Abstract
The study was conducted to evaluate the effects of different starch sources on Bacillus spp. in intestinal tract and expression of intestinal development related genes of weanling piglets. Twenty-eight PIC male piglets were divided into four homogeneous groups according to initial body weight (similar birth and parity, weaned at 21 ± 1.5 days). Diets for the four treatments consisted of corn starch, wheat starch, tapioca starch and pea starch with the determined ratio for amylose to amylopectin of 0.21, 0.24, 0.12 and 0.52 respectively. Real-time quantitative polymerase chain reaction was applied to: (1) detect genomic DNA of Bacillus and to quantify the number of Bacillus in the intestinal tract chyme of piglets with the primers and probe which designed based on the 16S rRNA sequences of maximum species of Bacillus on GenBank; (2) measure the mRNA level of glucagon-like peptide 2 (GLP-2), insulin-like growth factors 1 (IGF-1) and epidermal growth factor (EGF) in duodenum, jejunum and ileum. Results showed that the number of Baciilus and the percentage based on all bacteria in the whole intestinal content of piglets fed pea starch was highest in all groups (P < 0.05). There was no significant differance on copy numbers for all bacteria and Bacillus in the whole intestinal tract of piglets between the corn starch group and wheat starch group (P > 0.05). In addition, the expression level of GLP-2, IGF-1 mRNA in jejunum and ileum of pea starch treatment (the high amylose/amylopectin ratio) were increased while the tapioca starch decreased their mRNA level significantly compared to other three treatments (P < 0.05). There was no significant difference for the mRNA level of EGF in each group. The present study revealed that high amylose/amylopectin ratio of starches significantly enhanced the numbers of Bacillus in all segments of intestine and the mRNA level of intestinal development related genes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The swine gastrointestinal tract (GIT) represents a dynamic ecosystem composing of a highly complex and diverse community of microaerophilic and anaerobic microbes that are involved in the fermentation of ingested feed and the components secreted by the host into the GIT [1, 2]. Regulating the composition and metabolic activity of the intestinal microbiota through the diets to improve gut metabolism and health is an increasing focus of nutritionists in the post-antibiotic era [1]. Recently, prebiotics and probiotics have been used as a strategy to manipulate the intestinal ecosystem for enhancing the growth performance and gut health of animals [3, 4]. Some Bacillus species have been as probiotics added to the swine feed for improvement in nutrient digestion and utilization in the latter part of the growing-finishing period. Bacillus supplementation in previous research may have been due to several functions of Bacillus microorganisms, which have been identified as potent producers of extracellular degrading enzymes, including amylases, cellulases, lipases and proteases [5]. The role of Bacillus organisms is that it may increase porcine intestinal health through regulating immune system and protect from pathogenic challenges [6]. Bacillus is a genus of Gram-positive rod-shaped bacteria. Bacillus species can be obligate aerobes or facultative anaerobes, which has the vital role in the intestinal microecology balance by consuming intestinal oxygen, creating the anaerobic environment. Pieper et al. [1] reported that different amylose/amylopectin ratio starch not only result in changing microbial ecophysiology but also affect the susceptibility of the host to opportunistic pathogens.
Starch is mostly composed of the dry matter in certain corps, and it is the major energy-yielding ingredient in diets for both animal and human [7]. It is a polysaccharide, and consists of the polymers amylose and amylopectin [8]. The starches are classified into rapidly digestible starch (RDS), slowly digestible starch (SDS) and resistant starch (RS) [9]. They have significant influence on the composition and activity of the intestine Microflora, tending to enhance bacteria associated with a healthy intestine and tending to reduce those potentially diseases [10].
Up to now, there have been few reports about the effects of different starch sources on Bacillus spp. in intestinal tract and expression of intestinal development related genes of weanling piglets. In this paper, in an effort to elucidate this problem, real-time quantitative PCR protocol was applied to quantify the absolute abundance of Bacillus species in the porcine intestinal chyme and expression of intestinal development related genes treated with different dietary starch source.
Materials and methods
Animals and management
A total of 28 PIC male piglets (similar birth and parity), weaned at 21 ± 1.5 days of age were used in this experiment. The piglets were housed in special metabolism pens (0.6 × 1.2 m) in a thermo regulated environment (ambient temperature 22°C and relative humidity 55%) in the Animal Center of Animal Nutrition Institute in Sichuan Agricultural University. Piglets were fed with the experimental diets for 24 days (3-d pre-experimental and 21-d experimental periods).
Experimental design and diets
The piglets were randomly assigned into four treatments (7 pigs/treatment) with four sources of starch (CS: corn starch, WS: wheat starch, TS: tapioca starch, PS: pea starch). The determined ratio of amylose to amylopectin for corn starch, wheat starch, tapioca starch and pea starch was 0.21, 0.24, 0.12 and 0.52 respectively. Experimental diets were formulated according to NRC (1998) nutrient requirement for piglets weighing 5 to 10 kg. The ingredients and nutrient levels of the four experiment diets were uniform except for the starch source. Diets contained neither antibiotics nor alternative antimicrobial substances.
Sample collection
At the end of the experiment, the chyme of the duodenum, jejunum, ileum and cecum and colon were removed immediately and stored at −80°C until further analysis. The intestinal mucosa from ileum, duodenum and jejunum were also collected and stored at −80° until use. All experimental procedures followed the actual law of animal protection which was approved by the Animal Care Advisory Committee of Sichuan Agricultural University.
DNA Extraction from chyme
Genomic DNA from Bacillus subtilis ATCC6633 was extracted with the TaKaRa minibest bacterial genomic DNA extraction kit (TaKaRa, Dalian, China) and bacterial DNA was isolated from the chyme samples using an E.Z.N.A.TM stool DNA isolation kit (Omega Bio-Tek, Doraville, USA) according to the manufacturer’s instructions.
Design and validation of primers for Bacillus
Primers and probe (Table 1) for quantitative detection of particular Bacillus were designed accorrding to 16S rRNA sequences of maximum species of each genus encountered in the swine intestinal tract downloaded from the GenBank database as well as EMBL and DDBJ. In order to avoid any non-specific amplification, the sequences of all the genera fetched from the database were submitted to DNAStar (MegAlign) programme (DNASTAR, Inc., Madison, WI). It was picked up that the Bacillus blocks of hyper variable regions comprised with all other genera. These sequences were then submitted to second round of alignment where the maximum number of species belonging one genus was aligned and the regions showing conservations were selected as Bacillus genus-specific primers and probes. In order to ensure furthermore that the oligonucleotide sequences were complementary pairing with the target genus only,they were checked with GenBank program BLAST (NCBI BLAST, http://blast.ncbi.nlm.nih.gov/Blast.cgi) and RDP program Check-Probe (Details about RDP data and analytical functions can be found at http://rdp.cme.msu.edu/). Primers (Table 2) for all bacteria were obtained from the published work [11]. All the primers and probe were commercially synthesized from Invitrogen (Shanghai, China).
Reference strains, culture conditions and genome extraction
A total of thirty strains including Bifidobacterium, Lactobacillus, Bacillus, Escherichia coli, Staphylococcus aureus and Streptococcus were used as reference strains in this study (Data not shown). The strains were cultured anaerobically or aerobically in respective culture supplemented with 1% glucose at 37°C for 12 to 48 h. Total genomic DNA from the different reference strains were extracted and purified by using the method described in related Kit Manual (E.Z.N.A. TM Bacterial DNA Kit, Omega Bio-Tek, Doraville, USA).
Standard curve generation
To quantify the copy numbers of Bacillus and all bacteria in test samples, two specific standard curves were generated by constructing the standard plasmids. In brief, DNA encoding Bacillus and all bacteria were respectively extracted from the standard stain of Bacillus (ATCC6633) and the test samples. PCR amplification was carried out using their specific primers YB-P1, YB-P2 and Eub338F, Eub518R (Table 1). After amplification, approximately 424 and 200 bp fragment were obtained respectively. The transformation of purified PCR products, screening of transformed bacteria and extraction of the positive recombinant plasmids containing the inserts were conducted according to the standard procedure. Then the standard plasmids for Bacillus and all bacteria were constructed successfully.
DNA concentrations of the standard plasmids were determined by spectrophotometer (Coulter DU 800, Beckman, USA). A series of 10-fold dilution (1 × 109 to 1 × 101 copies/μl) of the plasmids DNA for Bacillus and all bacteria were prepared and used to generate their respective standard curves with the the logarithum of Standard templates as the abscissa while the Ct as the ordinate. The copy numbers for Bacillus and all bacteria were calculated using the following formula: (DNA concentration in μg/μl × 6.0233 × 1023 copies/mol)/(DNA size(bp) × 660 × 106).
Quantitative PCR conditions and specificity validation of primers
All primers and probe used in this study are presented in Table 1. Quantitative Real-time PCR was carried out in a 25 μl reaction volume which was composed of 12.5 μl SYBR Premix Ex Taq (2×), 1 μl each of forward and reverse primers (100 nM), 9.5 μl ddH2O and 1 μl DNA template for detecting all Bacteria. The reaction was carried out using the following reaction cycles: initial predenaturation at 95°C for 20 s followed by 40 consecutive cycles of denaturation at 95° for 5 s, annealing for 30 s at 60°, extension at 72° for 50 s. Melting curve conditions were 95°C for 0 s, 55°C for 1 min and 95°C for 1 min (temperature change velocity: 0.5°C/s). For Bacillus detection the PrimerScriptTM PCR kit (Perfect Real Time) (TaKaRa, Dalian, China) was used with 100 nM of genus-specific primers and fluorescent probe. The reaction protocol was composed of 1 cycle of predenaturation at 95°C for 2 min; 50 cycles of denaturation at 95°C for 15 s; annealing at 60°C for 30 s and extension at 72°C for 50 s. The genomic DNA of each reference strain was detected using primers YB-F and YB-R and probe YB-P for specificity validation while primers Eub338F and Eub518R were used for verification feasibility.
Real-time PCR for quantification of GLP-2, IGF-1 and EGF
Total RNA were extracted from intestinal mucosa using RNAzol reagent (TaKaRa, Dalian, China) according to the manufacturer’s instructions. Reverse transcription reactions were performed using PrimeScript II 1st Strand cDNA Synthesis Kit (TaKaRa, Dalian, China) following the manufacturer’s instructions. Real-Time PCR for quantification of GLP-2, IGF-1and EGF were perfomed using β-actin as the housingkeeping gene. Sequences of primers for GLP-2, IGF-1and EGF were shown in Table 2. The relative quantification of target gene transcripts in comparison to the reference gene transcript was calculated as described previously [12].
Statistical analysis
The datas were analyzed using SPSS12.0. All results were expressed as means ± SD. One-way ANOVA procedure was performed to assess the statistical significance between treatments. P < 0.05 was considered significant.
Results
Specific verification of quantitative PCR products and specificity of the PCR
Primers and probe designed with Primer Express 3.0 for quantitative detection of particular Bacillus were firstly verificated to be specific by corresponding software from website. Then conventional PCR were carried out using DNA extracted from thirty reference strains including Bifidobacterium, Lactobacillus, Bacillus, Escherichia coli, Staphylococcus aureus and Streptococcus as the templates for verifying the specificity of the primers and the specificity of the PCR. Results showed that a band of approximately 92 bp was obtained after PCR amplification (data not shown) when used DNA extracted from Bacillus as the template while there were no specific amplification for other reference strains (Fig. 1) with the primers YB-F, YB-R, YB-P. For the primers Eub338F, Eub518R, there were specific amplification for all the reference strains. The above results demonstrated that there were 100% specificity for the primers (Table 1) and could be used in the following study.
Quantitative PCR standard curve and reproducibility
The standard curves with the logarithm of Standard templates as the abscissa while the Ct as the ordinate were obtained in this study. There was good linear relation between the copy numbers and the Ct value when the copy numbers within 1 × 109~1 × 104copies/μl. The regression equation was: Y = −2.991X + 51.76 (Y represented the threshold cycle (Ct), X represented the Log (copy number of 16S rDNA)10 ). The correlation coefficient of the standard curves were 0.999 and the amplification efficiencies calculated using the equation: E = 10[−1/slope] [14] were both above 115.94%, which indicated that the crossing threshold values for the standard curves were within an acceptable range. To verificate the validity of the standard curves, it was further confirmed by reproducibility test. Four different concentrations (1.5 × 109−1.5 × 106 copies/μl) of the standard plasmids DNA were applied to the PCR as template in triplicate to verify the reproducibility between experiments. The results showed that the coefficient of variation was statistically low, at <1.5%. The threshold cycle for each concentration ranged from 1.5 × 109 copies/μl to 1.5 × 106 copies/μl and was different between 0.1 and 0.3 cycles, which indicated that the assay was highly reproducible. DNA copies of samples were calculated using the above regression equation.
Calculation of copy numbers for Bacillus and all bacteria
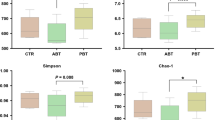
Real-time PCR analysis was performed to determine the copy numbers of Bacillus and all bacteria in the content of intestine of all piglets. Quantitative test results for the copy numbers of Baciilus and all bacteria in the chyme treated with different starch were presented in Figs. 2 and 3. Results showed that there was no obvious difference (P > 0.05) on the copy numbers of all bacteria in the chyme of proximal intestine (duodenum, jejunum and ileum) for the different treatments (Fig. 2). However, the copy numbers of all bacteria in the cecal and colon content of piglets fed PS was significantly lower than other treatments (P < 0.05) while there was no difference (P > 0.05) among other three treatments. It can be seen from Fig. 3 that the copy numbers of Baciilus in the whole intestinal tract were affected significantly. The copy numbers of Baciilus in the chyme of the whole intestinal tract (duodenum, jejunum, ileum, cecum and colon) for treatment with PS was the highest in all groups (P < 0.05) while there was no difference (P > 0.05) among other three treatments. Throughout the entire digestive tract, the copy numbers of all bacteria and Bacillus were lowest in the chyme of duodenum and gradually increasing from proximal intestine to distal intestine.
The proportion of Bacillus based on all bacteria
Results for the proportion of bacillus based on all bacteria in the chyme treated with different starch were presented in Fig. 4. Different starch source affected obviously the proportion of Bacillus (based on all bacteria) in the chyme of intestinal tract. The proportion of Bacillus in the chyme of the whole intestinal tract (duodenum, jejunum, ileum, cecum and colon) for PS treatment elevated significantly compared with other treatments (P < 0.05) while there was no difference among other three treatments (P > 0.05). In addition, the proportion of Bacillus in the chyme of the whole intestinal tract for TS treatment was the lowest.
mRNA level of intestinal development related genes GLP-2, IGF-1and EGF
Results for the mRNA level of intestinal development related genes treated with different starch sources were shown in Fig. 5. As was shown in Fig. 5, different starch source affected obviously the mRNA level of intestinal development related genes. For the expression level of GLP-2, IGF-1 in jejunum and ileum, the pea starch treatment (the high amylose/amylopectin ratio) increased their mRNA level significantly compared to other three treatments (P < 0.05) while the tapioca starch decreased their mRNA level significantly (P < 0.05). As to EGF, there was no significant difference for the mRNA level of it in each group (P > 0.05).
Discussion
As is known to all that starch is the main carbohydrate source of animal diets and the main energy source needed by animals. The digestive function and physiological effect of different corn starchs were various due to their respective composition and structure. Generally, amylose and amylopectin were the main forms. The nutritional value of starch depends on the ratio of amylose/amylopectin to a great extent [15]. In this experiment, to compare the effects of different starch sources on Bacillus spp. in intestinal tract and expression of intestinal development related genes of weanling piglets, the corn starch (CS), wheat starch (WS), tapioca starch (TS), pea starch (PS) were selected as the only starch source of diets. The feed components and nutrient composition (GE, CP, St, lysine, methionine + cysteine and tryptophan contents) of the four experiment diets were uniform except for the starch source which had the different ratio of amylose/amylopectin. In addition, there was no difference for the feed processing and management condition for piglets, so the difference between the treatments for Bacillus spp. in intestinal tract and expression of intestinal development related genes could be attributed to the starch source.
In recent years, real-time PCR is becoming the most suitable method in molecular biology for the detection and quantification of mRNA and was widely used by researchers [16, 17]. It has also become a potentially powerful method to quantify the population of gastrointestinal tract microbial [18, 19]. Moreover, with the development of biotechnology, the specific primer-probe combination is becoming an available method for detecting the counts of intestinal bacterial species [19, 20]. In order to study the copy numbers of Bacillus in the chyme of intestinal tract of piglets, an fluorescence quantitative PCR protocol was developed, optimized and applied in the present study. Results of comparison analysis on the sequences of the Primers and probe used in the study using the bioinformatics software and database indicated that there was high specificity for the primers and probe. To further conform their′s specificity, conventional PCR were carried out using DNA extracted from reference strains as the templates. PCR results verified that there were 100% specificity for the primers and probe and could be used for detection Bacillus and all bacteria. To quantify the copy numbers of Bacillus and all bacteria in the chyme of intestinal tract of weanling piglets treated with different starch sources, the standard curve with the the logarithum of Standard templates as the abscissa while the Ct as the ordinate was obtained which had good linear relation between the copy numbers and the Ct value when the copy numbers within 1 × 109 ~ 1 × 104copies/μl, the correlation coefficient was 0.999 and the amplification efficiencies was 115.94%. Besides, the reproducibility test showed that the crossing threshold values for the standard curve were within an acceptable range. So the standard curve we obtained could be used for quantitation of the copy numbers of Bacillus.
The applications of Bacillus species as probiotics have been developed in farming and aquaculture as alternatives to antibiotics. Certain Bacillus species execute a positive stimulation of immune system and promote the synthesis of antimicrobials, thereby potentially prevent or moderate intestine inflection. Certain dietary non-digestible carbohydrates can allow specific changes in the composition or activity of the intestinal microbiota [21] and selectively stimulate the growth of the health-promoting bacteria (Bifidobacterium and Lactobacilli) in the intestine [22, 23]. Results of our study revealed that the copy numbers of Baciilus in the whole intestinal tract were affected significantly and treatment with PS (high ratio of amylose/amylopectin) was highest (P < 0.05). Throughout the entire digestive tract, the copy numbers of all bacteria and Bacillus were lowest in the chyme of duodenum and gradually increasing from proximal intestine to distal intestine. This may be relative to the anti-digestibility of the PS. It has been reported that resistant starch has strong anti-digestibility, but it could be used by microorganism in colon and generate much short-chain fatty acid which was capable of maintaining a low acid conditions and promote the proliferation of beneficial bacteria [24, 25]. Another study also found that pigs fed potato starch had an increased concentration of short-chain fatty acid in the large intestine with increasing amount of RS. The possible mechanism was that resistant starch could act as the fermentation substrate for bacteria in large intestine and thereby promote the growth of bacteria. We also found that the proportion (based on all bacteria) of Bacillus in the chyme of the whole intestinal tract for PS treatment elevated significantly compared with other treatments (P < 0.05) while there was no difference (P > 0.05) among other three treatments. This variation may be due to the different of the amylose/amylopectin ratio starch. It has been shown that the higher the amylose content, the more difficult the starch is to degrade [26]. The digestibility of starch is generally inverse proportion to its amylose content [27]. There is a higher amylose contents in the pea starch diets, and the previous result showed that it was a slowly digestible progress in the porcine GIT [28]. One explanation could be that high amylose/amylopectin ratio starch can promote the growth of indigenous Bacillus spp.
When it comes to the intestinal development related genes, our results indicated that the mRNA level of GLP-2, IGF-1 in jejunum and ileum were increased in the pea starch treatment (the high amylose/amylopectin ratio) while the tapioca starch decreased their mRNA level significantly. GLP-2 is supposed to play an important role in the regulation of the size and absorptive capacity of the gut. It was reported that GLP-2 increased mucosal thickness in mice [29] and mucosal weight, activity of jejunal maltase-glucoamylase, sucrase-isomaltase mRNA abundance of Neonatal Piglets [30]. Insulin-like growth factor-1 (IGF-1) is an important regulator of gastrointestinal tract and plays a fundamental role in postnatal mammalian growth, development, and metabolism [31]. In particular, orally administered IGF-I is beneficial for the integrity and function of the small intestine [32]. So we suggested that the the pea starch may promote the development of gastrointestinal tract by enhancing the expression level of GLP-2, IGF-1.
In conclusion, we developed a new real-time Taq-man PCR assays that allowed us for rapid, convenient, reproducible, and steady quantification of the Bacillus group in the intestinal content of piglets. Additionally, the present study revealed that high amylose/amylopectin ratio of starches not only significantly enhanced the numbers of Bacillus and the percentage of its (based on all bacteria) in chyme of the all intestine segment, but also the mRNA level of the intestinal development related genes of GLP-2, IGF-1. That was to say, high amylose/amylopectin ratio of starches may be more beneficial for the healthy intestinal tract.
References
Pieper R, Bindelle J, Rossnagel B, Van Kessel A, Leterme P (2009) Effect of carbohydrate composition in barley and oat cultivars on microbial ecophysiology and proliferation of Salmonella enterica in an in vitro model of the porcine gastrointestinal tract. Appl Environ Microbiol 75(22):7006–7016
Konstantinov SR, Favier CF, Zhu WY, Williams BA, Klüß J, Souffrant W-B, WMd Vos, Akkermans ADL, Smidt H (2004) Microbial diversity studies of the porcine gastrointestinal ecosystem during weaning transition. Anim Res 53(4):317–324
Estrada A, Drew MD, VanKessel A (2001) Effect of the dietary supplementation of fructooligosaccharides and Bifidobacterium longum to early-weaned pigs on performance and fecal bacterial populations. Can J Anim Sci 81:141–148
Macfarlane GT, Cummings JH (1999) Probiotics and prebiotics: can regulating the activities of intestinal bacteria benefit health? BMJ 318(7189):999–1003
Macfarlane GT, Cummings JH (1999) Probiotics and prebiotics: can regulating the activities of intestinal bacteria benefit health? West J Med 171(3):187–191
Hong HA, Ducle H, Cutting SM (2005) The use of bacterial spore formers as probiotics. FEMS Microbiol Rev 29(4):813–835
Wiseman J (2006) Variations in starch digestibility in non-ruminants. Anim Feed Sci Technol 130(1–2):66–77
Biliaderis CG (1991) The structure and interactions of starch with food constituents. Can J Physiol Pharmacol 69(1):60–78
Englyst KN, Englyst HN, Hudson GJ, Cole TJ, Cummings JH (1999) Rapidly available glucose in foods: an in vitro measurement that reflects the glycemic response. Am J Clin Nutr 69(3):448–454
Conlon MA, Bird AR (2009) Interactive and individual effects of dietary non-digestible carbohydrates and oils on DNA damage, SCFA and bacteria in the large bowel of rats. Br J Nutr 101(8):1171–1177
Fierer N, Jackson JA, Vilgalys R, Jackson RB (2005) Assessment of soil microbial community structure by use of taxon-specific quantitative PCR assays. Appl Environ Microbiol 71(7):4117–4120
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29(9):e45
Erkens T, Van Poucke M, Vandesompele J, Goossens K, Van Zeveren A, Peelman LJ (2006) Development of a new set of reference genes for normalization of real-time RT-PCR data of porcine backfat and longissimus dorsi muscle, and evaluation with PPARGC1A. BMC Biotechnol 6:41
Amann RI, Ludwig W, Schleifer KH (1995) Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59(1):143–169
Camp LK, Southern LL, Bidner TD (2003) Effect of carbohydrate source on growth performance, carcass traits, and meat quality of growing-finishing pigs. J Anim Sci 81(10):2488–2495
Larsen K, Madsen LB, Bendixen C (2011) Porcine UCHL1: genomic organization, chromosome localization and expression analysis. Mol Biol Rep. doi:10.1007/s11033-011-0836-9
Sun C, Shao HL, Zhang XW, Zhao XF, Wang JX (2011) Molecular cloning and expression analysis of signal transducer and activator of transcription (STAT) from the Chinese white shrimp Fenneropenaeus chinensis. Mol Biol Rep. doi:10.1007/s11033-011-0681-x
Deng SX, Cheng AC, Wang MS, Yan B, Yin NC, Cao SY, Zhang ZH, Cao P (2008) The pathogenesis of Salmonella enteritidis in experimentally infected ducks: a quantitative time-course study using taqman polymerase chain reaction. Poult Sci 87(9):1768–1772
Deng SX, Cheng AC, Wang MS, Cao P (2007) Gastrointestinal tract distribution of Salmonella enteritidis in orally infected mice with a species-specific fluorescent quantitative polymerase chain reaction. World J Gastroenterol 13(48):6568–6574
De Medici D, Croci L, Delibato E, Di Pasquale S, Filetici E, Toti L (2003) Evaluation of DNA extraction methods for use in combination with SYBR green I real-time PCR to detect Salmonella enterica serotype enteritidis in poultry. Appl Environ Microbiol 69(6):3456–3461
Roberfroid M (2007) Prebiotics: the concept revisited. J Nutr 137(3 Suppl 2):830S–837S
Ebersbach T, Jorgensen JB, Heegaard PM, Lahtinen SJ, Ouwehand AC, Poulsen M, Frokiaer H, Licht TR (2010) Certain dietary carbohydrates promote Listeria infection in a guinea pig model, while others prevent it. Int J Food Microbiol 140(2–3):218–224
Kaplan H, Hutkins RW (2000) Fermentation of fructooligosaccharides by lactic acid bacteria and bifidobacteria. Appl Environ Microbiol 66(6):2682–2684
Cummings JH, Beatty ER, Kingman SM, Bingham SA, Englyst HN (1996) Digestion and physiological properties of resistant starch in the human large bowel. Br J Nutr 75(5):733–747
Stephen AM, Cummings JH (1980) The microbial contribution to human faecal mass. J Med Microbiol 13(1):45–56
Brown IL, McNaught KJ, Moloney E (1995) Hi-maize™: new directions in starch technology and nutrition. Food Aust 47(6):272–275
Rooney LW, Pflugfelder RL (1986) Factors affecting starch digestibility with special emphasis on sorghum and corn. J Anim Sci 63(5):1607–1623
Sun T, Lærke HN, Jørgensen H, Knudsen KEB (2006) The effect of extrusion cooking of different starch sources on the in vitro and in vivo digestibility in growing pigs. Anim Feed Sci Technol 131(1):67–86
Drucker DJ, Erlich P, Asa SL, Brubaker PL (1996) Induction of intestinal epithelial proliferation by glucagon-like peptide 2. Proc Natl Acad Sci USA 93(15):7911–7916
Petersen YM, Elnif J, Schmidt M, Sangild PT (2002) Glucagon-like peptide 2 enhances maltase-glucoamylase and sucrase-isomaltase gene expression and activity in parenterally fed premature neonatal piglets. Pediatr Res 52(4):498–503
Xiao S, Li S, Zhang J, Zhang S, Dai L, Bao Y, Jiang N, Gao Y, Zhao Z, Mo D, Chen Y (2009) Cloning and characterization of class 1 and class 2 insulin-like growth factor-I mRNA in Songliao black pig. Mol Biol Rep 36(2):415–421
Alexander AN, Carey HV (1999) Oral IGF-I enhances nutrient and electrolyte absorption in neonatal piglet intestine. Am J Physiol 277(3 Pt 1):G619–G625
Acknowledgments
This work was granted by Program for Changjiang Scholars and Innovative Research Team in University of China with grant. No. IRTO555, China Ministry of Education.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Han, GQ., Xiang, ZT., Yu, B. et al. Effects of different starch sources on Bacillus spp. in intestinal tract and expression of intestinal development related genes of weanling piglets. Mol Biol Rep 39, 1869–1876 (2012). https://doi.org/10.1007/s11033-011-0932-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-011-0932-x