Abstract
Background
The importance of identifying the pathogenic fungi rapidly has encouraged the development of differential media for the presumptive identification of yeasts. In this study two differential media, CHROMagar Candida and bismuth sulphite glucose glycine yeast agar, were evaluated for the presumptive identification of yeast species.
Methods
A total number of 270 yeast strains including 169 Candida albicans, 33 C. tropicalis, 24 C. glabrata, 18 C. parapsilosis, 12 C. krusei, 5 Trichosporon spp., 4 C. kefyr, 2 C. lusitaniae, 1 Saccharomyces cerevisiae and 1 Geotrichum candidum were included. The strains were first identified by germ tube test, morphological characteristics on cornmeal tween 80 agar and Vitek 32 and API 20 C AUX systems. In parallel, they were also streaked onto CHROMagar Candida and bismuth sulphite glucose glycine yeast agar plates. The results were read according to the color, morphology of the colonies and the existance of halo around them after 48 hours of incubation at 37°C.
Results
The sensitivity and specificity values for C. albicans strains were found to be 99.4, 100% for CHROMagar Candida and 87.0, 75.2% for BiGGY agar, respectively. The sensitivity of CHROMagar Candida to identify C. tropicalis, C. glabrata and C. krusei ranged between 90.9 and 100% while the specificity was 100%. The sensitivity rates for BiGGY agar were 66.6 and 100% while the specificity values were found to be 95.4 and 100% for C. tropicalis and C. krusei, respectively.
Conclusions
It can be concluded that the use of CHROMagar Candida is an easy and reliable method for the presumptive identification of most commonly isolated Candida species especially C. albicans, C. tropicalis and C. krusei. The lower sensitivity and specificity of BiGGY agar to identify commonly isolated Candida species potentially limits the clinical usefulness of this agar.
Similar content being viewed by others
Background
The incidence of fungal infections is increasing because of a rising number of immunocompromised patients, widespread use of broad-spectrum antibiotics and invasive devices or procedures [1–3]. Although Candida albicans remains the most frequently isolated yeast pathogen, other Candida species such as C. glabrata, C. krusei, and C. tropicalis are emerging as opportunistic pathogens and these species are reported to be less susceptible than C. albicans to antifungal agents [4–6]. C. parapsilosis has become the second most frequently recovered Candida species from blood cultures in Europe, Canada, and Latin America and ranked third in the United States [7, 8]. For these reasons, yeast infections require rapid diagnosis and early adapted antifungal therapy.
Ideally, laboratories should be able to detect and identify the major Candida species in clinical specimens [9]. Most laboratories start the yeast identification process with the germ tube test and continue with more extensive testing. Reference identification procedures that use biochemical and morphological studies are not often practical for the clinical laboratory because they are labor intensive and take several days. The conventional methods of yeast identification which mainly consist of assimilation/fermentation characteristics are reported to be cumbersome and often beyond the expertise range in non-specialized clinical microbiology laboratories [10]. Packaged kit systems and automated systems are widely used, but they are expensive and are limited by the size of their databases [11].
The importance of identifying the pathogen as quickly as possible has encouraged the development of differential media for the presumptive identification of yeasts. Several chromogenic media for isolation and identification of Candida species are available [12]. These media are based on the formation of various colored colonies with different morphology which result from the cleavage of chromogenic substrates by species specific enzymes [13]. In this study, the performance of two differential media, CHROMagar Candida (CA) and bismuth sulphite glucose glycine yeast (BiGGY) agar were evaluated for the identification of yeast species in comparison with the standard methods.
Methods
Isolates
A total number of 270 yeast strains including 169 Candida albicans, 33 C. tropicalis, 24 C. glabrata, 18 C. parapsilosis, 12 C. krusei, 5 Trichosporon spp. 4 C. kefyr, 2 C. lusitaniae, 1 C. guilliermondii, 1 Geotrichum candidum, and 1 Saccharomyces cerevisiae isolated from various clinical specimens (98 urine, 82 tongue-oral swabs, 47 blood, 18 sputum and bronchoalveoler lavage, 12 wound and abscess aspirate, 5 catheter, 4 vaginal secretion, 3 peritoneal aspirate and 1 cerebrospinal fluid) were used in this study. Quality control strains were C. albicans ATCC 90028, C. parapsilosis ATCC 90018, and C. krusei ATCC 6258. The strains were first identified by germ tube test, morphological characteristics on cornmeal tween 80 agar, then by Vitek 32 yeast identification (bioMérieux, France) and API 20 C AUX (bioMérieux, France) systems.
Culture Media
CA (CHROMagar Company, France) and BiGGY agar (Nickersen agar) (Oxoid Company, England) were purchased as powdered media. The plates were prepared according to the manufacturers' instructions. Briefly, 47.7 grams of dehydrated CA and 42 grams of dehydrated BiGGY agar were reconstituted in 1 litre of distilled water. They were brought to boil by repeated heatings and then cooled in a water bath at 45°C by swirling and stirring. Then 20 ml's of the media were dispensed into petri dishes. They were kept in the dark at 4°C and used within three days.
CHROMagar Candida contains peptone (10.2 g/L), chloramphenicol (0.5 g/L) and chromogenic mix (22.0 g/L), agar (15.0 g/L), pH: 6.1. The composition of BiGGY agar is yeast extract (1.0 g/L), glycine (10.0 g/L), glucose (10.0 g/L), agar (13.0 g/L), sodium sulphite (3.0 g/L), bismuth ammonium citrate (5.0 g/L), pH: 6.8 ± 0.2.
C. albicans, C. tropicalis and C. krusei were reported to be identifiable on CA by the manufacturer and Pfaller et al. [9] proposed that CA could also identify C. glabrata and described the characteristic appearance. C. albicans, C. tropicalis, C. krusei and C. kefyr were reported to be identifiable on BiGGY agar by the manufacturer.
Method
Isolates were subcultured twice on Sabouraud dextrose agar prior to inoculation of chromogenic media. A single yeast colony was streaked onto the plates to give isolated colonies. The plates were incubated at 37°C without CO2 in the dark. The results were read by two different people according to the color, morphology of colonies and the existence of halo around them after 48 hours. They were identified according to the manufacturers' instructions and as described by Odds and Bernaerts [14].
Statistical Analysis
The sensitivity, specificity, positive and negative predictive values (PPV, NPV) of two media for the species, which has been reported to be identifiable, were calculated by using Epi Info version 6.0 [15].
Results
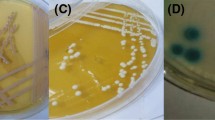
All the strains did grow and the properties of the colonies were clearly distinguished after 48 hours of incubation. The distribution of the colony colors on CA and BiGGY agar within each yeast species is listed in Table 1. The colonies of various Candida species on CA and BiGGY agar are shown in figure 1 and 2, respectively.
The sensitivity and specificity of CA for C. albicans strains were found to be 99.4 and 100%. These rates were detected as 87.0 and 75.2% for BiGGY agar, respectively. The sensitivity of CHROMagar Candida to identify C. tropicalis, C. glabrata and C. krusei ranged between 90.9 and 100% while the specificity was 100% (Table 2). The sensitivity and specificity of BIGGY agar obtained for C. tropicalis and C. krusei are shown in Table 3. These rates for C. glabrata are not calculated because no reference color and morphology is described for this species by the manufacturer.
Discussion
CA and BiGGY agar are two chromogenic media which allow the presumptive differentiation of yeasts. CA contains various substrates for the enzymes of yeast species. It has been demonstrated that β-N-acetylgalactosaminidase which was produced by C. albicans enables the chromogenic substrates to be incorporated into the medium and the isolates of these species were seen as green colored colonies [16]. BiGGY agar contains bismuth sulphite and the growth on this medium produces brown to black colonies because of the extracellular reduction of bismuth sulphite to bismuth sulphide.
CA is reported to give green colonies of C. albicans and blue colonies of C. tropicalis. In this study 168 of 169 C. albicans isolates grew as distinctive light green colonies on CA, only one isolate was seen as off white colonies. Although we didn't have any C. dubliniensis isolates, differentiation of this species from C. albican s seems to be a problem in CA. Tintelnot et al [17] and Willinger et al [18] reported that some of C. dubliniensis isolates yielded a dark green color. It has been suggested that dark bluish-green coloration might be taken as an indication of the presence of C. dubliniensis but could not be used as a criteria for identification. Jabra-Rizk et al [18] compared the results of original CA and newly reformulated CA for these two species. They concluded that the difference in colony color between C. albicans and C. dubliniensis was slightly more enchanced on the new CA. They, in accordance with Pfaller et al. [9] suggested that the dark green color was detected to be more pronounced if the plates were incubated longer than 48 h.
We obtained the sensitivity and specificity of CA for C. albicans as 99.4 and 100%. Our results are in parallel with various studies at which these values were found to be 97–100% and 100% [14, 20–25].
Thirty two of 33 C. tropicalis strains produced blue-violet, smooth (S type) colonies with halo diffusing into the surrounding agar on CA. One strain was observed as dark pink colonies. It has been reported that some strains of Pichia spp. gave an identical appearance [14]. However; San-Millan et al. [23] emphasized that they didn't observe this appearance in their study. The same authors also found false positive results due to S. cerevisiae isolates. This may be true due to the fact that dark pink colonies with halo may resemble C. tropicalis colonies on CA. We had only one S. cerevisiae isolate and its color was somewhat different from blue-purple color of C. tropicalis. The sensitivity and specificity calculated for C. tropicalis was 97 and 100%. The results of this study are in agreement with those described by many authors whose sensitivity and specificity rates ranged between 66.7–99.0% and 93.8–100%, respectively [14, 20, 22–24].
Rapid identification of C. krusei and C. glabrata isolates with chromogenic media has a special importance because C. glabrata is less sensitive than other species to ketoconazole and fluconazole and C. krusei exhibits innate resistance to fluconazole [26]. All our C. krusei isolates produced rough, fuzzy, spreading, big pink colonies on CA. Cooke et al. [27] reported that the usefulness of colony color in identification of C. krusei appeared to be limited as several other yeasts gave pink and purple colonies on this medium. Our findings are in contrast with the authors' in the fact that although there are many yeast species giving the same colony color, the morphology of the colonies of C. krusei are distinctly different (fuzzy, rough, large, pink). Odds and Bernaerts [14]. proposed that C. krusei like colonies are also formed by isolates of C. norvegensis. This is an important point to remember however, this species is very rarely encountered in clinical specimens. The sensitivity and specificity we obtained for this species was 100%. These values are concordant with the reults of many studies [14, 22, 23].
In this study, twenty of C. glabrata isolates produced pink, glossy colonies with pale edges but 4 of them were grown as off white-cream colonies. Pfaller et al [9] and Willinger et al [24] concluded that CA also allowed the identification of C. glabrata however other authors showed that many other species such as C. kefyr, C. lusitaniae, C. guilliermondii, C. famata, C. rugosa, C. utilis, Cryptococcus neoformans, S. cerevisiae produced similar colonies which might lead to a high degree of confusion [11, 14, 16, 28]. In our study many other species also produced the same color with C. glabrata which is very confusing, however, it would be more helpful if special attention is given to the existence of pale edges on the pink colonies. Most of the C. glabrata strains showed this property in our study.
The isolates of C. parapsilosis, C. guilliermondii, C. kefyr and C. lusitaniae had various tones from off white to pink on CA. These colors were difficult to define because they were similar pastel tones and they were not typical for any species.
Most of the C. albicans strains showed light brown while 13 and 20 of C. tropicalis isolates produced light and dark brown color when grown on BiGGY agar. It was hard to differentiate the colors of these two species. In our study two species showed typical, distinctive appearance on BiGGY agar. One was C. krusei which produced typical large, rough, dark brown colonies with surrounding yellow zone and the other was C. parapsilosis which grew as light brown-geenish, gray, cream colored colonies. C. glabrata strains grew weakly on BiGGY agar after 48 hours. Brown color was only observed at the first streaks of the cultures, where the colonies were very crowded. Other areas especially single colonies were very small and colorless. The sensitivity and specificity were detected as 87.0 and 75.2% for C. albicans, 66.6 and 95.4% for C. tropicalis and 100% for C. krusei. To our knowledge, a study related to the efficacy of BiGGY agar was not reported in the literature.
Odds and Bernaerts [14] emphasized that these chromogenic media were not proposed as substitutes for thorough identification protocols. They can be used alone or with some other identification procedures for rapid identification and diagnosis in Clinical Microbiology laboratories. When the current price of both media were compared, the estimated cost of one liter of CA medium was calculated to be 7.7 times of the same amount of BiGGY agar. This difference may be offset by further identification costs for BiGGY agar.
Conclusions
As a result, it can be concluded that the use of CA is an easy and reliable method for the presumptive identification of most commonly isolated Candida species especially C. albicans, C. tropicalis and C. krusei. The lower sensitivity and specificity of BiGGY agar to identify commonly isolated Candida species potentially limits the clinical usefulness of this agar.
References
Fraser VJ, Jones M, Dunkel J, Storfer S, Medoff G, Dunagan WC: Candidemia in a tertiary care hospital: epidemiology, risk factors, and predictors of mortality. Clin Infect Dis. 1992, 15: 415-421.
Moran GP, Sullivan DJ, Coleman DC: Emergence of non-Candida albicans species as pathogens. In Candida and Candidiasis. Edited by: Calderone RA. 2003, 37-53. Washington D.C.: American Society for Microbiology, 1,
Jarwis WR: Epidemiology of nosocomial fungal infections, with emphasis on Candida species. Clin Infect Dis. 1995, 20: 1526-1530.
Abi-Said D, Anaissie E, Uzun O, Raad F, Pinzcowski H, Vartivarian S: The epidemiology of hematogenous candidiasis caused by different Candida species. Clin Infect Dis. 1997, 24: 1122-1128.
Rex JH, Pfaller MA, Barry AL, Nelson PW, C.D. Webb or the NIAID Mycoses Study Group and the Candidemia Study Group : Antifungal susceptibility testing of isolates from a randomized, multicenter trial of fluconazole versus amphotericin B for the treatment of nonneutropenic patients with candidemia. Antimicrob Agents Chemother. 1995, 39: 40-44.
Wingard JR: Importance of Candida species other than C. albicans as pathogens in oncology patients. Clin Infect Dis. 1995, 20: 115-125.
Pfaller MA, Jones RN, Doern GV, Sader HS, Messer SA, Houston A, Coffman S, Hollis RJ: Bloodstream infections due to Candida species: SENTRY antimicrobial surveillance program in North America and Latin America, 1997–1998. Antimicrob Agents Chemother. 2000, 44: 747-751. 10.1128/AAC.44.3.747-751.2000
Pfaller MA, Messer SA, Hollis RJ, Jones RN, Doern GV, Branddt ME, Hajjeh RA: Trends in species distribution and susceptibility to fluconazole among blood stream isolates of Candida species in the United States. Diagn Microbiol Infect Dis. 1999, 33: 217-222. 10.1016/S0732-8893(98)00160-6
Pfaller MA, Houston A, Coffmann S: Application of CHROMagar Candida for rapid screening of clinical specimens for Candida albicans, Candida tropicalis, Candida krusei, and Candida (Torulopsis) glabrata. J Clin Microbiol. 1996, 34: 58-61.
Freydiere AM, Guinet R, Boiron P: Yeast identification in the clinical microbiology laboratory: phenotypical methods. Medical Mycology. 2001, 39: 9-33.
Koehler AP, Chu KC, Houang ETS, Cheng AFB: Simple, reliable, and cost-effective yeast identification scheme for the clinical laboratory. J Clin Microbiol. 1999, 37: 422-426.
Letscher-Bru V, Meyer MH, Galoisy AC, Waller J, Candolfi E: Prospective evaluation of the new chromogenic medium Candida ID, in comparison with Candiselect, for isolation of molds and isolation and presumptive identification of yeast species. J Clin Microbiol. 2002, 40: 1508-1510. 10.1128/JCM.40.4.1508-1510.2002
Bauters TG, Nelis HJ: Comparison of chromogenic and fluorogenic membrane filtration methods for detection of four Candida species. J Clin Microbiol. 2002, 40: 1838-1839. 10.1128/JCM.40.5.1838-1839.2002
Odds FC, Bernaerts R: CHROMagar Candida, a new differential isolation medium for presumptive identification of clinically important Candida species. J Clin Microbiol. 1994, 32: 1923-1929.
Dean AG, Dean JA, Coulombier D, Brendel KA, Smith DC, Burton AH, Dicher RC, Sullivan K, Fagan RF, Arner TG: Epi Info, version 6.0: a word processing database, and statistics program epidemiology on microcomputers. Centers for Disease Control and Prevention. Atlanta, Georgia. U.S.A. 1994,
Beighton D, Ludford R, Clark DT, Brailsford SR, Pankhurst CL, Tinsley GF, Fiske J, Lewis D, Daly B, Khalifa N, Marren V, Lynch E: Use of CHROMagar Candida medium for isolation of yeasts from dental samples. J Clin Microbiol. 1995, 33: 3025-3027.
Tintelnot K, Haase G, Seibold M, Bergmann F, Staemmler M, Franz T, Naumann D: Evaluation of phenotypic markers for selection and identification of Candida dubliniensis. J Clin Microbiol. 2000, 38: 1599-1608.
Willinger B, Hillowoth C, Selitsch B, Manafi M: Performance of Candida ID, a new chromogenic medium for presumptive identification of Candida species, in comparison to CHROMagar Candida. J Clin Microbiol. 2001, 39: 3793-3795. 10.1128/JCM.39.10.3793-3795.2001
Jabra-Rizk MA, Brenner TM, Romagnoli M, Baqui AAMA, Merz WG, Falkler WA Jr, Meiller TF: Evaluation of a reformulated CHROMagar Candida. J Clin Microbiol. 2001, 39: 2015-2016. 10.1128/JCM.39.5.2015-2016.2001
Yücesoy M, Ergor G, Yulug N: Evaluation of CHROMagar Candida medium for the identification of Candida. Mikrobiyol Bult. 2001, 35: 549-557.
Ainscough S, Kibbler CC: An evaluation of the cost-effectiveness of using CHROMagar for yeast identification in a routine microbiology laboratory. J Med Microbiol. 1998, 47: 623-628.
Bernal S, Martin Mazuelos E, Garcia M, Aller AI, Martinez MA, Gutierrez MJ: Evaluation of CHROMagar Candida medium for the isolation and presumptive identification of species of Candida of clinical importance. Diagn Microbiol Infect Dis. 1996, 24: 201-204. 10.1016/0732-8893(96)00063-6
San-Millan R, Ribacoba L, Ponton J, Quindos G: Evaluation of a commercial medium for identification of Candida species. Eur J Clin Microbiol Infect Dis. 1996, 15: 153-158.
Willinger B, Manafi M: Evaluation of CHROMagar Candida for rapid screening of clinical specimens for Candida species. Mycoses. 1999, 42: 61-65. 10.1046/j.1439-0507.1999.00406.x
Hoppe JE, Frey P: Evaluation of six commercial tests and the germ tube test for presumptive identification of Candida albicans. Eur J Clin Microbiol Infect Dis. 1999, 18: 188-191. 10.1007/s100960050256
Bouchara JP, Declerck P, Cimon B, Planchenault C, De Gentile L, Chabasse D: Routine use of CHROMagar Candida medium for presumptive identification of Candida yeast species and detection of mixed fungal populations. Clin Microbiol Infect. 1996, 2: 202-208.
Cooke VM, Miles RJ, Price RG, Midgley G, Khamri W, Richardson AC: New chromogenic agar medium for the identification of Candida spp. Appl Environ Microbiol. 2002, 68: 3622-3627. 10.1128/AEM.68.7.3622-3627.2002
Freydiére AM: Evaluation of CHROMagar Candida plates. J Clin Microbiol. 1996, 34: 20-48.
Author information
Authors and Affiliations
Corresponding author
Additional information
Author's Contributions
M.Y: Designed and coordinated the study, drafted the manuscript, did the laboratory work and evaluated the results with SM.
S.M: Did the laboratory work and evaluated the results with MY.
All authors read and agreed to its content. The paper has not already have been published in a journal and is not not under consideration by any other journal.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Yücesoy, M., Marol, S. Performance of CHROMAGAR candida and BIGGY agar for identification of yeast species. Ann Clin Microbiol Antimicrob 2, 8 (2003). https://doi.org/10.1186/1476-0711-2-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1476-0711-2-8