differential gene expression, suggesting functional cross-talk giving rise to multi-protein networks. Subsequent transcriptome analysis of post-mortem human brain samples from diseased and control individuals also suggested the involvement of ~ 100 RBPs influencing gene expression changes. These RBPs were found to regulate various processes including transcript splicing, mRNA transport, localization, stability, and translation. They were also found to form an extensive interactive network. Further, hnRNP, SRSF, and PCBP family RBPs, Matrin3, U2AF2, KHDRBS1, PTBP1, and also PABPN1 were found to be the hub proteins of the RBP network.
Conclusions
Extensive RBP networks involving a few hub proteins could result in transcriptome-wide dysregulation of post-transcriptional modifications, potentially driving multiple psychiatric disorders. Understanding the functional involvement of RBP networks in psychiatric disorders would provide insights into the molecular basis of psychiatric co-morbidities.
Similar content being viewed by others
Background
Mental illnesses are a major cause of morbidity and mortality across age groups globally [1]. Depression, schizophrenia, and bipolar disorders account for an estimated 264 million, 20 million, and 45 million cases of psychiatric disorders respectively [2]. Existing treatments for these disorders help only a small subset of patients. Thus, there is a need for a better understanding of the biological basis of these disorders and identify novel therapeutic targets. Notably, psychiatric disorders are known to manifest as co-morbid conditions, indicating a potentially shared underlying biology. Patients with schizophrenia are known to have higher risk of anxiety and depression, substance use disorders, post-traumatic stress disorder, as well as obsessive–compulsive disorder [3, 4]. Similarly, bipolar disorder, attention-deficit/hyperactivity disorder (ADHD) [5], and substance use disorders [6] are known to be co-morbid. The molecular basis of these psychiatric disorders and co-morbidities are yet to be understood [7]. Recent research has highlighted the importance of understanding the biological basis of disease co-morbidities [8]. Thus, delineating the molecular regulation of psychiatric co-morbidities would be imperative for more efficient treatment strategies.
Several studies have revealed extensive gene expression dysregulation associated with psychiatric conditions. A number of genes are also known to be commonly dysregulated across psychiatric disorders [9, 10]. A better understanding of the regulatory mechanisms resulting in gene expression dysregulation could provide further insights into multiple genes and their networks affected by common regulatory factors. In this regard, most studies on molecular regulation in mental illnesses have focused on transcription factors, microRNAs, long non-coding RNAs, alternative splicing, and epigenetic modifications [11,12,13,14]. However, recent research has highlighted the dominant role of RNA-binding proteins (RBPs) on gene regulatory processes [15, 16]. Though the involvement of RBPs in disorders such as cancers [17], genetic diseases [18], cardiovascular diseases [19], diabetes [20], and neurodegenerative diseases [21] has been established, their potential involvement in mental illnesses is only beginning to be understood. A recent report has suggested the involvement of dysregulated RBP-binding sites in psychiatric conditions [22]. Hence, further understanding of the involvement of RBPs in gene expression dysregulation in mental illnesses could provide new insights into the molecular basis of these disorders.
RBPs are known to function as multi-protein complexes. RBP networks composed of multi-protein clusters are known to regulate common targets. Such RBP clusters could form master-regulatory modules of cellular processes [23]. The complexity and functional implications of these RBP networks are now beginning to be understood [24]. Owing to the progress in our understanding of the importance of RBPs in disease conditions, therapeutic approaches such as RBP-PROTACs are being developed to manipulate RBP functioning [25]. However, since manipulating the functions of individual RBPs could potentially affect multiple other RBPs and RBP networks, a holistic understanding of the RBP networks would be of high scientific and clinical importance.
In light of these research reports, the present study focused on identification of RBPs involved in psychiatric disorders, and the analysis of their potential interactions could give rise to RBP networks and combinatorial gene regulatory modules. To this end, the RBPs associated with psychiatric disorders and their potential interactions were identified. Subsequently, publicly available transcriptome data were analysed to identify other RBPs which could be functionally related. Further, transcriptome data from human post-mortem brain samples were retrieved from public repositories and analysed to identify RBPs implicated in gene expression dysregulation in psychiatric conditions (major depression, schizophrenia, and bipolar disorder). These analyses suggested a significant number of RBPs potentially involved in psychiatric disorders. Next, the interactive networks of the identified RBPs were analysed, which revealed extensive connections among them. Subsequently, hub proteins having more pronounced inter-connections were identified, which showed hnRNP, PCBP, SRSF family RBPs, Matrin3, and PTBP to be among the hub RBPs. Thus, the present study identified RBPs and their networks, which could be driving transcriptomic dysregulation in multiple psychiatric disorders. In future, large-scale human studies to understand their functional implications in mental health could be of importance, towards a holistic understanding of molecular regulation of psychiatric co-morbidities.
Materials and methods
Identification of RBPs associated with psychiatric disorders and their potential interactions
In order to identify RBPs associated with psychiatric disorders, the genes curated onto PsyGeNET database (v2.0) (http://www.psygenet.org/web/PsyGeNET/menu) [26] were compared with the known RBPs from Transite database (v1.2.1) (transite.mit.edu/) [27]. PsyGeNET is an expert-curated database of disease-gene associations in psychiatric disorders, comprising 1549 genes and 117 diseases. Transite provides a computational platform for the analysis of RBP-mediated gene expression changes in transcriptomic studies, encompassing a database of ~ 150 RBPs from human and mouse, with known target motifs. Thus, comparing the psychiatric disease-associated genes from PsyGeNET with RBPs from Transite yielded the RBPs associated with psychiatric disorders. Subsequently, potential interactions between these RBPs were analysed via STRING (https://string-db.org/) (v11.5).
Analysis of transcriptome-wide differential gene expression and identification of potential RBPs driving gene expression changes
Due to combinatorial functioning, modulating individual RBPs could affect a multitude of other RBPs. In order to understand such potential interaction networks, the transcriptomes of human cell lines subjected to genetic manipulation (knock-down/knockout/overexpression) of the psychiatric disease-associated RBPs were analysed. The transcriptome studies related to the gene expression patterns associated with genetic manipulation of the considered RBPs were identified on GEO database (https://www.ncbi.nlm.nih.gov/geo/). The studies on human cell lines with at least two replicates per condition (control and knock-down/knockout/overexpression) were considered. Differential gene expression patterns of RNA-seq and microarray studies were analysed using OneStopRNAseq (v1.0.0; https://mccb.umassmed.edu/OneStopRNAseq/index.php) [28] and GEO2R tools (https://www.ncbi.nlm.nih.gov/geo/geo2r/), respectively. OneStopRNAseq is a comprehensive platform for the analysis of RNA-seq data, including quality check (via FastQC v0.11.5 and MultiQC v1.6), alignment (via STARv. 2.7.5a), read summarization (via featureCountsv.2.0.0), and differential gene expression (via DESeq2v 1.28.1). Human genome hg38 (gencode. v34.primary_assembly) was used as reference. The differentially expressed genes (DEGs) were identified with a false discovery rate (FDR) threshold of 0.05 and minimum log2 fold change threshold of 0.585. The microarray datasets were analysed through GEO2R (using default settings). GEO2R identifies statistically significant DEGs via GEOquery and limma packages within R framework. The gene expression levels in knockout/knock-down/overexpression (experimental) groups were compared to those of control datasets. Similarly, transcriptome studies of human post-mortem brain samples involving at least two psychiatric disorders were identified on GEO database (shown in Table 2), followed by differential gene expression analysis as described above.
Further, the RBPs having enriched binding sites within 5’ and 3’ untranslated regions (UTRs) of the DEGs were identified using Transite (https://transite.mit.edu/) (v1.2.1) [27]. Transite performs global computational identification of RBPs involved in post-transcriptional regulation (PTGR) of gene expression using RNA-seq or microarray data. Transcript Set Motif Analysis (TSMA) was employed to identify statistically significant overrepresentation of RBP target motifs in the UTRs of DEGs. TSMA was performed via k-mer and matrix-based approaches. k-mer-based approach identifies RBPs by comparing the hexamer/heptamer (k-mer) sequences between the foreground (DEGs) and background (all genes identified in the transcriptomic study) datasets. Matrix-based approach identifies the potential RBP-binding sites by scoring the sequence positions within the foreground and background datasets. Thus, the platform identifies statistically significant RBP target motifs potentially contributing to RBP-mediated PTGR.
Analysis of protein–protein interaction network, identification of significant modules, and hub proteins
Protein interaction network was obtained using STRING database (https://string-db.org/) (v11.5). Experimentally deciphered as well as the predicted interactions were visualized using default settings, against Homo sapiens database. The network from STRING was exported to Cytoscape (v3.8) [29]. The significant modules within the network were identified using MCODE (v2.0.0) plug-in of Cytoscape, using default settings (degree cut-off = 2, node score cut-off = 0.2, k-core = 2, maximum depth = 100). The hub proteins of the network were identified using Cytoscape through the cytoHubba (v0.1) plug-in [30]. Top 20 hub proteins were identified and ranked using maximal clique centrality (MCC) method.
Analysis of gene ontology and expression levels
The functional aspects affected by the selected RBPs were identified by analysing the enriched gene ontology terms of biological processes, associated with them. ShinyGO (v0.75) web server (http://bioinformatics.sdstate.edu/go/; [31]) was used to perform gene ontology analysis, using default settings. The gene expression levels in different brain regions were obtained using the human protein atlas (v.21.0) (https://www.proteinatlas.org/).
Results
Identification of RBPs associated with psychiatric disorders and analysis of their potential interactions
Comparison of the genes associated with psychiatric diseases (from PsyGeNET database) with human/mouse RBPs (from Transite database) yielded ten common RBPs: QKI, ELAVL2, EIF2S1, SRSF3, IGF2BP2, EIF4B, SNRNP70, FMR1, DAZAP1, and MBNL1 (Fig. 1a). The gene names and associated disorders for each gene are provided in Additional file 3: Table S1. Overall, these RBPs were associated with schizophrenia, major depressive disorder, bipolar disorder, and mixed anxiety. The analysis of interactions between these RBPs showed that they could potentially form an interactive network (Fig. 1b). Thus, it could be inferred that combinatorial functioning of RBPs could influence psychiatric disorders.
Psychiatric disorders-associated RBPs. a Psychiatric disease-associated genes from PsyGeNET database (http://www.psygenet.org [26]) were compared with RBPs curated onto Transite (https://transite.mit.edu/ [27]), to identify RBPs associated with psychiatric disorders. This comparison identified ten RBPs to be associated with psychiatric disorders. b The identified RBPs were also found to form a potential interactive network
Identification of RBPs driving differential gene expression in response to in vitro genetic manipulation of psychiatric disorder-associated RBPs
Owing to the combinatorial functioning, modulating the activity of individual RBPs could influence other RBPs and larger networks. In the present study, transcriptome-wide gene expression changes in response to manipulation of individual RBPs were analysed for the potential involvement of other RBPs in modulating gene expression through 5′ and 3′UTRs of DEGs. For this purpose, the transcriptomic datasets associated with gene expression changes in response to knock-down (KD)/knockout (KO)/overexpression of RBPs were selected. The details of transcriptome datasets considered for this study are given in Table 1. These transcriptome datasets were analysed to identify DEGs. (The number of DEGs found in each dataset is given in Table 1.) Subsequently, the RBPs having overrepresented target motifs within the 5′ and 3′ UTRs of DEGs were identified. A compiled list of RBPs with significant overrepresentation (p < 0.05) of target motifs, potentially influencing the transcriptome, are given in Additional file 1. The number of RBPs having enriched binding sites in the UTRs of DEGs in response to depletion of the individual RBPs is shown in Fig. 2. 3′UTRs of the DEGs were found to have a higher number of RBP target motifs across the datasets (Fig. 2). Also, depletion of SRSF3 and DAZAP1 was found to potentially cause widespread gene expression changes via 3′UTRs as well as 5′UTRs, mediated by multiple other RBPs (Fig. 2).
Effect of the depletion of psychiatric disorder-associated RBPs on global gene expression regulation by RBPs. Transcriptomic changes in human cell lines subjected to genetic manipulation (knock-down/knockout) of the selected RBPs were analysed to identify up/downregulated genes (differentially expressed genes/DEGs). The untranslated regions (UTRs) of DEGs were screened to identify overrepresented target motifs of known RBPs, using Transite (https://transite.mit.edu/) [27]. A number of RBPs with overrepresented binding sites in the UTRs of DEGs (y-axis) associated with the selected RBPs (x-axis) are shown. Overall, 3’UTRs were found to be highly enriched for RBP target motifs
RBPs influencing gene expression changes in diseased post-mortem human brain samples
Transcriptomic data from diseased post-mortem human brain samples (schizophrenia, bipolar, and major depressive disorders; Table 2) were analysed to identify DEGs with respect to control individuals. The DEGs were further studied to detect overrepresented RBP-binding sites within their 5’ and 3’ UTRs, which could influence differential gene expression. Thirteen RBPs were found to be common in all the three disorders—CPEB3/CPEB4, DAZAP1, ELAVL1/ELAVL3, hnRNPC, hnRNPCL1, PCBP4/PCBP1/PCBP3, PCBP4/PCBP3, PTBP1/PTBP2/ROD1, RBMS2/RBMS1, SF3B4, TARDBP, U2AF2, and YBX2/CSDA (Fig. 3), indicating their potential involvement in molecular dysregulation of multiple disorders. The dataset-wise number of RBPs found to be influencing differential gene expression in diseased samples is given in Table 2.
Number of RBPs potentially driving differential gene expression in diseased post-mortem human brain samples. Differentially expressed genes within the transcriptome datasets of post-mortem human brain samples (schizophrenia/SCZ, major depressive disorder/MDD, and bipolar disorder/BD) were analysed to identify the potential RBPs driving gene expression changes in disease conditions. Thirteen RBPs (CPEB3/CPEB4, DAZAP1, ELAVL1/ELAVL3, hnRNPC, hnRNPCL1, PCBP4/PCBP1/PCBP3, PCBP4/PCBP3, PTBP1/PTBP2/ROD1, RBMS2/RBMS1, SF3B4, TARDBP, U2AF2, and YBX2/CSDA) were found to be involved in the three disorders considered, indicating their potential central roles in multiple disorders
RBPs common between in vitro and human brain datasets
In order to get insights into the overall interactive networks of the identified RBPs, common RBPs between those found to be involved in psychiatric disorders, as well as those driving gene expression changes in in vitro datasets were identified. This comparison yielded 132 RBPs (Additional file 2). The functions of these RBPs were analysed through gene ontology terms (biological processes) significantly associated with them. As shown in Fig. 4, the identified RBPs were found to be involved in multiple aspects of transcript regulation including RNA splicing, stabilization, degradation, transport, as well as localization. Thus, it could be inferred that the RBP networks potentially involved in dysregulated gene expression in diseased conditions could affect multiple processes at the post-transcriptional level.
Gene ontology analysis of the identified RBPs. Biological processes associated with the common RBPs were identified via gene ontology (GO) enrichment analysis (ShinyGO v0.75; http://bioinformatics.sdstate.edu/go/) [31]. Significantly enriched GO terms were identified with FDR cut-off of 0.05, using human database
Analysis of interaction network of the identified RBPs, significant modules, and identification of hub proteins
The RBPs found to be common between in vitro and human brain datasets were analysed for their potential interactions, via STRING. This analysis revealed a protein interaction network containing 120 nodes and 1447 edges (Fig. 5a). Further identification of significant modules in this network showed the presence of four prominent modules of interacting proteins. Module-1 consisted of 37 nodes and 615 edges, comprising hnRNP family proteins, SRSF, and PABP family RBPs among others (Fig. 5b). Module-2 consisted of 5 nodes and 8 edges, consisting of the RBPs QKI, RBM4, KHDRBS3, PTBP2, and RBFOX2 (Fig. 5c). The two other modules consisted of 3 nodes and 3 edges each. ESRP2, KHDRBS2, and RBM24 constituted module-3, while module-4 consisted of CPEB family proteins (Fig. 5d, e). Thus, the identified RBPs were found to have potentially extensive inter-connections, with significant modules containing hnRNP, CPEB, and RBM family proteins.
Protein–protein interaction networks of the identified RBPs, and significant modules within the network. a The interaction network was obtained through STRING database (v11.5) (https://string-db.org/), using default settings. b-e Subsequently, significant modules within the network were identified using Cytoscape (v3.8.0) [29], through the MCODE plug-in. Four modules were identified, having a high degree of connectivity with other proteins in the network
Further, the network was analysed to identify top twenty hub proteins, which revealed hnRNP and SRSF family proteins, PTBP1, PABPN1, PCBP2, MATR3, and PCBP1 to be among the hub proteins of the network (Fig. 6a and Table 3). Additionally, the expression levels of the hub proteins were analysed in different regions of human brain. Interestingly, the expression levels of hnRNPA1 were found to be relatively high in most regions considered, except olfactory bulb and hippocampal formation (Fig. 6b). The expression levels of most genes were higher in cerebral cortex, while they were substantially low in olfactory bulb. Hypothalamus was found to have a higher expression of the hnRNP family RBPs, except hnRNPH1 and hnRNPDL.
Top 20 hub RBPs in the protein interaction network and their expression levels. a The hub RBPs were identified using cytoHubba plug-in [30] of Cytoscape (v3.8.0), ranked by maximal clique centrality (MCC) method. b Gene expression levels in different regions of the brain were obtained from the human protein atlas (https://www.proteinatlas.org/)
Discussion
Psychiatric conditions such as schizophrenia, major depression, and bipolar disorders often manifest as co-morbidities [3, 4]. Understanding the genetic and regulatory basis of psychiatric co-morbidities could provide insights into their molecular underpinnings and open-up potential therapeutic strategies. The available research reports on gene expression dysregulation in psychiatric conditions have focused on microRNA-mediated processes, alternative splicing, epigenetics, and non-coding RNAs [11,12,13,14]. Recent studies have established RBPs as a dominant class of regulatory proteins influencing multiple aspects of transcript regulation [15]. However, their potential involvement in psychiatric disorders is only beginning to be understood. RBPs form protein–protein interaction networks which could act as master regulators of gene expression [23, 50]. The present study employed computational approaches to identify RBPs potentially driving gene expression dysregulation in psychiatric conditions and co-morbidities. As a result, RBP network consisting of hub proteins including hnRNP, SRSF, PCBP family members, as well as Matrin3, PTBP, and PABPN1, was identified to be potentially involved in psychiatric co-morbidities.
To begin with, PsyGeNET, a database of genes associated with psychiatric disorders [26], was screened to find the RBPs implicated in various psychiatric disorders, identifying ten RBPs, viz. QKI, ELAVL2, EIF2S1, SRSF3, IGF2BP2, EIF4B, SNRNP70, FMR1, DAZAP1, and MBNL1. These RBPs are known to regulate multiple aspects of neural biology, thereby influencing disease conditions. The mRNA levels of QKI isoforms QKI5, 6, and 7 have been reported to be strongly perturbed in several cortical regions and hippocampus, in schizophrenic patients [51]. QKI has been implicated in astrocyte maturation in mouse brain, wherein it was found to be important in stabilizing its target transcripts [52]. QKI proteins are now known to regulate multiple aspects of gene expression regulation, including alternative splicing, mRNA stability, transcription, and translation [53]. ELAVL2 is known to be associated with schizophrenia and has been reported to be involved in regulation of MMP-9 (Matrix metalloproteinase-9, implicated in several disorders including those of nervous system) mRNA stability in hippocampal neurons [54]. FMRP, encoded by FXR1, is widely studied for its involvement in brain function and plasticity. Mutations in FXR1 are known to be associated with several psychiatric disorders including autism spectrum disorders, attention deficit and hyperactivity disorder, obsessive–compulsive disorder, and substance abuse [55, 56]. SRSF3 (serine-/arginine-rich splicing factor 3) is a member of SR family RBPs, involved in multiple regulatory processes including alternative splicing and polyadenylation, mRNA export, translation, and also microRNA processing [57]. SRSF3 is also implicated in bipolar disorder, wherein its mRNA levels were found to be elevated in peripheral white blood cells [58]. Aberrant mRNA translation is associated with a number of psychiatric conditions including schizophrenia, major depression, and bipolar disorder [59]. The translation initiation factor EIF4B was reported to be associated with major depressive disorder, wherein its levels were found to be reduced in prefrontal cortices of MDD patients [60]. It was also reported to be potentially involved in neuronal plasticity and synaptic changes [61]. The expression of EIF2, another initiation factor, was found to be dysregulated in schizophrenia [62]. EIF2S1 is a main initiation factor controlling protein synthesis, with important roles in cellular stress response associated with mitochondrial dysfunction in psychiatric disorders [63]. IGF2BP2 was reported to be involved in susceptibility to schizophrenia, indicating potential similarities in genetic bases of schizophrenia and diabetes [64]. DAZAP1 is a conserved hnRNP protein potentially involved in mRNA localization, alternative splicing, and translation [65] and is also a potential marker of bipolar disorder and schizophrenia [66]. On PsyGeNET database, SNRNP70, a major component of spliceosome, was associated with alcohol dependence. This protein was reported to regulate local protein synthesis in the synapses and also influence axonal growth and synapse formation [67]. MBNL1 is another protein involved in splicing, and a marker of schizophrenia [68]. Thus, multiple RBPs regulating transcript splicing, localization, and stability were found to be associated with psychiatric disorders.
RBPs are known to form interactive clusters and chains, and co-bind to their targets. Such clusters could contain cooperating/competing RBPs, giving rise to complex regulatory modules [23]. In order to get insights into multi-RBP networks modulating gene expression, it would be important to identify the RBPs potentially interacting with psychiatric disorder-associated RBPs. To this end, transcriptome studies related to gene expression changes in response to genetic manipulation (knockout/knock-down/overexpression) of the selected RBPs were identified, followed by their analysis to index all the RBPs which could be influencing differential gene expression. As a result, downregulation of individual RBPs associated with psychiatric disorders was found to result in global gene expression changes potentially driven by multiple other RBPs. 75–100 RBPs were found to influence transcriptomic changes in response to the depletion of EIF2S1, SRSF3, IGF2BP2, FMR1, DAZAP1, and MBNL1, mainly through the 3’UTRs of DEGs.
Subsequently, publicly available transcriptome data from diseased and control human brain samples were analysed to identify RBPs potentially driving differential gene expression in multiple psychiatric disorders, which could be an underlying molecular feature of psychiatric co-morbidities. Next, the RBPs obtained via the two approaches (analyses of in vitro and human brain transcriptomes) were compared to identify those RBPs commonly present in both the sets, which yielded 132 RBPs. These RBPs were found to be involved in multiple processes of post-transcriptional regulation, including splicing, mRNA transport, localization, and stabilization. Since mRNA transport and localization could have a profound impact on neuronal processes and plasticity [69], RBPs controlling these processes could potentially contribute to local differential gene expression within the neurons and synapses. For example, RBPs such as TDP-43 (Transactive Response DNA-Binding Protein 43) and SMN (survival motor neuron) are known to influence local mRNA translation within axons and dendrites [70].
An analysis of potential interactions between the identified RBPs showed an extensive network. Four discrete modules were identified within this network. The larger module (with 37 nodes and 615 edges) was composed of hnRNP, SRSF, PCBP, and ELAVL family RBPs, implicating their widespread interactions. In agreement with this observation, further analysis of the network also identified hnRNP, SRSF, and PCBP family RBPs to be among the hub proteins. hnRNP family RBPs associate with the pre-mRNA and control their splicing, stability, and translation. Multiple hnRNPs have been implicated in neurological disorders and cancers. For example, hnRNPA1, A2, F, H, and K are known to regulate splicing, translation, and stability of the target transcripts, and are associated with amyotrophic lateral sclerosis [71]. hnRNPA2 and hnRNPC1 were found to be associated with Alzheimer’s disease (AD), while hnRNPC1 is involved in fragile X syndrome [71]. These proteins interact with other RBPs, and disrupting such interactions could contribute to diseases like spinal muscular dystrophy and ALS [71]. hnRNPA1, which was the top ranked hub protein, is known to be associated with several neurological diseases including ALS, SMN, multiple sclerosis (MS), AD, and Huntington’s disease [72]. Mutations and altered expression of hnRNPA1 could lead to dysregulated splicing, translation, and transport of the target transcripts [72]. PABPN1 is associated with oculopharyngeal muscular dystrophy (OPMD), characterized by (GCN)n mutation [73]. The SRSF proteins are involved in regulating alternative splicing [74]. SRSF1, 3, 6, and 9 were found to be among the hub proteins of the RBP network. SRSF1 is known to be important in functioning of T cells and is implicated in autoimmune diseases [75]. SRSF3 is involved in alternative splicing and polyadenylation, mRNA export, and also miRNA processing. Further, it is also associated with bipolar disorder, tauopathies, and AD [57], while SRSF6 was reported to be potentially associated with Huntington’s disease [76]. PCBP1 and 2 are also potentially associated with neurological conditions. The target transcripts of PCBP1 were reported to be associated with neuropathies [77], while PCBP2 was found to be downregulated in ALS [78]. Matrin3 is an established RBP associated with ALS [79], which was also detected to be a hub RBP in the network. Thus, the hub proteins identified in the RBP network in this study are known to be involved in a number of neurological conditions, which suggests a shared molecular basis underlying these disorders.
The potential dysregulation of RBP activity in psychiatric disorders is recently being examined. SF3B4 (splicing factor 3B subunit 4) was associated with ADHD, bipolar disorder, and major depression [22]. Also, EFTUD2 (Elongation Factor Tu GTP-Binding Domain-Containing 2) was associated with ADHD, bipolar disorder, and schizophrenia, providing insights into shared molecular dysregulation in multiple psychiatric disorders [22]. Thus, the present study catalogued RBPs which could be potentially driving psychiatric co-morbidities. Future studies in this regard could elucidate the functional importance of these RBPs in disease conditions.
Conclusions
The present study involved the identification of RBPs associated with psychiatric disorders and also their interaction networks. The RBPs potentially driving gene expression changes in diseased human brain samples were also identified. Subsequently, hnRNP, PCBP, and SRSF family RBPs and a few other RBPs were found to form highly inter-connected hubs, representing their interactions with multiple other RBPs. These RBPs functioning as multi-protein networks could regulate multiple post-transcriptional regulatory processes. Disruption of one or a few RBPs could lead to dysfunction of the larger modules and networks, leading to multiple psychiatric conditions. The number of human studies conducted in this regard so far is limited, which could be a potential limitation of the present study. In future, large-scale functional studies to delineate the involvement of these RBPs in psychiatric co-morbidities could provide insights into gene regulatory processes underlying such conditions, which can help identify drug targets and design effective treatment strategies.
Availability of data and materials
Not applicable.
Abbreviations
- AD:
-
Alzheimer’s disease
- ADHD:
-
Attention-deficit/hyperactivity disorder
- ALS:
-
Amyotrophic lateral sclerosis
- BD:
-
Bipolar disorder
- CPEB:
-
Cytoplasmic polyadenylation element-binding protein
- DAZAP1:
-
DAZ-associated protein 1
- DEG:
-
Differentially expressed gene
- DLPFC:
-
Dorsolateral prefrontal cortex
- EFTU:
-
Elongation Factor Tu GTP-Binding Domain-Containing 2
- EIF2S1:
-
Eukaryotic translation initiation factor 2
- EIF4B:
-
Eukaryotic translation initiation factor 4B
- ELAVL2:
-
ELAV like neuron-specific RNA-binding protein 2
- ESRP2:
-
Epithelial splicing regulatory protein 2
- FDR:
-
False discovery rate
- FMR1:
-
Fragile X mental retardation 1
- FMRP:
-
Fragile X mental retardation protein
- GO:
-
Gene ontology
- HNRNP:
-
Heterogeneous nuclear ribonucleoprotein
- IGF2BP2:
-
Insulin-like growth factor 2 mRNA-binding protein 2
- KD:
-
Knock-down
- KHDRBS1:
-
KH RNA-binding domain containing, signal transduction-associated 1
- KHDRBS3:
-
KH RNA-binding domain containing, signal transduction-associated 3
- KO:
-
Knockout
- MATR3:
-
Matrin3
- MBNL1:
-
Muscleblind-like splicing regulator 1
- MCC:
-
Maximum clique centrality
- MDD:
-
Major depressive disorder
- OPMD:
-
Oculopharyngeal muscular dystrophy
- PABP:
-
Poly(A)-binding protein
- PABPN1:
-
Poly(A)-binding protein nuclear 1
- PCBP:
-
Poly(rC)-binding protein
- PFC:
-
Prefrontal cortex
- PTBP:
-
Polypyrimidine tract-binding protein
- PTGR:
-
Post-transcriptional regulation
- RBM:
-
RNA-binding motif protein
- RBP:
-
RNA-binding protein
- SCZ:
-
Schizophrenia
- SF3B4:
-
Splicing factor 3B subunit 4
- SNRNP70:
-
Small nuclear ribonucleoprotein U1 subunit 70
- SRSF:
-
Serine- and arginine-rich splicing factor
- TDP-43:
-
Transactive response DNA-binding protein 43
- TSMA:
-
Transcript Set Motif Analysis
- U2AF2:
-
U2 small nuclear RNA auxiliary factor 2
- UTRs:
-
Untranslated regions
References
James SL, Abate D, Abate KH, Abay SM, Abbafati C, Abbasi N et al (2018) Global, regional, and national incidence, prevalence, and years lived with disability for 354 Diseases and Injuries for 195 countries and territories, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet. https://doi.org/10.1016/S0140-6736(18)32279-7
World Health Organization. Mental disorders 2019. https://www.who.int/news-room/fact-sheets/detail/mental-disorders (accessed April 19, 2022).
Samsom JN, Wong AHC (2015) Schizophrenia and depression co-morbidity: What we have learned from animal models. Front Psychiatry 6:13. https://doi.org/10.3389/fpsyt.2015.00013
Buckley PF, Miller BJ, Lehrer DS, Castle DJ (2009) Psychiatric comorbidities and schizophrenia. Schizophr Bull 35:383–402. https://doi.org/10.1093/schbul/sbn135
Schiweck C, Arteaga-Henriquez G, Aichholzer M, Edwin Thanarajah S, Vargas-Cáceres S, Matura S et al (2021) Comorbidity of ADHD and adult bipolar disorder: A systematic review and meta-analysis. Neurosci Biobehav Rev 124:100–123. https://doi.org/10.1016/j.neubiorev.2021.01.017
Grunze H, Schaefer M, Scherk H, Born C, Preuss UW (2021) Comorbid bipolar and alcohol use disorder—a therapeutic challenge. Front Psychiatry 12:660432. https://doi.org/10.3389/fpsyt.2021.660432
Hyman SE (2018) The daunting polygenicity of mental illness: Making a new map. Philos Trans R Soc B Biol Sci 373:20170031. https://doi.org/10.1098/rstb.2017.0031
Sánchez-Valle J, Tejero H, Fernández JM, Juan D, Urda-García B, Capella-Gutiérrez S et al (2020) Interpreting molecular similarity between patients as a determinant of disease comorbidity relationships. Nat Commun 11:1–13. https://doi.org/10.1038/s41467-020-16540-x
Hernandez LM, Kim M, Hoftman GD, Haney JR, de la Torre-Ubieta L, Pasaniuc B et al (2021) Transcriptomic insight into the polygenic mechanisms underlying psychiatric disorders. Biol Psychiatry 89:54–64. https://doi.org/10.1016/j.biopsych.2020.06.005
Ramaker RC, Bowling KM, Lasseigne BN, Hagenauer MH, Hardigan AA, Davis NS et al (2017) Post-mortem molecular profiling of three psychiatric disorders. Genome Med 9:72. https://doi.org/10.1186/s13073-017-0458-5
Martins HC, Schratt G (2021) MicroRNA-dependent control of neuroplasticity in affective disorders. Transl Psychiatry 11:263. https://doi.org/10.1038/s41398-021-01379-7
Egervari G, Kozlenkov A, Dracheva S, Hurd YL (2019) Molecular windows into the human brain for psychiatric disorders. Mol Psychiatry 24:653–673. https://doi.org/10.1038/s41380-018-0125-2
Reble E, Dineen A, Barr CL (2018) The contribution of alternative splicing to genetic risk for psychiatric disorders. Genes Brain Behav 17:e12430. https://doi.org/10.1111/gbb.12430
Rusconi F, Battaglioli E, Venturin M (2020) Psychiatric disorders and lncrnas: a synaptic match. Int J Mol Sci 21:3030. https://doi.org/10.3390/ijms21093030
Hentze MW, Castello A, Schwarzl T, Preiss T (2018) A brave new world of RNA-binding proteins. Nat Rev Mol Cell Biol 19:327–341. https://doi.org/10.1038/nrm.2017.130
Harvey RF, Smith TS, Mulroney T, Queiroz RML, Pizzinga M, Dezi V et al (2018) Trans-acting translational regulatory RNA binding proteins. Wiley Interdiscip Rev RNA 9:e1465. https://doi.org/10.1002/wrna.1465
Wang ZL, Li B, Luo YX, Lin Q, Liu SR, Zhang XQ et al (2018) Comprehensive genomic characterization of RNA-binding proteins across human cancers. Cell Rep 22:286–298. https://doi.org/10.1016/j.celrep.2017.12.035
Gebauer F, Schwarzl T, Valcárcel J, Hentze MW (2021) RNA-binding proteins in human genetic disease. Nat Rev Genet 22:185–198. https://doi.org/10.1038/s41576-020-00302-y
De Bruin RG, Rabelink TJ, Van Zonneveld AJ, Van Der Veer EP (2017) Emerging roles for RNA-binding proteins as effectors and regulators of cardiovascular disease. Eur Heart J 38:1380–1388. https://doi.org/10.1093/eurheartj/ehw567
Nutter CA, Kuyumcu-Martinez MN (2018) Emerging roles of RNA-binding proteins in diabetes and their therapeutic potential in diabetic complications. Wiley Interdiscip Rev RNA 9:208. https://doi.org/10.1002/wrna.1459
Conlon EG, Manley JL (2017) RNA-binding proteins in neurodegeneration: Mechanisms in aggregate. Genes Dev 31:1509–1528. https://doi.org/10.1101/gad.304055.117
Park CY, Zhou J, Wong AK, Chen KM, Theesfeld CL, Darnell RB et al (2021) Genome-wide landscape of RNA-binding protein target site dysregulation reveals a major impact on psychiatric disorder risk. Nat Genet 53:166–173. https://doi.org/10.1038/s41588-020-00761-3
Quattrone A, Dassi E (2019) The architecture of the human RNA-binding protein regulatory network. IScience 21:706–719. https://doi.org/10.1016/j.isci.2019.10.058
Sternburg EL, Karginov FV (2020) Global approaches in studying RNA-binding protein interaction networks. Trends Biochem Sci 45:593–603. https://doi.org/10.1016/j.tibs.2020.03.005
Ghidini A, Cléry A, Halloy F, Allain FHT, Hall J (2021) RNA-PROTACs: degraders of RNA-binding proteins. Angew Chemie Int Ed 60:3163–3169. https://doi.org/10.1002/anie.202012330
Gutierrez-Sacristan A, Grosdidier S, Valverde O, Torrens M, Bravo A, Piñero J et al (2015) PsyGeNET: a knowledge platform on psychiatric disorders and their genes. Bioinformatics 31:3075–3077. https://doi.org/10.1093/bioinformatics/btv301
Krismer K, Bird MA, Varmeh S, Kong YW, Cannell IG, Yaffe MB (2020) Transite: a computational motif-based analysis platform that identifies RNA-binding proteins modulating changes in gene expression ll. Cell Rep 32:108064. https://doi.org/10.1016/j.celrep.2020.108064
Li R, Hu K, Liu H, Green MR, Zhu LJ (2020) Onestoprnaseq: a web application for comprehensive and efficient analyses of rna-seq data. Genes (Basel) 11:1165. https://doi.org/10.3390/genes11101165
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D et al (2003) Cytoscape: a software Environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. https://doi.org/10.1101/gr.1239303
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY (2014) cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst Biol 8:S11. https://doi.org/10.1186/1752-0509-8-S4-S11
Ge SX, Jung D, Jung D, Yao R (2020) ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 36:2628–2629. https://doi.org/10.1093/bioinformatics/btz931
Miro J, Bougé AL, Murauer E, Beyne E, Da CD, Claustres M et al (2020) First identification of rna-binding proteins that regulate alternative exons in the dystrophin gene. Int J Mol Sci 21:7803. https://doi.org/10.3390/ijms21207803
Berto S, Usui N, Konopka G, Fogel BL (2016) ELAVL2-regulated transcriptional and splicing networks in human neurons link neurodevelopment and autism. Hum Mol Genet 25:2451–2464. https://doi.org/10.1093/hmg/ddw110
ENCODE Project Consortium (2012) An integrated encyclopedia of DNA elements in the human genome ENCODE Encyclopedia of DNA Elements. Nature 489:57–74
Ke H, Zhao L, Zhang H, Feng X, Xu H, Hao J et al (2018) Loss of TDP43 inhibits progression of triple-negative breast cancer in coordination with SRSF3. Proc Natl Acad Sci USA 115:E3426–E3435. https://doi.org/10.1073/pnas.1714573115
Song X, Wan X, Huang T, Zeng C, Sastry N, Wu B et al (2019) SRSF3-regulated RNA alternative splicing promotes glioblastoma tumorigenicity by affecting multiple cellular processes. Cancer Res 79:5288–5301. https://doi.org/10.1158/0008-5472.CAN-19-1504
He X, Zhang P (2015) Serine/arginine-rich splicing factor 3 (SRSF3) regulates homologous recombination-mediated DNA repair. Mol Cancer 14:158. https://doi.org/10.1186/s12943-015-0422-1
Okholm TLH, Sathe S, Park SS, Kamstrup AB, Rasmussen AM, Shankar A et al (2020) Transcriptome-wide profiles of circular RNA and RNA-binding protein interactions reveal effects on circular RNA biogenesis and cancer pathway expression. Genome Med. https://doi.org/10.1186/s13073-020-00812-8
Utami KH, Skotte NH, Colaço AR, Yusof NABM, Sim B, Yeo XY et al (2020) Integrative analysis identifies key molecular signatures underlying neurodevelopmental deficits in fragile X syndrome. Biol Psychiatry 88:500–511. https://doi.org/10.1016/j.biopsych.2020.05.005
Appocher C, Mohagheghi F, Cappelli S, Stuani C, Romano M, Feiguin F et al (2017) Major hnRNP proteins act as general TDP-43 functional modifiers both in Drosophila and human neuronal cells. Nucleic Acids Res 45:8026–8045. https://doi.org/10.1093/nar/gkx477
Seachrist DD, Hannigan MM, Ingles NN, Webb BM, Weber-Bonk KL, Yu P et al (2020) The transcriptional repressor BCL11A promotes breast cancer metastasis. J Biol Chem 295:11707–11719. https://doi.org/10.1074/jbc.ra120.014018
Tabaglio T, Low DHP, Teo WKL, Goy PA, Cywoniuk P, Wollmann H, et al. MBNL1 alternative splicing isoforms play opposing roles in cancer. Life Sci Alliance 2018;1: e201800157. https://doi.org/10.26508/lsa.201800157.
Fish L, Pencheva N, Goodarzi H, Tran H, Yoshida M, Tavazoie SF (2016) Muscleblind-like 1 suppresses breast cancer metastatic colonization and stabilizes metastasis suppressor transcripts. Genes Dev 30:386–398. https://doi.org/10.1101/gad.270645.115
Lanz TA, Reinhart V, Sheehan MJ, Rizzo SJS, Bove SE, James LC et al (2019) Postmortem transcriptional profiling reveals widespread increase in inflammation in schizophrenia: a comparison of prefrontal cortex, striatum, and hippocampus among matched tetrads of controls with subjects diagnosed with schizophrenia, bipolar or major depressive disorder. Transl Psychiatry. https://doi.org/10.1038/s41398-019-0492-8
Hagenauer MH, Schulmann A, Li JZ, Vawter MP, Walsh DM, Thompson RC, et al. Inference of cell type content from human brain transcriptomic datasets illuminates the effects of age, manner of death, dissection, and psychiatric diagnosis. PLoS One 2018;13: e0200003. https://doi.org/10.1371/journal.pone.0200003.
Abdolmaleky HM, Gower AC, Wong CK, Cox JW, Zhang X, Thiagalingam A et al (2019) Aberrant transcriptomes and DNA methylomes define pathways that drive pathogenesis and loss of brain laterality/asymmetry in schizophrenia and bipolar disorder. Am J Med Genet Part B Neuropsychiatr Genet 180:138–149. https://doi.org/10.1002/ajmg.b.32691
Chen C, Meng Q, Xia Y, Ding C, Wang L, Dai R, et al. The transcription factor POU3F2 regulates a gene coexpression network in brain tissue from patients with psychiatric disorders. Sci Transl Med 2018;10: eaat8178. https://doi.org/10.1126/scitranslmed.aat8178.
Chen C, Cheng L, Grennan K, Pibiri F, Zhang C, Badner JA et al (2013) Two gene co-expression modules differentiate psychotics and controls. Mol Psychiatry 18:1308–1314. https://doi.org/10.1038/mp.2012.146
Harris LW, Wayland M, Lan M, Ryan M, Giger T, Lockstone H et al (2008) The cerebral microvasculature in schizophrenia: a laser capture microdissection study. PLoS ONE 3:e3964. https://doi.org/10.1371/journal.pone.0003964
Dassi E (2017) Handshakes and fights: the regulatory interplay of RNA-binding proteins. Front Mol Biosci 4:1–8. https://doi.org/10.3389/fmolb.2017.00067
Haroutunian V, Katsel P, Dracheva S, Davis KL (2006) The human homolog of the QKI gene affected in the severe dysmyelination “quaking” mouse phenotype: downregulated in multiple brain regions in schizophrenia. Am J Psychiatry 163:1834–1837. https://doi.org/10.1176/ajp.2006.163.10.1834
Sakers K, Liu Y, Llaci L, Lee SM, Vasek MJ, Rieger MA et al (2021) Loss of Quaking RNA binding protein disrupts the expression of genes associated with astrocyte maturation in mouse brain. Nat Commun 12:1–14. https://doi.org/10.1038/s41467-021-21703-5
Neumann DP, Goodall GJ, Gregory PA. The Quaking RNA‐binding proteins as regulators of cell differentiation. Wiley Interdiscip Rev 2022;e1724.
Zybura-Broda K, Wolder-Gontarek M, Ambrozek-Latecka M, Choros A, Bogusz A, Wilemska-Dziaduszycka J et al (2018) HuR (Elavl1) and HuB (Elavl2) stabilize matrix metalloproteinase-9 mRNA during seizure-induced Mmp-9 expression in neurons. Front Neurosci 12:224. https://doi.org/10.3389/fnins.2018.00224
Hagerman RJ, Protic D, Rajaratnam A, Salcedo-Arellano MJ, Aydin EY, Schneider A (2018) Fragile X-associated neuropsychiatric disorders (FXAND). Front Psychiatry. https://doi.org/10.3389/fpsyt.2018.00564
Fernández E, Rajan N, Bagni C (2013) The FMRP regulon: From targets to disease convergence. Front Neurosci. https://doi.org/10.3389/fnins.2013.00191
More DA, Kumar A (2020) SRSF3: Newly discovered functions and roles in human health and diseases. Eur J Cell Biol 99:151099. https://doi.org/10.1016/j.ejcb.2020.151099
Watanuki T, Funato H, Uchida S, Matsubara T, Kobayashi A, Wakabayashi Y et al (2008) Increased expression of splicing factor SRp20 mRNA in bipolar disorder patients. J Affect Disord 110:62–69. https://doi.org/10.1016/j.jad.2008.01.003
Laguesse S, Ron D (2020) Protein translation and psychiatric disorders. Neuroscientist 26:21–42. https://doi.org/10.1177/1073858419853236
Jernigan CS, Goswami DB, Austin MC, Iyo AH, Chandran A, Stockmeier CA et al (2011) The mTOR signaling pathway in the prefrontal cortex is compromised in major depressive disorder. Prog Neuro-Psychopharmacol Biol Psychiatry 35:1774–1779. https://doi.org/10.1016/j.pnpbp.2011.05.010
Bettegazzi B, Bellani S, Roncon P, Guarnieri FC, Bertero A, Codazzi F et al (2017) EIF4B phosphorylation at Ser504 links synaptic activity with protein translation in physiology and pathology. Sci Rep 7:1–16. https://doi.org/10.1038/s41598-017-11096-1
Chu TT, Liu Y, Kemether E (2009) Thalamic transcriptome screening in three psychiatric states. J Hum Genet 54:665–675. https://doi.org/10.1038/jhg.2009.93
Cuperfain AB, Zhang ZL, Kennedy JL, Gonçalves VF (2018) The complex interaction of mitochondrial genetics and mitochondrial pathways in psychiatric disease. Mol Neuropsychiatry 4:52–69. https://doi.org/10.1159/000488031
Zhang X, Hui L, Liu Y, Wang ZQ, You Y, Miao LN et al (2013) The type 2 diabetes mellitus susceptibility gene IGF2BP2 is associated with schizophrenia in a Han Chinese Population. J Clin Psychiatry 74:e287–e292. https://doi.org/10.4088/JCP.12m07846
Choudhury R, Roy SG, Tsai YS, Tripathy A, Graves LM, Wang Z (2014) The splicing activator DAZAP1 integrates splicing control into MEK/Erk-regulated cell proliferation and migration. Nat Commun 5:3078. https://doi.org/10.1038/ncomms4078
Zucchi FCR, Yao Y, Ward ID, Ilnytskyy Y, Olson DM, Benzies K, et al. Maternal stress induces epigenetic signatures of psychiatric and neurological diseases in the offspring. PLoS ONE 2013;8:e56967. https://doi.org/10.1371/journal.pone.0056967.
Nikolaou N, Gordon P, Hamid F, Taylor R, Makeyev E, Houart C. Cytoplasmic pool of spliceosome protein SNRNP70 regulates the axonal transcriptome and development of motor connectivity. BioRxiv 2020.
Dean B, Keriakous D, Scarr E, Thomas EA (2007) Gene expression profiling in Brodmann’s area 46 from subjetcs with schizophrenia. Aust N Z J Psychiatry 41:308–320. https://doi.org/10.1080/00048670701213245
Fila M, Diaz L, Szczepanska J, Pawlowska E, Blasiak J. Mrna trafficking in the nervous system: A key mechanism of the involvement of activity-regulated cytoskeleton-associated protein (arc) in synaptic plasticity. Neural Plast 2021;2021. https://doi.org/10.1155/2021/3468795.
Thelen MP, Kye MJ (2020) The role of RNA binding proteins for local mRNA translation: implications in neurological disorders. Front Mol Biosci 6:161. https://doi.org/10.3389/fmolb.2019.00161
Geuens T, Bouhy D, Timmerman V (2016) The hnRNP family: insights into their role in health and disease. Hum Genet 135:851–867. https://doi.org/10.1007/s00439-016-1683-5
Clarke JP, Thibault PA, Salapa HE, Levin MC (2021) A comprehensive analysis of the role of hnRNP A1 function and dysfunction in the pathogenesis of neurodegenerative disease. Front Mol Biosci 8:659610. https://doi.org/10.3389/fmolb.2021.659610
Richard P, Trollet C, Stojkovic T, De Becdelievre A, Perie S, Pouget J et al (2017) Correlation between PABPN1 genotype and disease severity in oculopharyngeal muscular dystrophy. Neurology 88:359–365. https://doi.org/10.1212/WNL.0000000000003554
Zhou Z, Fu XD (2013) Regulation of splicing by SR proteins and SR protein-specific kinases. Chromosoma 122:191–207. https://doi.org/10.1007/s00412-013-0407-z
Cassidy MF, Herbert ZT, Moulton VR (2022) Splicing factor SRSF1 controls distinct molecular programs in regulatory and effector T cells implicated in systemic autoimmune disease. Mol Immunol 141:191–207. https://doi.org/10.1016/j.molimm.2021.11.008
Mason MA, Gomez-Paredes C, Sathasivam K, Neueder A, Papadopoulou AS, Bates GP (2020) Silencing Srsf6 does not modulate incomplete splicing of the huntingtin gene in Huntington’s disease models. Sci Rep 10:14057. https://doi.org/10.1038/s41598-020-71111-w
Geuens T, De Winter V, Rajan N, Achsel T, Mateiu L, Almeida-Souza L et al (2017) Mutant HSPB1 causes loss of translational repression by binding to PCBP1, an RNA binding protein with a possible role in neurodegenerative disease. J Intensive Care 5:5. https://doi.org/10.1186/s40478-016-0407-3
Yoshimura M, Honda H, Sasagasako N, Mori S, Hamasaki H, Suzuki SO et al (2021) PCBP2 is downregulated in degenerating neurons and rarely observed in TDP-43-positive inclusions in sporadic amyotrophic lateral sclerosis. J Neuropathol Exp Neurol 80:220–228. https://doi.org/10.1093/jnen/nlaa148
Salem A, Wilson C, Rutledge B, Dilliott A, Farhan S, Choy W, et al. Matrin3: Disorder and ALS Pathogenesis. Front Mol Biosci 2022.
Acknowledgements
The authors are thankful to SASTRA Deemed to be University for infrastructural facilities. Data from ENCODE Project Consortium were used in this study.
Funding
Research in the laboratory of SJ is funded by Department of Science and Technology, Govt. of India (Grant No. SRG/2019/000382), and TRR Grant of SASTRA Deemed to be University. NM receives and acknowledges INSPIRE Fellowship from the Department of Science and Technology, Govt. of India (IF170376).
Author information
Authors and Affiliations
Contributions
NMJ and SJ were involved in conceptualization, analysis, interpretation, and manuscript writing and revision. Both authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1
. RBPs influencing DEGs.
Additional file 2
. Common RBPs.
Additional file 3
. RBPs and psychiatric disorders disorders.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Nishanth, M.J., Jha, S. Genome-wide landscape of RNA-binding protein (RBP) networks as potential molecular regulators of psychiatric co-morbidities: a computational analysis. Egypt J Med Hum Genet 24, 2 (2023). https://doi.org/10.1186/s43042-022-00382-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43042-022-00382-x