Abstract
In recent years, significant progress in genomic technologies has revolutionized the field of aquaculture. These advancements have facilitated the utilization of DNA markers, particularly single nucleotide polymorphisms (SNPs), to enhance the genetic characteristics of aquatic species, leading to overall improvements in economically important traits. A SNP array or panel is a DNA microarray designed with probes for SNP locations, allowing the identification of specific alleles in a DNA sample through hybridization with fragmented DNA. SNP arrays are known for their efficiency, cost-effectiveness, and automation, making them a high-throughput method for genotyping. Thus, genome scale SNP genotyping, aided by SNP arrays and genotyping-by-sequencing (GBS), has transformed aquaculture genetics. Recently, multi-species arrays allow researchers to study closely related species simultaneously, reducing costs and enabling comparative genomics and resource sharing. In this paper, we reviewed the global advancements in SNP array development for key aquaculture species and highlighted their applications in genetic selection and fisheries management. SNP panels, commonly used in genome-wide association studies (GWAS), leverage population linkage disequilibrium (LD) to pinpoint genetic variants associated with production or performance traits. Using genomic estimated breeding values (GEBVs) derived from SNP data offers enhanced selection accuracy compared to traditional pedigree based methods, especially when dealing with challenging traits in aquatic species. Overall, the study indicates that high-density SNP panels offer a consistent and reliable tool for genotyping across diverse breeding populations. This technology has demonstrated versatility and efficiency, being applied in genomic selection, genome characterization, population genomics, and QTL mapping in aquatic species. The growing accumulation of genomic information and the abundance of SNPs in aquaculture species have driven the demand for efficient and cost effective genotyping techniques in genetic improvement programs and fisheries management.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The field aquaculture has grown significantly in recent years, contributing to food and nutritional security for an increasing global population [1]. The aquaculture intensification owes a lot to the development of new methodologies such as quantitative approaches and algorithms. Selective breeding is the most commonly used method for increasing production in aquaculture species [2, 3]. Studies have shown that that numerous traits have been targeted successfully using genetic selection for production enhancement. An average genetic gain of approximately 12.5% has been observed in selectively bred aquaculture species [3]. Additionally, genetic and genomic tools are currently being applied to decode the biological information of key aquaculture species, thus enhancing their performance and production capabilities. Numerous experimental and review papers have been published, examining the use of DNA markers and their extensive application in aquatic species and resource management [4,5,6,7,8,9].
In recent years, advancements in technology, particularly in next-generation sequencing (NGS) and genomics, have significantly contributed to the improvement of aquaculture production and the conservation of biodiversity. These breakthroughs have made the process of whole genome assembly for diverse aquatic species more efficient, allowing for gene characterization and facilitating functional studies [6, 10, 11]. Selective breeding programs in aquaculture are progressively integrating genomic technologies to enhance genetic gain sustainably. Earlier reports have highlighted the importance of genomic tools in assessing genetic diversity and relationships within various aquatic species [6]. Single nucleotide polymorphisms (SNPs) are the preferred markers because of their widespread presence across the genome, and their typically bi-allelic nature, which allows for automated genotyping. Previous studies have shown that SNPs are highly effective for conducting genome-wide association studies (GWAS) for complex traits, as their high density distribution facilitates the detection of linkage disequilibrium (LD) [7]. Advances in NGS and bioinformatics tools have simplified SNP discovery in aquatic species, aiding genetic improvement efforts [12, 13]. These SNPs can be used for constructing linkage maps, identifying quantitative trait loci (QTLs) linked to specific traits, assigning parentage, and facilitating genetic selection [14,15,16,17]. It was suggested to have 100–500 SNPs at least for successful parentage analysis [18].
The development of Next Generation Sequencing (NGS) technology and computational tools have made the process of genome sequencing, re-sequencing, and genome-wide marker discovery more accessible and cost effective for aquatic species. Previous researchers have extensively reviewed the advancements in genome sequencing for significant fish species [19]. In 2009, the Genome 10 K Project was initiated with the ambitious aim of sequencing and constructing genomes for approximately 10,000 vertebrate species, including 4,000 teleosts [20]. Many genomes of important aquatic species have been sequenced and are readily available via public databases [18, 21]. In the last decade, this genomic information has been integrated into studies focusing on genomic selection and population dynamics in cultured species. Earlier studies have shown that genomic selection (GS) holds significant promise in aquaculture, which improves selection accuracy, genetic gain, and addresses inbreeding concerns in aquaculture [22]. Incorporating dense genome-wide SNP data into breeding programs using GS enhances the accuracy of trait selection [11, 23]. Therefore, GWA studies facilitate the identification of linkage among specific SNPs and traits.
Genome-scale SNP genotyping is a useful tool for studying the genetic makeup and improving fish species [24, 25]. It helps in creating SNP arrays or chips, which are used in various applications, including population structure evaluation, genome-wide association studies, genomic selection, linkage-map development, and gene mapping. The GBS technique has resulted in customized SNP arrays designed specifically for aquatic species that vary in density from low-medium to high-density options. The development of customized SNP arrays/chips designed specifically for aquatic species has shown in Table 1. Although the use of LD-SNP panels can lead to variable genomic prediction accuracy, high-density genotyping platforms offer a significant advantage in accelerating genetic enhancements within breeding programs. However, the detection and genotyping of SNPs can be challenging in non-model species that lack available whole genome information. In this paper, we have discussed the SNP array or panels developed in aquaculture species and their potential applications, as well as the challenges and future perspectives of the technology. Incorporating comprehensive genome-wide SNP data into breeding programs of aquatic species, along with extensive pedigree and phenotypic information, improves selection accuracy for important traits through genomic selection. Overall, this approach has found extensive use in genetic studies of fish species, making it a valuable tool for aquaculture research.
SNP array or chip development
The SNP array is regarded as a highly efficient, cost-effective, and automated method for genotyping due to its high-throughput capabilities. Through a collaborative endeavour between the Whitehead Institute for Biomedical Research and Affymetrix, Inc., United States (Now ThermoFisher Scientific) the first SNP chip was created, marking the inception of genome-wide association studies [44]. Currently, high-throughput genotyping primarily relies on Genotyping-by-Sequencing (GBS) [45], which is a low cost, high-throughput genotyping method that depends on restriction enzymes to reduce genome complexity and allow discovery of genome-wide SNPs with a lower error rate. GBS has recently gained attention in fish species due to advancement in sequencing platforms, longer read lengths, coverage of data, and more accessible reference genomes [46]. These developments create opportunities for the use of DNA markers for a better understand genetic basis of production and performance traits. Different techniques employ various types and quantities of restriction enzymes, with GBS being a cost-effective option that provides greater coverage depth per locus compared to whole genome sequencing, making it feasible for larger sample sizes [46, 47]. Previous researchers have conducted reviews on GBS and its applications in fisheries and aquaculture [45, 48,49,50].
A SNP array is designed to identify specific alleles in a sample by hybridizing with fragmented DNA to detect SNP positions [51]. Several commercial SNP array platforms are available, including the GeneChip® CustomExpress® Arrays (Affymetrix, Inc., USA), Illumina iSelect HD or HTS Custom BeadChip (Illumina, USA), SureScan SNP-Microarray platform (Agilent Technologies, Inc., USA), and Sequenom MassARRAY® system (iPLEX GOLD) (Sequenom, Inc., USA). Affymetrix predominantly utilizes Axiom genotyping principle, allowing the customization of SNP arrays. Molecular inversion probe technology (MIP) allows for versatile multiplexing, accommodating anywhere from 1,500 to 5,00,000 SNPs in an individual assay, making it the most flexible option in this regard. The Illumina chip incorporates a high throughput screening (HTS) custom panel, harnessing the capabilities of the Infinium iSelect microarray platform. This panel allows for the interrogation of an extensive set of up to 700,000 custom targets, including SNPs, indels, and CNVs. With its remarkable versatility, this platform empowers comprehensive target analysis across a diverse spectrum of species. Sequenom MassARRAY iPLEX Gold leverages a straightforward and precise primer extension chemistry combined with cutting-edge MALDI-TOF mass spectrometry. The widely utilized iPLEX Gold assay, enables assay design with a high degree of flexibility, accommodating multiplexing levels of up to 40plex and featured in numerous publications.
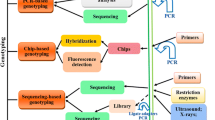
As previously discussed, there are various computational algorithms or tools being developed for extracting SNP genotypes from next-generation sequencing (NGS) data [52]. The typical procedure for SNP identification pipelines for NGS data, involves several steps, such as pre-processing of raw data, read alignment, SNP calling, filtering, and validation [53]. SNP arrays do have limitations, including the need for preexisting genomic information and the ability to genotype only known SNP positions. The diagram below illustrates the typical procedure for creating SNP array panels (Fig. 1). Several computational algorithms are accessible for SNP genotype calling based on FASTQ files. These algorithms can be broadly categorized into heuristic methods (such as CLC-Genomic Workbench, VarScan2, Galaxy, GSMapper, and DNSTAR) and probabilistic approaches (including GATK, SAM-tools, SNVer, Atlas SNP2, and SOAPsnp).
Various tools are available for copy number variation (CNV) analysis from the SNP array data. Illumina's CNVPartition tool utilizes Gaussian distributions (bivariate) in the process of SNP/ CNV analysis, which is accessible freely within the GenomeStudio platform. CamCNV is an R-package designed for the identification of rare CNVs, requiring a minimum of three probes for detection [54]. A Bayesian Gaussian mixture algorithm, such as PlatinumCNV is used for SNP array based copy number polymorphism genotyping [55]. Affymetrix also offers dedicated tools for CNV analysis, accessible through the Affymetrix Genotyping Console (GTC), an interface using the Affymetrix Array Power Tools. R-GADA (Genome Alteration Detection Analysis) offers a versatile and comprehensive pipeline within the R tools. It can effectively genotype CNVs, visualize results graphically, and conduct association analysis for various array types, including Illumina, Affymetrix, or array comparative-genomic hybridization (aCGH) arrays [56]. Illumina and Affymetrix share most criteria when selecting SNP markers for arrays, differing primarily in sequence length (20 bp for Affymetrix, 50 bp for Illumina) and assessment factors. SNP selection involves several critical factors, including considerations of depth and frequency, SNP types, and variations within probe parameters and sequences. The depth at which SNPs are read is of paramount importance, as inadequate depth can introduce errors, while excessive depth can lead to SNP calls from repetitive genomic regions. When developing SNP arrays, preference is often given to transition SNPs such as A/G and T/C, while transversion SNPs, INDELs, and allelic SNPs (those with multiple variants) are typically excluded. This exclusion is primarily because A/T and C/G SNPs need the use of two distinct probes for accurate genotyping, in contrast to other SNP types, which can be genotyped with just a single probe [57, 58]. In the development of SNP array probes, the evaluation of surrounding sequences adjacent to target SNPs is crucial, often employing metrics such as Affymetrix P-convert or Illumina ADT values. A primary objective is to achieve a balanced distribution of SNPs throughout the chromosomes, with consideration for their potential inclusion within exonic regions [59, 60].
Progress of SNP array developed in an important aquaculture species
Genome sequencing has brought about a revolution in genetic research, enabling us to study genetic variations, perform GWAS, explore gene environment interactions, and analyze evolution in organisms, including commercially important fish and shellfish [18, 61, 62]. Over the past two decades, 594 fish genomes have been sequenced, which cover about 1.85% of the estimated 32,000 fish species (https://www.ncbi.nlm.nih.gov/genome, accessed on 5th September 2023). The majority of fish genome sequences are available in draft form. Genomics has been extremely helpful in fisheries management by answering key questions like stock identification, population structure, and responses to environmental changes. This data has been instrumental in identifying SNPs linked to production and performance traits, ultimately paving the way for the creation of SNP arrays or panels in diverse aquatic species. We have summarized the advancements in SNP arrays across aquatic species worldwide (Table 1 and their applications in candidate aquaculture species, such as GWAS, linkage map construction, analysis of population structure, and selective breeding programs.
Atlantic Salmon (Salmo salar)
In the early 1970s, selective breeding programs were initiated in Norway to enhance the genetic traits of Atlantic salmon. The development of single nucleotide polymorphism (SNP) markers and arrays for Atlantic salmon has been made possible by the availability of genomic information [29, 63, 64]. Initially, a low-density (LD) SNP panel was developed, comprising roughly 6,000 polymorphic SNPs [65]. In a GWA study using Cermaq Canada broodstock with around ~ 480 fishes, researchers utilized a panel of 6.5 K SNPs to reveal their association with growth and the timing of sexual maturation in S. salar. The most robust associations were observed for "grilsing" with markers on Ssa10, Ssa02, Ssa12, Ssa13, and Ssa25, as well as markers on Ssa01, Ssa21, and Ssa28 for late maturation [66]. A HD SNP panel with around 130 K SNPs for S. salar was developed using RRSeq, RNASeq, and RADSeq methods. It was utilized to genotype 96 samples from farmed Scottish, farmed Norwegian, and wild fish categories [29]. Researchers validated around 160,000 SNPs using a 200 K SNP panel for S. salar in diverse populations across North America, Europe, and Chile [29, 67]. A GWA study on bacterial-kidney disease (BKD) resistance in North American S. salar utilized a 50 K SNPs chip to genotype 576 individuals [68]. Indeed, this study highlights the invaluable nature of the data for guiding genetic improvement efforts aimed at enhancing resistance to BKD in S. salar populations. A 220 K SNP chip for S. salar was used to recognize key chromosomes associated with sex determination. Ssa02 (heritability = 0.42) and Ssa21 (heritability = 0.26) exhibited the highest genetic variance, with Ssa02 being a strong candidate for sex determination and Ssa21 indicating an alternate lineage in wild populations [69]. To assess genetic relationships and the impact of chromosomal fusions on adaptation, 728 salmon (juvenile) were genotyped using a 50 K SNP panel [70]. These findings indicate that chromosomal fusion and genomic variations associated with life history play a significant part in the process of local adaptation (within the river) in S. salar. Using a 57 K SNP chip, genotyping of 642 salmon was conducted to identify SNPs linked to omega-3 fatty acid muscle content [71]. Significant associations with the DHA/DPA ratio, particularly in the vicinity of elovl2, which plays a significant role in the conversion of DPA to DHA, were found and traced to chr21.
A revolutionary high-density chip, called the 930-K XHD Ssal array, was developed for fish research, specifically for Atlantic Salmon.This innovative tool, created using Aquagen lines consisting of 29 individuals, played a crucial role in discovering significant insights such as the crucial influence of the VGLL3 on age at sexual maturity [72] and the identification of the CDH as the primary factor determining Atlantic salmon resistance to IPNV [73]. In S. salar, resequencing of 20 individuals of whole genome from three sources produced a catalog of 9.7 million SNPs, which were further refined to create a focused 200 K SNP panel [67].
A study was conducted to evaluate the accuracy of genotype imputation in a salmon breeding program that utilized both 25 K and 78 K SNP panels [74]. The imputation accuracy was observed to be ranged from 0.62 to 0.90. Genomic prediction with imputed genotypes was shown to be slightly less accurate than true genotypes but significantly better than pedigree-based methods for body weight and sea-lice resistance. In another study, a 50 K SNP panel was used to genotype 1,756 fish from 248 families of North American S. salar [75]. These fish were exposed to salmon lice, with resistance showing a heritability of 0.21 ± 0.03. A significant SNP on chromosome Ssa04, related to cell survival and inflammation, was identified as highly suggestive of salmon lice resistance. Similarly, a GWAS analysis was performed on 11,166 male S. salar from a single strain to investigate age at sexual maturity [76]. Genotypes were imputed at 50-K SNPs, revealing significant associations on 28 of the 29 chromosomes, where robust signals were observed in the gene regions of six6 (Chr9) and vgll3 (Chr25).
Common carp (Cyprinus carpio)
Common carp is the third most widely farmed fish species globally, contributing around 7% to the total aquaculture production [1]. The reference genome of the common carp stands as an important resource for essential genetic and molecular studies. Recent research has highlighted the progress of genomic resources and genetic improvement in the common carp [77]. For the first time, a 250 K SNP array originally developed for C. carpio demonstrated remarkable efficiency in genotyping other related fish species, such as goldfish and zebrafish [27]. The SNP array is strategically placed across the reference genome of C. carpio, and was assessed by genotyping 1,072 collected samples. Over 74% of these sites were found to be polymorphic, while over 99.8% of the qualified SNPs showed high reliability. This carp SNP array is a pioneering high throughput genotyping platform specifically tailored for C. carpio. In 2016, it was used for the development of a high density linkage map for Yellow River carp [78]. Similarly, in another study, this 250 K SNP panel was utilized for efficient genotyping, yielding a robust high density linkage map specific to common carp, with a total of 11 K unique loci and an average locus interval of 0.3 cM [79]. To ensure accurate pedigree assignment in common carp, a set of SNP markers was selected from the 250 K SNP panel. The 48 K SNP panel, created with selected markers and the Fluidigm genotyping platform, was used to genotype a genetically diverse Yellow River carp population, including half sibling and full sibling families [80].
In a study involving 203 common carp samples, GWAS was conducted to screen for traits related to fat and fatty acid content. Using a carp 250 K SNP array, this analysis revealed 9 significant SNPs and 15 transcripts linked with the muscular polyunsaturated fatty acid content. After passing quality control, 193 samples were retained, and 108,684 polymorphic SNPs were selected for association analysis Tag SNPs were filtered out if they showed high linkage disequilibrium (LD R2 > 0.9), and finally, 29,026 tag SNPs were selected for association analysis [81]. Using 250 K SNP panel, a study analyzed 2,198 individuals from 14 global carp populations to understand genetic structure and environmental adaptation. The analysis revealed that linkage disequilibrium (LD) block lengths ranged from 3.9 kb to 36.6 kb among the populations [82]. The work confirmed the successful use of the 250 K SNP panel in investigating the genetic basis of an unusual scale mutation within a sample of 82 Yellow River carp specimens [83].
To pinpoint specific genomic regions and potential candidate genes linked to abdominal fat and muscle fat content traits, GWAS was performed using the available 250 K SNP panel using carp F2 generation [84]. A GWA study with a 250-K SNP panel was performed on Yellow River carp fishes (433 Numbers) to reveal genes associated with the head size traits. Candidate genes like parvalbumin, igf1, srpk2, fsrp5, igf1r, grb10, igf3, notch2, and sfrp2 were found near significant SNPs [85]. Further, this SNP panel was used to understand the involvement of genetic factors responsible for abnormal skin coloration and identify their linkage with this trait(s). Interestingly, this work revealed significant SNPs (18) which is located on Chr11, and found five key genes related to pigmentation such as apoeb, mitf, lrp8, ap1m1, and oca2 [86].
A 250 K SNP array was used for QTL identification and GWA study in Mirror carp exposed to koi-herpesvirus infection to detect genetic regions and genes associated with virus resistance [87]. Numerous immune-related genes, such as tnfa, rootletin, galectin-8, hif1a, and palladin, were identified near significant QTLs or SNPs. This study suggests that information obtained can be useful in implementing MAS in common carp strains to imrpove their resistance to koi herpesvirus. Another study on amino acid metabolism in common carp revealed sixty-two genes associated with tyrosine, proline, and glycine content, through GWAS analysis with the 250 K array [88].
Rainbow trout (Oncorhynchus mykiss)
Rainbow trout is the most commonly cultivated species among salmonid fishes in aquaculture. Its popularity is due to the availability of well documented genomic data resources [88]. In 2014, researchers successfully established the first rainbow trout reference genome, which encompassed a total size of 1.9 gigabases (Gb) (scaffold N50 value of 384 kb). From a pool of approximately 2.12 million potential SNPs, a HD 57 K array was developed for O. mykiss, which became commercially available in 2015 [26, 89]. This array includes 50 K top-notch SNPs that provide extensive coverage across the genome. Further, it has proven highly effective in various applications such aspopulation structure analysis, GS, and selective breeding programs of trout [90,91,92,93].
A 57 K SNP array designed for O. mykiss was employed to identify polymorphisms in several salmonid species [89]. The rainbow trout SNP array's ability to share 28,882 SNPs among six salmonid populations from four different genera and detected 525 polymorphic SNPs across all 4 genera, highlights its flexibility as a common genotypic tool for multiple salmonids. In a study involving 7,893 rainbow trout from 102 families, Bacterial cold water disease (BCWD) resistance was assessed based on survival phenotypes. Using a 57 K SNP array, genotypes were available for 1,473 fish from 50 families [94, 95]. Notably, for BCWD resistance, GEBVs obtained from various GS models were observed outperformed as compared to EBVs from the pedigree-based BLUP model.
Genome sequencing (30 X coverage) of INRAE isogenic lines resulted in the development of HD 665 K SNP array for O. mykiss in the French commercial line [96]. Among the initial 576 K SNPs, around 38 K SNPs were retained from earlier the MD 57 K SNP array/chip. In this study, an average variant spacing of 60 bp underscored significant polymorphism in the genome of O. mykiss. The high-density chip (HD) demonstrated its utility by revealing faster LD decay at 2 to 10 kb intervals compared to typical 50–100 kb distances seen with medium-density chips.
Atlantic cod (Gadus morhua)
Atlantic cod is an important species in the North Atlantic region, both ecologically and economically. Recently, there has been a significant expansion in the genomic resources available for this species. The genetic differentiation between Baltic cod populations in distinct salinity environments, such as those in the Bay of Gdansk, Eastern site, and the Kiel Bight, Western site, was verified using a cod-specific SNP array (Illumina) containing 10 K SNPs [97]. The overall FST value across all populations stood at 0.039 (P < 0.01), signifying a modest degree of differentiation. Additionally, the cod SNP arrays were used to analyze and reveal significant variations in differentiation patterns across the genome. This provided insights into the complex biological structures within the Norwegian coastal-cod and Norwegian coastal-south populations [98].
Genome sequence data currently available is harnessed to identify genomic SNP variants, leading to the creation of a SNP array tailored specifically for the Norwegian cod population [99]. The utility of this SNP panel is exemplified through its application in the analysis of data derived from the National Cod Breeding Program (NCBP). The newly developed SNP- array is used to genotype approximately 2,500 individuals distributed across the two datasets.
Channel catfish (Ictalurus punctatus)
The catfish, particularly channel catfish holds a pivotal role in aquaculture within the United States and China [100]. Initially, a 250 K SNP panel was developed using SNPs derived from channel catfish, blue catfish, and inter-specific SNPs. Its utility in catfish genetic research was confirmed by genotyping wild populations and backcross families. Furthermore, an HD genetic linkage map was developed using the 250-K SNP panel in the hybrid catfish system [28, 101]. This map incorporated 26,238 SNPs distributed across 29 linkage groups, resulting in 12,776 distinct marker positions. A GWAS on heat stress in catfish utilized the 250 K catfish SNP array. The approach involved utilizing interspecific backcross offspring, which were obtained by mating female channel catfish with male F1-hybrid catfish (♀ channel catfish × ♂ blue catfish) [102]. In another GWA study, a 250-K SNP panel was used for the evaluation of body weight, with ~ 556 back-cross progenies resulting from the mating of male F1-hybrids with females [103]. Remarkably, a genomic location of around 1.0 Mb in linkage-group-5 exhibited a significant association with body weight.
A 690 K high density SNP array was created to provide uniform genome coverage, whichincludes 98.6% of the catfish reference genome [104]. The 250 K SNP panel was used for linkage mapping, which resulted in mapping to 29 linkage groups (LGs) with a highest marker density. In a population of an I. punctatus, a 55 K SNP panel was used to identify significant SNPs linked with harvest weight and carcass weight [105]. Genomic evaluation (ssGBLUP) led to significant improvement in predictive accuracy, increasing around 28% for harvest weight and 36% for carcass weight compared to traditional pedigree based methods. A GWA study using the 250 K SNP panel identified QTL related to low DO tolerance in the blue catfish × channel catfish system [106]. The associations were found in four linkage groups, with genes in these regions involved in various pathways (e.g., MAPK, VEGF, etc.) revealing the intricate genetic basis of hypoxia tolerance in catfish.
Japanese flounder (Paralichthys olivaceus)
Japanese flounder is a significant fish in aquaculture across Asian countries and substantial genomic data has been generated for this species [107]. A 50 K SNP panel or array named "Yuxin No. 1" was developed to enhance selection accuracy for selected bacterial disease resistance in Japanese flounder aquaculture [43]. A high quality SNP panel was developed using a genome re-sequencing approach from 1,099 fish. It demonstrated strong genotyping performance, with over 74% of loci showing high call rates and strong polymorphic SNP clusters. The array significantly improved the accuracy of GEBV by 21.2% using wGBLUP compared to ABLU), showcasing its effectiveness for GS in Japanese flounder.
Nile tilapia (Oreochromis niloticus)
Nile tilapia is the one of the most important aquaculture species in the world, contributing approximately 7.5% of the total aquaculture production. To develop SNPs markers, 32 Genomar strains were re-sequenced and selected for their polymorphisms and genomic locations using the Orenil1.1-assembly [31]. They SNP performance was assessed by genotyping 4,991 fishes, resulting in 43,588 high quality SNPs. This endeavor culminated in the creation and validation of a 58 K SNP panel/array known as the Onil50 array, and additionally, an integrated genetic and physical linkage-map has been constructed, revealing sex specific recombination patterns in LGs.
A comprehensive SNP array comprising approximately 65 K SNPs was meticulously crafted by leveraging whole-genome sequence data sourced from a GIFT breeding-nucleus population. This innovative array was further enriched with polymorphic SNPs derived from diverse wild-fish populations and various farmed Nile tilapia species, enhancing its utility and genetic diversity [36]. These SNP panel were successfully used for the differentiation of Asia and African tilapia populations.
In a study on Nile tilapia's resistance to Tilapia lake virus (TiLV), a GWA study was conducted using a 65 K SNP panel, and important QTL was discovered on chr-Oni22, containing genes relevant to the host response to viral infections, such as lgals17, vps52, and trim29 [108]. This discovery offers insights into TiLV resistance mechanisms in Nile tilapia. Similarly, a study on feed associated traits in GIFT tilapia used the 65 K SNP panel [109]. This work identified significant QTLs for body-weight gain and feed-intake on chr 7 and 5, respectively, harboring key genes such as ntrk3a, ghrh, and eif4e3. Genomic data improved prediction accuracy by up to 34%, surpassing pedigree records alone, and marker density effects were explored.
European sea bass (Dicentrarchus labrax L.)
European sea bass is an economical important fish species in Europe. Over the years, genomic resources have been developed and genome as well as SNP markers are available in the database [110,111,112]. Currently, two SNP panels are available for this species, which include a 3 K SNP panel (Illumina iSelect) [113] and a 'DlabCHIP' 57 K panel (Thermofisher Axiom™; Unpublished). These arrays, the 3 K (Illumina) and the 57 K DlabCHIP (Axiom) offer efficient genotyping, with the latter showing excellent performance in French sea bass populations. It was suggested that the 57 K SNP panel is expected to become a standard tool for GS in European sea bass selective breeding programs.
In GWAS on VNN resistance in D. labrax, the Axiom™ 57 K SNP (DlabChip) was used. A total 7 QTLs, including one on linkage group 12 (LG 12), were identified [114]. In a study on D. labrax, a GWAS using a 57 K SNP panel identified 5 significant SNPs related to stress indicators, body weight, and growth performance in 865 offspring, including 332 challenged with Vibrio anguillarum [115]. The study also estimated moderate to high genomic heritability for these traits using the REML approach.
Pacific oyster (Crassostrea gigas)
The Pacific oyster is economical significant bivalve species. Notably, it exhibits one of the most extensive DNA variations amongst all the animal species. A 190 K SNP array for Pacific oysters was created using Affymetrix Axiom technology, making it the first HD SNP panel designed specifically for molluscan groups [116]. The chip incorporated around 190 K SNPs chosen from ~ 54 million SNPs recognized by the re-sequencing method from diverse regions (n = 472), with a remarkable 70.4% of these SNPs proving to be polymorphic. SNP arrays for Pacific oysters enable testing of GS for polygenic traits. A study on growth-associated traits found moderate heritability and a polygenic genetic architecture. Genomic prediction models outperformed pedigree-based methods, with GBLUP achieving 25–30% higher accuracy for shell traits [117].
A MD 15-K SNP panel was designed for both European flat-oyster (Ostrea edulis) and Pacific oyster (C. gigas) and further tested on 219 samples, primarily from European populations [30]. In the European flat-oyster, growth traits were found to have a largely polygenic genetic basis with two key QTLs on chromosome 4 [118]. GS using LD SNP array, as low as 100 SNPs, maintained high prediction accuracies (> 0.83) for all the traits, suggesting cost effective breeding potential.
Sea cucumber (Apostichopus japonicus)
Sea cucumber is highly valuable in the global market due to its nutritional content and exclusive pharmaceutical compounds. A 24 K SNP panel known as ‘HaishenSNP24K’ was developed for sea cucumber using the HD Marker genotyping approach and MOLO algorithm [40]. It achieved high genotyping call rates (> 96%), accuracy (> 95%), and showed strong polymorphism in A. japonicus populations.
Large yellow croaker (Larimichthys crocea)
The large yellow croaker is a highly valued marine species. Recently, 82 fish from various Chinese locations were resequenced, which revealed 9.3 million SNPs. These SNPs were used to create a tailored 600 K SNP panel for this species [119].
Challenges, future perspectives, and concluding remarks
In the last decade, NGS (Next Generation Sequencing) technologies have become more popular for sequencing purposes, particularly for transcriptome or whole genome sequencing in various fish species. This is due to the reduced cost and decreased time required for sequencing. NGS technologies generate vast amounts of sequence information, allowing for the creation of marker databases, including Single Nucleotide Polymorphisms (SNPs). Various genotyping approaches customized for NGS, such as GBS, Exome seq, and RAD seq, have been instrumental in discovering a significant volume of SNPs in numerous fish species.
However, choosing the right SNPs for SNP panel or array design is essential to the effective use of SNP arrays. SNPs sourced from gene enriched regions are the preferred candidates for the development of SNP panels. When considering depth for SNP calling, if it is too low, there is a risk of calling SNPs due to errors in sequencing depth. Conversely, high depth of sequencing resulted in calling SNPs from repetitive regions. In certain scenarios, when SNPs are called from multiple sequence sources, it is important to consider the source of the SNP to ensure accurate analysis and interpretation. Genome duplication adds complexity to the identification of authentic bi-allelic SNPs during bioinformatics analyses.
The primary consideration when including SNPs in the array is whether they meet the probe design requirements of the chosen platform. The second aspect to consider is the dosage of the SNPs. A third optional factor to consider is the distribution of SNPs across homologous chromosomes and functional regions, with a focus on genic areas, to mitigate the impact of ascertainment bias. This analysis is particularly beneficial when a fully annotated reference genome is accessible, and it can be further enhanced by incorporating SNP data from wild or closely related species.
GWAS studies produce vast SNP datasets, offering the potential for structural variant genotyping. GBS methods aid genome-wide SNP discovery but have complexities and computational demands, especially for species without a reference genome. Custom SNP arrays or panels can include specific SNPs from regions of interest, and their numbers can be adjusted as needed on both Illumina and Affymetrix platforms. SNP arrays have their limitations, including the necessity for pre-existing genomic data and their ability to genotype only known SNP positions. The design and optimization of SNP arrays could be a time-consuming procedure. Ascertainment bias in genotyping arrays often arises from non-random polymorphism sampling or limited sample sizes in SNP discovery panels, which can exclude rare alleles.
Recently, High-Density (HD) SNP panels have been developed in several aquatic fishes. These arrays are valuable for studying economically important traits and implementing Genome Selection (GS) in aquaculture breeding programs. They can enhance genetic progress, particularly for challenging or costly measured traits. GWA studies identify vital traits linked with SNPs. Conversely, complex traits may involve multiple QTLs. RADseq offers versatility for genomic selection (GS), genome analysis, population genomics, and QTL mapping.
In conclusion, SNP arrays or panels are essential for genetic studies and breeding in aquaculture species, and their demand is growing due to the wealth of genomic resources and SNP discoveries. High-density SNP arrays provide standardized and reproducible genotyping data, although challenges like cost and ascertainment bias exist. NGS-based genotyping approaches are gaining traction for targeted genotyping. HD markers offer an affordable method for in-solution SNP array development, facilitating genetic progress in aquaculture through comprehensive trait analysis and genomic selection.
Availability of data and materials
No datasets were generated or analysed during the current study.
Abbreviations
- SNPs:
-
Single nucleotide polymorphisms
- GWAS:
-
Genome-wide association studies
- GEBVs:
-
Genomic estimated breeding values
- QTL:
-
Quantitative Trait Locus
- GS:
-
Genomic selection
- LD:
-
Linkage disequilibrium
- NGS:
-
Next Generation Sequencing
- GBS:
-
Genotyping-by-Sequencing
References
FAO. The state of world fisheries and aquaculture 2022 2. Towards Blue Transformation. Rome: FAO; 2022.
Gjedrem T, Baranski M. Selective breeding in aquaculture: an introduction, vol. 10: Springer Science & Business Media; 2010. https://springerlink.fh-diploma.de/book/10.1007/978-90-481-2773-3.
Gjedrem T, Robinson N. Advances by selective breeding for aquatic species: a review. Agric Sci. 2014;5(12):1152.
Verspoor E, Stradmeyer L, Nielsen JL. The Atlantic salmon: genetics, conservation and management. John Wiley & Sons; 2008. https://onlinelibrary.wiley.com/doi/book/10.1002/9780470995846.
Chauhan T, Rajiv K. Molecular markers and their applications in fisheries and aquaculture. Adv Biosci Biotechnol. 2010;1(04):281–91.
Houston RD, Bean TP, Macqueen DJ, Gundappa MK, Jin YH, Jenkins TL, Selly SLC, Martin SA, Stevens JR, Santos EM. Harnessing genomics to fast-track genetic improvement in aquaculture. Nat Rev Genet. 2020;21(7):389–409.
Rasal KD, Chakrapani V, Pandey AK, Rasal AR, Sundaray JK, Ninawe A, Jayasankar P. Status and future perspectives of single nucleotide polymorphisms (SNPs) markers in farmed fishes: way ahead using next generation sequencing. Gene Rep. 2017;6:81–6.
Wenne R. Single nucleotide polymorphism markers with applications in conservation and exploitation of aquatic natural populations. Animals. 2023;13(6):1089.
Sundaray JK, Rasal KD, Chakrapani V, Swain P, Kumar D, Ninawe AS, Nandi S, Jayasankar P. Simple sequence repeats (SSRs) markers in fish genomic research and their acceleration via next-generation sequencing and computational approaches. Aquacult Int. 2016;24:1089–102.
McAndrew B, Napier J. Application of genetics and genomics to aquaculture development: current and future directions. J Agric Sci. 2011;149(S1):143–51.
Abdelrahman H, ElHady M, Alcivar-Warren A, Allen S, Al-Tobasei R, Bao L, Beck B, Blackburn H, Bosworth B, Buchanan J. Aquaculture genomics, genetics and breeding in the United States: current status, challenges, and priorities for future research. BMC Genomics. 2017;18(1):1–23.
Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA, Selker EU, Cresko WA, Johnson EA. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE. 2008;3(10):e3376.
Davey JW, Hohenlohe PA, Etter PD, Boone JQ, Catchen JM, Blaxter ML. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat Rev Genet. 2011;12(7):499–510.
Yue GHJF. Recent advances of genome mapping and marker-assisted selection in aquaculture. Fish Fish. 2014;15(3):376–96.
Das P, Sahoo L, Das SP, Bit A, Joshi CG, Kushwaha B, Kumar D, Shah TM, Hinsu AT, Patel N. De novo assembly and genome-wide SNP discovery in Rohu Carp, Labeo rohita. Front Genet. 2020;11:386.
Nandanpawar P, Sahoo L, Sahoo B, Murmu K, Chaudhari A, Das P. Identification of differentially expressed genes and SNPs linked to harvest body weight of genetically improved rohu carp, Labeo rohita. Front Genet. 2023;14:1153911.
Su J, Shang X, Wan Q, Su J. SNP-based susceptibility–resistance association and mRNA expression regulation analyses of tlr7 to grass carp Ctenopharyngodon idella reovirus. J Fish Biol. 2018;92(5):1505–25.
Flanagan SP, Jones AG. The future of parentage analysis: from microsatellites to SNPs and beyond. Mol Ecol. 2019;28(3):544–67. https://doi.org/10.1111/mec.14988.
Lu G, Luo M. Genomes of major fishes in world fisheries and aquaculture: status, application and perspective. Aquaculture Fisheries. 2020;5(4):163–73.
Bernardi G, Wiley EO, Mansour H, Miller MR, Orti G, Haussler D, O’Brien SJ, Ryder OA, Venkatesh B. The fishes of Genome 10K. Mar Genomics. 2012;7:3–6.
Star B, Nederbragt AJ, Jentoft S, Grimholt U, Malmstrøm M, Gregers TF, Rounge TB, Paulsen J, Solbakken MH, Sharma A. The genome sequence of Atlantic cod reveals a unique immune system. Nature. 2011;477(7363):207–10.
Song H, Dong T, Yan X, Wang W, Tian Z, Sun A, Dong Y, Zhu H, Hu H. Genomic selection and its research progress in aquaculture breeding. Rev Aquac. 2023;15(1):274–91.
Kumar G, Kocour M. Applications of next-generation sequencing in fisheries research: A review. Fish Res. 2017;186:11–22.
Robledo D, Fernandez C, Hermida M, Sciara A, Alvarez-Dios JA, Cabaleiro S, Caamano R, Martinez P, Bouza C. Integrative transcriptome, genome and quantitative trait loci resources identify single nucleotide polymorphisms in candidate genes for growth traits in Turbot. Int J Mol Sci. 2016;17(2):243.
Peñaloza C, Manousaki T, Franch R, Tsakogiannis A, Sonesson AK, Aslam M, Allal F, Bargelloni L, Houston R. Tsigenopoulos: development and testing of a combined species SNP array for the European seabass (Dicentrarchus labrax) and gilthead seabream (Sparus aurata). Genomics. 2021;113(4):2096–107.
Palti Y, Gao G, Liu S, Kent M, Lien S, Miller M, Rexroad C III, Moen T. The development and characterization of a 57 K single nucleotide polymorphism array for rainbow trout. Mol Ecol Resour. 2015;15(3):662–72.
Xu J, Zhao Z, Zhang X, Zheng X, Li J, Jiang Y, Kuang Y, Zhang Y, Feng J, Li C. Development and evaluation of the first high-throughput SNP array for common carp (Cyprinus carpio). BMC Genomics. 2014;15(1):1–10.
Liu S, Sun L, Li Y, Sun F, Jiang Y, Zhang Y, Zhang J, Feng J, Kaltenboeck L, Kucuktas H. Development of the catfish 250K SNP array for genome-wide association studies. BMC Res Notes. 2014;7:1–12.
Houston RD, Taggart JB, Cézard T, Bekaert M, Lowe NR, Downing A, Talbot R, Bishop SC, Archibald AL, Bron JE. Development and validation of a high density SNP genotyping array for Atlantic salmon (Salmo salar). BMC Genomics. 2014;15(1):1–13.
Gutierrez AP, Turner F, Gharbi K, Talbot R, Lowe NR, Peñaloza C, McCullough M, Prodöhl PA, Bean TP, Houston RD. Development of a medium density combined-species SNP array for Pacific and European oysters (Crassostrea gigas and Ostrea edulis). G3. 2017;7(7):2209–18.
Joshi R, Árnyasi M, Lien S, Gjøen HM, Alvarez AT, Kent M. Development and validation of 58K SNP-array and high-density linkage map in Nile tilapia (O. niloticus). Front Genet. 2018;9:472.
Barría A, Christensen KA, Yoshida G, Jedlicki A, Leong JS, Rondeau EB, Lhorente JP, Koop BF, Davidson WS, Yáñez JM. Whole genome linkage disequilibrium and effective population size in a coho salmon (Oncorhynchus kisutch) breeding population using a high-density SNP array. Front Genet. 2019;10:498.
Nugent CM, Leong JS, Christensen KA, Rondeau EB, Brachmann MK, Easton AA, Ouellet-Fagg CL, Crown MT, Davidson WS, Koop BF. Design and characterization of an 87k SNP genotyping array for Arctic charr (Salvelinus alpinus). PLoS ONE. 2019;14(4):e0215008.
Lu S, Zhou Q, Chen Y, Liu Y, Li Y, Wang L, Yang Y, Chen S. Development of a 38 K single nucleotide polymorphism array and application in genomic selection for resistance against Vibrio harveyi in Chinese tongue sole, Cynoglossus semilaevis. Genomics. 2021;113(4):1838–44.
Peñaloza C, Manousaki T, Franch R, Tsakogiannis A, Sonesson A, Aslam ML, Allal F, Bargelloni L, Houston R, Tsigenopoulos C. Development and validation of a combined species SNP array for the European seabass (Dicentrarchus labrax) and gilthead seabream (Sparus aurata). bioRxiv 2020:2020.2012. 2017.423305.
Peñaloza C, Robledo D, Barría A, Trịnh TQ, Mahmuddin M, Wiener P, Benzie JA, Houston RD. Development and validation of an open access SNP array for Nile tilapia (Oreochromis niloticus). G3. 2020;10(8):2777–85.
Mastrochirico-Filho VA, Ariede RB, Freitas MV, Borges CH, Lira LV, Mendes NJ, Agudelo JF, Cáceres P, Berrocal MH, Sucerquia GA. Development of a multi-species SNP array for serrasalmid fish Colossoma macropomum and Piaractus mesopotamicus. Sci Rep. 2021;11(1):19289.
Palombo V, De Zio E, Salvatore G, Esposito S, Iaffaldano N, D’Andrea M. Genotyping of two Mediterranean trout populations in central-southern Italy for conservation purposes using a rainbow-trout-derived SNP array. Animals. 2021;11(6):1803.
García-Ballesteros S, Villanueva B, Fernández J, Gutiérrez JP, Cervantes I. Genetic parameters for uniformity of harvest weight in Pacific white shrimp (Litopenaeus vannamei). Genet Sel Evol. 2021;53(1):1–9.
Lv J, Wang Y, Ni P, Lin P, Hou H, Ding J, Chang Y, Hu J, Wang S, Bao Z. Development of a high-throughput SNP array for sea cucumber (Apostichopus japonicus) and its application in genomic selection with MCP regularized deep neural networks. Genomics. 2022;114(4):110426.
Barría A, Peñaloza C, Papadopoulou A, Mahmuddin M, Doeschl-Wilson A, Benzie JA, Houston RD, Wiener P. Genetic differentiation following recent domestication events: a study of farmed Nile tilapia (Oreochromis niloticus) populations. Evol Appl. 2023;16:1220–35.
Xu S, Wang Y, Gao C, Babu VS, Li J, Liu Q, Xiao Z, Xu Y, Zhao C, Lin L. Effects of dissolved oxygen on intestinal bacterial community and immunity of Atlantic salmon Salmo salar. J Oceanol Limnol. 2023;41(1):364–75.
Zhou Q, Chen YD, Lu S, Liu Y, Xu WT, Li YZ, Wang L, Wang N, Yang YM, Chen SL. Development of a 50K SNP array for Japanese flounder and its application in genomic selection for disease resistance. Engineering. 2021;7(3):406–11.
Wang DG, Fan J-B, Siao C-J, Berno A, Young P, Sapolsky R, Ghandour G, Perkins N, Winchester E, Spencer J. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science. 1998;280(5366):1077–82.
Li Y-H, Wang H-P. Advances of genotyping-by-sequencing in fisheries and aquaculture. Rev Fish Biol Fisheries. 2017;27(3):535–59.
Robledo D, Palaiokostas C, Bargelloni L, Martínez P, Houston R. Applications of genotyping by sequencing in aquaculture breeding and genetics. Rev Aquac. 2018;10(3):670–82.
Andrews KR, Good JM, Miller MR, Luikart G, Hohenlohe PA. Harnessing the power of RADseq for ecological and evolutionary genomics. Nat Rev Genet. 2016;17(2):81–92.
Barría A, Christensen KA, Yoshida GM, Correa K, Jedlicki A, Lhorente JP, Davidson WS, Yáñez JM. Genomic predictions and genome-wide association study of resistance against Piscirickettsia salmonis in coho salmon (Oncorhynchus kisutch) using ddRAD sequencing. G3. 2018;8(4):1183–94.
Liu J, Luo W, Qin N, Ding P, Zhang H, Yang C, Mu Y, Tang H, Liu Y, Li WJT, et al. A 55 K SNP array-based genetic map and its utilization in QTL mapping for productive tiller number in common wheat. Theor Appl Genet. 2018;131:2439–50.
Zhou S, Zhang J, Che Y, Liu W, Lu Y, Yang X, Li X, Jia J, Liu X, Li L. Construction of Agropyron Gaertn. genetic linkage maps using a wheat 660K SNP array reveals a homoeologous relationship with the wheat genome. Plant Biotechnol J. 2018;16(3):818–27.
LaFramboise T. Single nucleotide polymorphism arrays: a decade of biological, computational and technological advances. Nucleic Acids Res. 2009;37(13):4181–93.
Balagué-Dobón L, Cáceres A, González JR. Fully exploiting SNP arrays: a systematic review on the tools to extract underlying genomic structure. Brief Bioinform. 2022;23(2):bbac043.
Sundaray JK, Dixit S, Rather A, Rasal KD, Sahoo L. Aquaculture omics: an update on the current status of research and data analysis. Mar Genomics. 2022;64:100967.
Dennis J, Walker L, Tyrer J, Michailidou K, Easton DF. Detecting rare copy number variants from Illumina genotyping arrays with the CamCNV pipeline: segmentation of z-scores improves detection and reliability. Genet Epidemiol. 2021;45(3):237–48.
Kumasaka N, Fujisawa H, Hosono N, Okada Y, Takahashi A, Nakamura Y, Kubo M, Kamatani N. PlatinumCNV: a Bayesian Gaussian mixture model for genotyping copy number polymorphisms using SNP array signal intensity data. Genet Epidemiol. 2011;35(8):831–44.
Pique-Regi R, Cáceres A, González JR. R-Gada: a fast and flexible pipeline for copy number analysis in association studies. BMC Bioinformatics. 2010;11:1–12.
Dalton-Morgan J, Hayward A, Alamery S, Tollenaere R, Mason AS, Campbell E, Patel D, Lorenc MT, Yi B, Long Y. A high-throughput SNP array in the amphidiploid species Brassica napus shows diversity in resistance genes. Funct Integr Genomics. 2014;14:643–55.
Clevenger J, Chu Y, Chavarro C, Agarwal G, Bertioli DJ, Leal-Bertioli SC, Pandey MK, Vaughn J, Abernathy B, Barkley NA. Genome-wide SNP genotyping resolves signatures of selection and tetrasomic recombination in peanut. Mol Plant. 2017;10(2):309–22.
Deulvot C, Charrel H, Marty A, Jacquin F, Donnadieu C, Lejeune-Hénaut I, Burstin J, Aubert G. Highly-multiplexed SNP genotyping for genetic mapping and germplasm diversity studies in pea. BMC Genomics. 2010;11:1–10.
Chagné D, Crowhurst RN, Troggio M, Davey MW, Gilmore B, Lawley C, Vanderzande S, Hellens RP, Kumar S, Cestaro A. Genome-wide SNP detection, validation, and development of an 8K SNP array for apple. PLoS ONE. 2012;7(2):e31745.
Randhawa SS, Pawar R. Fish genomes: Sequencing trends, taxonomy and influence of taxonomy on genome attributes. J Appl Ichthyol. 2021;37(4):553–62.
Yanez JM, Barria A, Lopez ME, Moen T, Garcia BF, Yoshida GM, Xu P. Genome-wide association and genomic selection in aquaculture. Rev Aquac. 2023;15(2):645–75.
Houston R, Macqueen D. Atlantic salmon (Salmo salar L.) genetics in the 21st century: taking leaps forward in aquaculture and biological understanding. Anim Genet. 2019;50(1):3–14.
Gao G, Waldbieser GC, Youngblood RC, Zhao D, Pietrak M, Allen M, Stannard J, Buchanan J, Long R, Milligan M. The generation of the first chromosome-level de-novo genome assembly and the development and validation of a 50K SNP array for North American Atlantic salmon. bioRxiv 2022:2022.2009. 2028.509896.
Lien S, Gidskehaug L, Moen T, Hayes BJ, Berg PR, Davidson WS, Omholt SW, Kent MP. A dense SNP-based linkage map for Atlantic salmon (Salmo salar) reveals extended chromosome homeologies and striking differences in sex-specific recombination patterns. BMC Genomics. 2011;12(1):1–10.
Gutierrez AP, Yáñez JM, Fukui S, Swift B, Davidson WS. Genome-wide association study (GWAS) for growth rate and age at sexual maturation in Atlantic salmon (Salmo salar). PLoS ONE. 2015;10(3):e0119730.
Yáñez JM, Naswa S, López M, Bassini L, Correa K, Gilbey J, Bernatchez L, Norris A, Neira R, Lhorente J. Genomewide single nucleotide polymorphism discovery in Atlantic salmon (Salmo salar): validation in wild and farmed American and European populations. Mol Ecol Resour. 2016;16(4):1002–11.
Holborn MK, Ang KP, Elliott J, Powell F, Boulding EG. Genome wide association analysis for bacterial kidney disease resistance in a commercial North American Atlantic salmon (Salmo salar) population using a 50 K SNP panel. Aquaculture. 2018;495:465–71.
Gabián M, Morán P, Fernández AI, Villanueva B, Chtioui A, Kent MP, Covelo-Soto L, Fernández A, Saura M. Identification of genomic regions regulating sex determination in Atlantic salmon using high density SNP data. BMC Genomics. 2019;20:1–12.
Wellband K, Mérot C, Linnansaari T, Elliott J, Curry RA, Bernatchez L. Chromosomal fusion and life history-associated genomic variation contribute to within-river local adaptation of Atlantic salmon. Mol Ecol. 2019;28(6):1439–59.
Horn SS, Ruyter B, Meuwissen TH, Moghadam H, Hillestad B, Sonesson AK. GWAS identifies genetic variants associated with omega-3 fatty acid composition of Atlantic salmon fillets. Aquaculture. 2020;514:734494.
Barson NJ, Aykanat T, Hindar K, Baranski M, Bolstad GH, Fiske P, Jacq C, Jensen AJ, Johnston SE, Karlsson S. Sex-dependent dominance at a single locus maintains variation in age at maturity in salmon. Nature. 2015;528(7582):405–8.
Moen T, Torgersen J, Santi N, Davidson WS, Baranski M, Ødegård J, Kjøglum S, Velle B, Kent M, Lubieniecki KP. Epithelial cadherin determines resistance to infectious pancreatic necrosis virus in Atlantic salmon. Genet Sel Evol. 2015;200(4):1313–26.
Tsai H-Y, Matika O, Edwards SM, Antolín-Sánchez R, Hamilton A, Guy DR, Tinch AE, Gharbi K, Stear MJ, Taggart JB. Genotype imputation to improve the cost-efficiency of genomic selection in farmed Atlantic salmon. G3. 2017;7(4):1377–83.
Holborn MK, Rochus CM, Ang KP, Elliott J, Leadbeater S, Powell F, Boulding EG. Family-based genome wide association analysis for salmon lice (Lepeophtheirus salmonis) resistance in North American Atlantic salmon using a 50 K SNP array. Aquaculture. 2019;511:734215.
Sinclair-Waters M, Ødegård J, Korsvoll SA, Moen T, Lien S, Primmer CR, Barson NJ. Beyond large-effect loci: large-scale GWAS reveals a mixed large-effect and polygenic architecture for age at maturity of Atlantic salmon. Genet Sel Evol. 2020;52(1):1–11.
Chen L, Xu J, Sun X, Xu P. Research advances and future perspectives of genomics and genetic improvement in allotetraploid common carp. Rev Aquac. 2022;14(2):957–78.
Peng W, Xu J, Zhang Y, Feng J, Dong C, Jiang L, Feng J, Chen B, Gong Y, Chen L. An ultra-high density linkage map and QTL mapping for sex and growth-related traits of common carp (Cyprinus carpio). Sci Rep. 2016;6(1):26693.
Zhang X, Luan P, Cao D, Hu G. A high-density genetic linkage map and fine mapping of QTL for feed conversion efficiency in common carp (Cyprinus carpio). Front Genet. 2021;12:778487.
Xu J, Feng J, Peng W, Liu X, Feng J, Xu P. Development and evaluation of a high-throughput single nucleotide polymorphism multiplex assay for assigning pedigrees in common carp. Aquac Res. 2017;48(4):1866–76.
Zhang H, Xu P, Jiang Y, Zhao Z, Feng J, Tai R, Dong C, Xu J. Genomic, transcriptomic, and epigenomic features differentiate genes that are relevant for muscular polyunsaturated fatty acids in the common carp. Front Genet. 2019;10:217.
Xu J, Jiang Y, Zhao Z, Zhang H, Peng W, Feng J, Dong C, Chen B, Tai R, Xu P. Patterns of geographical and potential adaptive divergence in the genome of the common carp (Cyprinus carpio). Front Genet. 2019;10:660.
Zhou Z, Chen L, Dong C, Peng W, Kong S, Sun J, Pu F, Chen B, Feng J, Xu P. Genome-scale association study of abnormal scale pattern in Yellow River carp identified previously known causative gene in European Mirror Carp. Mar Biotechnol. 2018;20:573–83.
Zheng X, Kuang Y, Lv W, Cao D, Sun Z, Sun X. Genome-wide association study for muscle fat content and abdominal fat traits in common carp (Cyprinus carpio). PLoS ONE. 2016;11(12):e0169127.
Chen L, Peng W, Kong S, Pu F, Chen B, Zhou Z, Feng J, Li X, Xu P. Genetic mapping of head size related traits in common carp (Cyprinus carpio). Front Genet. 2018;9:448.
Jiang Y, Li B, Yu M, Chang S, Li S, Xu J, Feng J, Zhang Q, Zhang H, Xu P. Genome-wide association study and gene editing reveals the causal gene responsible for abnormal red skin color in Yellow River carp. Aquaculture. 2022;560:738530.
Jia Z, Chen L, Ge Y, Li S, Peng W, Li C, Zhang Y, Hu X, Zhou Z, Shi L. Genetic mapping of koi herpesvirus resistance (KHVR) in Mirror carp (Cyprinus carpio) revealed genes and molecular mechanisms of disease resistance. Aquaculture. 2020;519:734850.
Zhang H, Zhu Y, Xu P, Zhao Z, Feng J, Wu B, Jiang Y, Dong C, Xu J. Multi-omics data reveal amino acids related genes in the Common Carp Cyprinus Carpio. Fishes. 2022;7(5):225.
Zhang H-Y, Zhao Z-X, Xu J, Xu P, Bai Q-L, Yang S-Y, Jiang L-K, Chen B-H. Population genetic analysis of aquaculture salmonid populations in China using a 57K rainbow trout SNP array. PLoS ONE. 2018;13(8):e0202582.
Vallejo RL, Leeds TD, Gao G, Parsons JE, Martin KE, Evenhuis JP, Fragomeni BO, Wiens GD, Palti Y. Genomic selection models double the accuracy of predicted breeding values for bacterial cold water disease resistance compared to a traditional pedigree-based model in rainbow trout aquaculture. Genet Sel Evol. 2017;49(1):1–13.
Gonzalez-Pena D, Gao G, Baranski M, Moen T, Cleveland BM, Kenney PB, Vallejo RL, Palti Y, Leeds TD. Genome-wide association study for identifying loci that affect fillet yield, carcass, and body weight traits in rainbow trout (Oncorhynchus mykiss). Front Genet. 2016;7:203.
Liu S, Palti Y, Martin KE, Parsons JE, Rexroad CE III. Assessment of genetic differentiation and genetic assignment of commercial rainbow trout strains using a SNP panel. Aquaculture. 2017;468:120–5.
D’Agaro E, Favaro A, Matiussi S, Gibertoni PP, Esposito S. Genomic selection in salmonids: new discoveries and future perspectives. Aquacult Int. 2021;29(5):2259–89.
Liu S, Martin KE, Gao G, Long R, Evenhuis JP, Leeds TD, Wiens GD, Palti Y. Identification of haplotypes associated with resistance to bacterial cold water disease in rainbow trout using whole-genome resequencing. Front Genet. 2022;13:936806.
Vallejo RL, Liu S, Gao G, Fragomeni BO, Hernandez AG, Leeds TD, Parsons JE, Martin KE, Evenhuis JP, Welch TJ, et al. Similar genetic architecture with shared and unique quantitative trait loci for bacterial cold water disease resistance in two rainbow trout breeding populations. Front Genet. 2017;8:156.
Bernard M, Dehaullon A, Gao G, Paul K, Lagarde H, Charles M, Prchal M, Danon J, Jaffrelo L, Poncet C, et al. Development of a high-density 665 K SNP array for rainbow trout genome-wide genotyping. Front Genet. 2022;13:941340.
Wenne R, Bernaś R, Kijewska A, Poćwierz-Kotus A, Strand J, Petereit C, Plauška K, Sics I, Árnyasi M, Kent MP. SNP genotyping reveals substructuring in weakly differentiated populations of Atlantic cod (Gadus morhua) from diverse environments in the Baltic Sea. Sci Rep. 2020;10(1):9738.
Johansen T, Besnier F, Quintela M, Jorde PE, Glover KA, Westgaard JI, Dahle G, Lien S, Kent MP. Genomic analysis reveals neutral and adaptive patterns that challenge the current management regime for East Atlantic cod Gadus morhua. Evol Appl. 2020;13(10):2673–88.
Hubert S, Higgins B, Borza T, Bowman S. Development of a SNP resource and a genetic linkage map for Atlantic cod (Gadus morhua). BMC Genomics. 2010;11(1):1–14.
Chen X, Zhong L, Bian C, Xu P, Qiu Y, You X, Zhang S, Huang Y, Li J, Wang M. High-quality genome assembly of channel catfish, Ictalurus punctatus. Gigascience. 2016;5(1):s13742-13016-10142–3745.
Liu S, Li Y, Qin Z, Geng X, Bao L, Kaltenboeck L, Kucuktas H, Dunham R, Liu Z. High-density interspecific genetic linkage mapping provides insights into genomic incompatibility between channel catfish and blue catfish. Anim Genet. 2016;47(1):81–90.
Jin Y, Zhou T, Geng X, Liu S, Chen A, Yao J, Jiang C, Tan S, Su B, Liu Z. A genome-wide association study of heat stress-associated SNP s in catfish. Anim Genet. 2017;48(2):233–6.
Li N, Zhou T, Geng X, Jin Y, Wang X, Liu S, Xu X, Gao D, Li Q, Liu Z. Identification of novel genes significantly affecting growth in catfish through GWAS analysis. Molecular Genetics, Genomics. 2018;293:587–99.
Zeng Q, Fu Q, Li Y, Waldbieser G, Bosworth B, Liu S, Yang Y, Bao L, Yuan Z, Li N. Development of a 690 K SNP array in catfish and its application for genetic mapping and validation of the reference genome sequence. Sci Rep. 2017;7(1):40347.
Garcia AL, Bosworth B, Waldbieser G, Misztal I, Tsuruta S, Lourenco DA. Development of genomic predictions for harvest and carcass weight in channel catfish. Genet Sel Evol. 2018;50:1–12.
Zhong X, Wang X, Zhou T, Jin Y, Tan S, Jiang C, Geng X, Li N, Shi H, Zeng Q. Genome-wide association study reveals multiple novel QTL associated with low oxygen tolerance in hybrid catfish. Mar Biotechnol. 2017;19:379–90.
Shao C, Bao B, Xie Z, Chen X, Li B, Jia X, Yao Q, Orti G, Li W, Li X. The genome and transcriptome of Japanese flounder provide insights into flatfish asymmetry. Nat Genet. 2017;49(1):119–24.
Barría A, Trịnh TQ, Mahmuddin M, Peñaloza C, Papadopoulou A, Gervais O, Chadag VM, Benzie JA, Houston RD. A major quantitative trait locus affecting resistance to Tilapia lake virus in farmed Nile tilapia (Oreochromis niloticus). Heredity. 2021;127(3):334–43.
Barría A, Benzie JA, Houston RD, De Koning D-J, De Verdal H. Genomic selection and genome-wide association study for feed-efficiency traits in a farmed Nile Tilapia (Oreochromis niloticus) population. Front Genet. 2021;12:737906.
Tine M, Kuhl H, Gagnaire P-A, Louro B, Desmarais E, Martins RS, Hecht J, Knaust F, Belkhir K, Klages S. European sea bass genome and its variation provide insights into adaptation to euryhalinity and speciation. Nat Commun. 2014;5(1):5770.
Palaiokostas C, Bekaert M, Taggart JB, Gharbi K, McAndrew BJ, Chatain B, Penman DJ, Vandeputte M. A new SNP-based vision of the genetics of sex determination in European sea bass (Dicentrarchus labrax). Genet Sel Evol. 2015;47(1):1–10.
Duranton M, Allal F, Fraïsse C, Bierne N, Bonhomme F, Gagnaire P-A. The origin and remolding of genomic islands of differentiation in the European sea bass. Nat Commun. 2018;9(1):2518.
Faggion S, Vandeputte M, Chatain B, Gagnaire P-A, Allal F. Population-specific variations of the genetic architecture of sex determination in wild European sea bass Dicentrarchus labrax L. Heredity. 2019;122(5):612–21.
Griot R, Allal F, Phocas F, Brard-Fudulea S, Morvezen R, Bestin A, Haffray P, François Y, Morin T, Poncet C. Genome-wide association studies for resistance to viral nervous necrosis in three populations of European sea bass (Dicentrarchus labrax) using a novel 57k SNP array DlabChip. Aquaculture. 2021;530:735930.
Oikonomou S, Samaras A, Tekeoglou M, Loukovitis D, Dimitroglou A, Kottaras L, Papanna K, Papaharisis L, Tsigenopoulos CS, Pavlidis M. Genomic selection and genome-wide association analysis for stress response, disease resistance and body weight in European seabass. Animals. 2022;12(3):277.
Qi H, Song K, Li C, Wang W, Li B, Li L, Zhang G. Construction and evaluation of a high-density SNP array for the Pacific oyster (Crassostrea gigas). PLoS ONE. 2017;12(3):e0174007.
Gutierrez AP, Matika O, Bean TP, Houston RD. Genomic selection for growth traits in Pacific oyster (Crassostrea gigas): potential of low-density marker panels for breeding value prediction. Front Genet. 2018;9:391.
Peñaloza C, Barria A, Papadopoulou A, Hooper C, Preston J, Green M, Helmer L, Kean-Hammerson J, Nascimento-Schulze JC, Minardi D. Genome-wide association and genomic prediction of growth traits in the European flat oyster (Ostrea edulis). Front Genet. 2022;13:926638.
Zhou T, Chen B, Ke Q, Zhao J, Pu F, Wu Y, Chen L, Zhou Z, Bai Y, Pan Y. Development and evaluation of a high-throughput single-nucleotide polymorphism array for large yellow croaker (Larimichthys crocea). Front Genet. 2020;11:571751.
Acknowledgements
The authors are thankful to the Director, Indian Council of Agricultural Research—CIFE, Mumbai, Maharashtra, India and the Director, Indian Council of Agricultural Research—CIFA, Bhubaneswar, Odhisa, India.
Funding
No funding is available.
Author information
Authors and Affiliations
Contributions
Conceptualization: Naresh Nagpure, Kiran D. Rasal; Methodology: Kiran D. Rasal, Manoj Brahmane; Investigation: Kiran D. Rasal, Pokanti Vinay Kumar; Writing—original draft preparation: Kiran D. Rasal, Pokanti Vinay Kumar, Prachi Asgolkar, Siyag Dhere Prabhugouda Siriyappagouder; Writing—review and editing: Kiran D. Rasal, Pokanti Vinay Kumar, Prachi Asgolkar, Prabhugouda Siriyappagouder, Siba Shinde, Siyag Dhere, Naresh Nagpure; Resources: Jitendra Kumar Sundaray, Kiran D. Rasal, Manoj Brahmane, Naresh Nagpure; Supervision: Naresh Nagpure.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors have given their consent for publication.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Rasal, K.D., Kumar, P.V., Asgolkar, P. et al. Single-Nucleotide Polymorphism (SNP) array: an array of hope for genetic improvement of aquatic species and fisheries management. Blue Biotechnology 1, 3 (2024). https://doi.org/10.1186/s44315-024-00004-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s44315-024-00004-8