Abstract
Background
Microsatellite instability (MSI) occurs in 15% of colorectal cancers (CRC). The genetic targets for mutation in the MSI phenotype include somatic mutations in the transforming growth factor beta receptor typeII (TGFbetaRII), BAX, hMSH3 and hMSH6. It is not clear how mutations of these genes mediate tumor progression in the MSI pathway, and the temporal sequence of these mutations remains uncertain. In this study, early stage CRCs were examined for frameshift mutations in these target genes, and compared with late stage tumors and CRC cell lines.
Methods
We investigated 6 CRC cell lines and 71 sporadic CRCs, including 61 early stage cancers and 10 late stage cancers. Mutations of repetitive mononucleotide tracts in the coding regions of TGFbetaRII, BAX, hMSH3, hMSH6, IGFIIR and Fas antigen were identified by direct sequencing.
Results
Thirteen (18.3%) of 71 CRC, including 9/61 (14.7%) early stage cancers and 4/10 (40%) late stage cancers, were identified as MSI and analyzed for frameshift mutations. No mutation in the target genes was observed in any of the 9 early stage MSI CRCs. In contrast, frameshift mutations of TGFbetaRII, BAX, hMSH3 and hMSH6 were present in 3/4 late stage MSI tumors. There is a statistical association (p = 0.014) between mutation in any one gene and tumor stage.
Conclusions
TGFbetaRII, BAX, hMSH3 and hMSH6 mutations are relatively late events in the genesis of MSI CRCs. The frameshift mutations in these target genes might mediate progression from early to late stage cancer, rather than mediating the adenoma to carcinoma transition.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
Microsatellite instability (MSI) is present in 15% of colorectal carcinomas (CRCs)[1]. Inactivation of the DNA mismatch repair (MMR) system leads to widespread somatic mutations at microsatellite loci. MSI tumors have been found to display microsatellite alterations not only in introns but also in coding exons. Genetic targets of this type of genomic instability include the transforming growth factor β receptor type II (TGFβRII), insulin-like growth factor II receptor (IGFIIR), BAX, Fas antigen, hMSH3 and hMSH6, all of which contain mononucleotide repeats in coding sequences [2, 3]. Although it has been reported that MSI in introns is an early event in sporadic colorectal carcinogenesis [4, 5], it is not known how mutations of these target genes are involved in tumor progression along the MSI pathway [2]. This study examined whether the frequency frameshift mutations in mononucleotide repeat regions within exons increases with stage in microsatellite unstable colorectal cancer. "Early stage cancer" was defined as intramucosal carcinoma, either carcinoma in situ or invasive cancer confined within the submucosa. For this study, "late stage cancer" was defined as any cancer invading the muscularis propria or serosa. Molecular events in early CRCs have not been well elucidated because of the limited availability of these cancers for detailed studies [5]. In this study, early stage MSI CRCs were examined for somatic frameshift mutations in the TGFβRII, IGFIIR and BAX, hMSH3 and hMSH6 genes, and compared with CRCS at more advanced stages and CRC cell lines.
Methods
Cell lines
Six CRC cell lines: HCT116, LoVo, SW480, LS174t, DLD1 and HT29, were obtained from the American Type Culture Collection (Rockville, MD), and maintained in tissue culture containing 10% fetal calf serum (GIBCO) at 37°C. LoVo, DLD1, SW480 and LS174t were derived from late stage colorectal cancer: LoVo [6] and DLD1 [7] were derived from Dukes' C tumor, and SW480 [8] and LS174t [9] were from Dukes' B tumor. Tumor stage of HCT116 [10] and HT29 [11] were not informative.
Tumor tissues
Seventy-one CRCs, including 61 early stage cancers and 10 late stage cancers, were collected for analysis. The histopathological diagnosis was determined according to the classification by the World Health Organization (WHO) [12] or by the General Rules of the Japanese Research Society for Cancer of Colon and Rectum [13]. Lymph node status was not taken into account. The tumor tissues were microdissected from formalin-fixed, paraffin-embedded tissues sections as previously described [14]. Normal tissues were obtained from histologically normal mucosa or normal lymph nodes of the same patients. This study was approved by the Osaka City University ethics committee. Informed consent was obtained from all patients.
DNA extraction
Genomic DNA from the cell lines was extracted with phenol-chloroform. Genomic DNA from tumor tissues was isolated using Proteinase K (Sigma, St. Louis, MO) at a final concentration of 100 μg/ml and were incubated for 5 h at 55°C [14].
Identification of MSI
DNA was amplified by polymerase chain reaction (PCR) using 32 P-end-labeled primers at microsatellite loci linked to the hMSH2 locus on 2p16 (CA-5 and D2S123), the hMLH1 locus on 3p23-21.3 (D3S1611 and D3S1561), the APC/MCC locus on 5q21 (D5S107 and D5S346), the p53 locus on 17p13 (D17S513 and p53 intron 1), and the DCC/SMAD4 locus on 18q21.3 (D18S35A and 18qDCC-TA) [14]. We utilized stringent criteria for the determination of MSI from a National Cancer Institute workshop [15]. MSI-high (referred to as MSI) was defined by a novel band shift or allele at >20% of the microsatellite loci tested when compared to non-neoplastic tissue from the same patient. Our assay for MSI was based on PCR amplification of a panel of 10 microsatellite primer pairs linked to tumor suppressor genes. In addition, poly (T)27 mononucleotide repeats in the noncoding region of β-catenin exon 16 was performed using the following primer sets: 5'-GGTACTGACTTTGCTTGCTT-3' and 5'-ACTTAACACTACGAGAGACT-3'. In each case, one of the primer pairs was end-labeled with [γ-32P]ATP and subjected to PCR. The resulting PCR products were analyzed for MSI by electrophoresis in 8% polyacrylamide gel containing 7.5 M urea and 0.5× TBE.
Frameshift mutation analysis
Mutations of repetitive mononucleotide tracts in the coding regions of TGFβRII, IGFIIR, BAX, hMSH3, hMSH6 and Fas antigen were identified using the following primer sets; 5'-CTTTATTCTGGAAGATGCTGC-3' and 5'-GAAGAAAGTCTCACCAGGC-3' for the poly(A)10 tract of TGFβRII, 5'-GCAGGTCTCCTGACTCAGAA-3' and 5'-GAAGAAGATGGCTGTGGAGC-3' for the poly(G)8 tract of IGFIIR, 5'-ATCCAGGATCGAGCAGGGCG-3' and 5'-ACTCGCTCAGCTTCTTGGTG-3' for the poly(G)8 tract of BAX, 5'-AGCTGGATGATGCTGTAAAT-3' and 5'-TTCCTCACCTGCAAAGTACT-3' for the poly(A)8 tract of hMSH3, 5'-CGCCCAGTAATTCTGTTG-3' and 5'-CATTTTCCTGCTCCTCTTC-3' for the poly(C)8 tract of hMSH6, and 5'-ACCCGGACCCAGAATACCAA-3' and 5'-GCAAGGGTCACAGTGTTCAC-3' for the poly(T)7 tract of Fas antigen. To confirm mutations observed as band shifts, DNA was excised from the gel, purified with the Microspin columns (Pharmacia Biotec, Inc., Uppsala, Sweden), and reamplified with the same primer. The amplified products were sequenced by the dideoxychain termination method, with the AmpliCycle sequencing kit (Perkin-Elmer, Branchburg, NJ).
Results
Frameshift mutations in CRC cell lines
It has been reported that HCT116, LoVo, LS174t and DLD1 have the MSI phenotype, whereas SW480 and HT29 do not [16]. In this study, frameshift mutations in the (A)10 tract of TGFβRII were found in all 4 MMR-deficient cell lines: HCT116, LoVo, LS174t and DLD1. All cell lines have only mutant alleles of TGFβRII, either (A)9 or (A)8. Microsatellite alterations in the (G)8 tract of BAX were present in HCT116, LoVo and LS174t. LoVo and LS174t cells had only (G)9 and (G)7 mutant BAX alleles; HCT116 had one (G)7 mutant allele and one (G)8wild-type allele. Deletions in the (A)8 tract of the coding sequence of hMSH3 were present in HCT116. Insertion and deletion mutations in the (C)8 tract of hMSH6 were detected in HCT116 and LS174t, respectively. In contrast, the MMR-proficient cell lines SW480 and HT29 demonstrated microsatellite stability, and did not have mutations in TGFβRII, BAX, hMSH3 or hMSH6. No alterations in the (G)8 tract of IGFIIR or the (T)7 tract of Fas antigen were found in any of the cell lines examined (Figure 1).
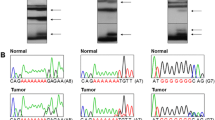
Frameshift mutations in CRC cell lines. Lane a: products from normal control DNA. The products of MSI cell lines are shown in Lanes b (HCT116), c (LoVo), e (LS174-T) and f (DLD1). Products of non-MSI cell lines are shown in Lanes d (SW480) and g (HT29). The genes are indicated at the left. Arrowheads indicate frameshift mutations in cell lines.
Frequencies of frameshift mutations in early and late stage sporadic MSI CRCs
Thirteen of 71(18.3%) sporadic CRCs, including 9/61 (14.7%) early stage CRCs and 4/10 (40%) late stage cancers were identified as MSI tumors (Table 1), and these tumors were analyzed for frameshift mutations. Mutations in the noncoding region of β-catenin, which contains a (T)27 sequence, were present in 7/9 MSI early stage CRCs (patients 1-9) and 3/4 MSI-H late stage CRCs (patients 10-13) (Figure 2A). No frameshift mutations in the target genes were present in any MSI early stage CRCs. In contrast, frameshift mutations of TGFβRII, BAX, hMSH3 and hMSH6 were present in 3/4 late stage CRCs (Table 1). A significant difference (p = 0.014) was found in the frequency of frameshift mutations between early stage and. late stage (Table 2). TGFβRII and BAX mutations were found in the late stage tumors of patients 11 and 10, respectively (Figure 2B). hMSH3 mutations were present in patients 11 and 12, and an hMSH6 mutation was found in patient 12. Each mutation was found to be a 1 bp deletion in the polynucleotide tract (Figure 3). No frameshift mutations in IGFIIR or Fas antigen were found in any CRC.
Discussion
Although CRCs with the MSI phenotype evolve because of loss of DNA MMR activity, there is little consensus on the sequence and timing of target gene inactivation in the evolution of tumors that occur through this pathway. It has been reported that the appearance of MSI, at least at noncoding microsatellites, is an early event in Lynch syndrome CRCs [4, 17], and that frameshift alterations in the functionally critical target genes may occur at an early stage in these tumors [18–20]. TGFβRII frameshift mutations are present in 80-90% of MSI CRCs, and it has been reported that these occur at the adenoma-carcinoma transition in MSI adenomas [21, 22]. The precise timing of the onset of MSI, and the occurrence of mutations at other genetic targets, have not been confidently determined.
In this study, we examined the timing of frameshift mutations of TGFβRII, IGFIIR, BAX, hMSH3, hMSH6 and Fas antigen in 9 early stage and 4 late stage MSI CRCs. Interestingly, no mutations were identified in any early stage MSI-H CRC at TGFβRII, BAX, hMSH3 or hMSH6 , but these were relatively frequent events in late stage MSI CRCs, and in the MSI CRC cell lines. There is a statistical association between mutation in any one gene and tumor stage. The frequency of frameshift mutations in mononucleotide repeat regions within exons might increase with stage in microsatellite unstable colorectal cancer. Our findings suggest that TGFβRII, BAX, hMSH3 and hMSH6 frameshift mutations are relatively later stage events in tumor progression for sporadic CRC with MSI. It has been reported that TGFβRII mutations are infrequent in the early stages of sporadic gastric cancers with MSI [23], but these occur more frequently in gastric tumors that are larger [24]. It has been reported that the loss of TGFβRII in intestinal epithelial cells promotes the invasion and malignant transformation of tumors initiated by APC mutations [25]. However, few studies have clarified how mutations of the target genes we have studied mediate tumor development in the MSI pathway. These findings suggested that the mutations of TGFβRII, BAX, hMSH3, and hMSH6 might play an important role in cancer progression from early to late stage tumors, rather than early in carcinogenesis. However, there still may be additional, currently unappreciated genes or DNA sequences involved in the process [18, 26–28]. Interestingly, no microsatellite mutations in IGFIIR and Fas antigen were found in any cell line or in any CRC. It has also been reported that no mutations in the IGFIIR genes were found in 27 MSI-H gastric tumors [29]. These findings suggest that mononucleotide repeat of IGFIIR and Fas antigen might be unimportant as target genes in MMR-deficient CRCs. A limitation of our study is the small number of tumor samples, while a significant difference between the early stage cancer and late stage cancers was recognized. Large numbers of patients with MSI cancer might be necessary in the future to conclude the correlation between tumor stage and frameshift mutations in target gene.
Conclusions
Our data confirm that MSI can be found very early in the evolution of a colorectal tumor with MSI, we propose that not all sequence alterations are equivalent, and that mutations in the coding microsatellites of TGFβRII and BAX mediate progression from early to late stage CRC with MSI. These finding raise interesting possibilities for diagnosis as well as treatment.
References
Boland CR: The molecular biology of gastrointestinal cancer: implications for diagnosis and therapy. Gastrointestinal endoscopy clinics of North America. 2008, 18 (3): 401-413. 10.1016/j.giec.2008.03.003. vii
Arnold CN, Goel A, Blum HE, Boland CR: Molecular pathogenesis of colorectal cancer: implications for molecular diagnosis. Cancer. 2005, 104 (10): 2035-2047. 10.1002/cncr.21462.
Yamamoto H, Gil J, Schwartz S, Perucho M: Frameshift mutations in Fas, Apaf-1, and Bcl-10 in gastro-intestinal cancer of the microsatellite mutator phenotype. Cell death and differentiation. 2000, 7 (2): 238-239. 10.1038/sj.cdd.4400651.
Pedroni M, Sala E, Scarselli A, Borghi F, Menigatti M, Benatti P, Percesepe A, Rossi G, Foroni M, Losi L, et al: Microsatellite instability and mismatch-repair protein expression in hereditary and sporadic colorectal carcinogenesis. Cancer research. 2001, 61 (3): 896-899.
Kambara T, Matsubara N, Nakagawa H, Notohara K, Nagasaka T, Yoshino T, Isozaki H, Sharp GB, Shimizu K, Jass J, et al: High frequency of low-level microsatellite instability in early colorectal cancer. Cancer research. 2001, 61 (21): 7743-7746.
Drewinko B, Romsdahl MM, Yang LY, Ahearn MJ, Trujillo JM: Establishment of a human carcinoembryonic antigen-producing colon adenocarcinoma cell line. Cancer research. 1976, 36 (2 Pt 1): 467-475.
Dexter DL, Barbosa JA, Calabresi P: N,N-dimethylformamide-induced alteration of cell culture characteristics and loss of tumorigenicity in cultured human colon carcinoma cells. Cancer research. 1979, 39 (3): 1020-1025.
Leibovitz A, Stinson JC, McCombs WB, McCoy CE, Mazur KC, Mabry ND: Classification of human colorectal adenocarcinoma cell lines. Cancer research. 1976, 36 (12): 4562-4569.
Tom BH, Rutzky LP, Jakstys MM, Oyasu R, Kaye CI, Kahan BD: Human colonic adenocarcinoma cells. I. Establishment and description of a new line. In vitro. 1976, 12 (3): 180-191. 10.1007/BF02796440.
Brattain MG, Fine WD, Khaled FM, Thompson J, Brattain DE: Heterogeneity of malignant cells from a human colonic carcinoma. Cancer research. 1981, 41 (5): 1751-1756.
Chen TR, Drabkowski D, Hay RJ, Macy M, Peterson W: WiDr is a derivative of another colon adenocarcinoma cell line, HT-29. Cancer genetics and cytogenetics. 1987, 27 (1): 125-134. 10.1016/0165-4608(87)90267-6.
Jass JR, Sobin LH, Watanabe H: The World Health Organization's histologic classification of gastrointestinal tumors. A commentary on the second edition. Cancer. 1990, 66 (10): 2162-2167. 10.1002/1097-0142(19901115)66:10<2162::AID-CNCR2820661020>3.0.CO;2-N.
General rules for clinical and pathological studies on cancer of the colon, rectum and anus. Part II. Histopathological classification. Japanese Research Society for Cancer of the Colon and Rectum. Jpn J Surg. 1983, 13 (6): 574-598. 10.1007/BF02469506.
Yashiro M, Carethers JM, Laghi L, Saito K, Slezak P, Jaramillo E, Rubio C, Koizumi K, Hirakawa K, Boland CR: Genetic pathways in the evolution of morphologically distinct colorectal neoplasms. Cancer research. 2001, 61 (6): 2676-2683.
Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN, et al: A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer research. 1998, 58 (22): 5248-5257.
Carethers JM, Pham TT: Mutations of transforming growth factor beta 1 type II receptor, BAX, and insulin-like growth factor II receptor genes in microsatellite unstable cell lines. In Vivo. 2000, 14 (1): 13-20.
Lynch HT, Lynch PM, Lanspa SJ, Snyder CL, Lynch JF, Boland CR: Review of the Lynch syndrome: history, molecular genetics, screening, differential diagnosis, and medicolegal ramifications. Clinical genetics. 2009, 76 (1): 1-18. 10.1111/j.1399-0004.2009.01230.x.
Imai K, Yamamoto H: Carcinogenesis and microsatellite instability: the interrelationship between genetics and epigenetics. Carcinogenesis. 2008, 29 (4): 673-680. 10.1093/carcin/bgm228.
Akiyama Y, Yagi OK, Ishikawa T, Nagasaki H, Saitoh K, Yuasa Y: Genetic alterations are frequent in APC but rare in the TGF-beta type II receptor gene in cancer in adenomas of the colon. Cancer Lett. 1998, 125 (1-2): 89-96. 10.1016/S0304-3835(97)00490-4.
Konishi M, Kikuchi-Yanoshita R, Tanaka K, Muraoka M, Onda A, Okumura Y, Kishi N, Iwama T, Mori T, Koike M, et al: Molecular nature of colon tumors in hereditary nonpolyposis colon cancer, familial polyposis, and sporadic colon cancer. Gastroenterology. 1996, 111 (2): 307-317. 10.1053/gast.1996.v111.pm8690195.
Grady WM, Rajput A, Myeroff L, Liu DF, Kwon K, Willis J, Markowitz S: Mutation of the type II transforming growth factor-beta receptor is coincident with the transformation of human colon adenomas to malignant carcinomas. Cancer research. 1998, 58 (14): 3101-3104.
Shin KH, Park YJ, Park JG: Mutational analysis of the transforming growth factor beta receptor type II gene in hereditary nonpolyposis colorectal cancer and early-onset colorectal cancer patients. Clin Cancer Res. 2000, 6 (2): 536-540.
Ogata S, Tamura G, Endoh Y, Sakata K, Ohmura K, Motoyama T: Microsatellite alterations and target gene mutations in the early stages of multiple gastric cancer. J Pathol. 2001, 194 (3): 334-340. 10.1002/path.895.
Pinto M, Oliveira C, Cirnes L, Carlos Machado J, Ramires M, Nogueira A, Carneiro F, Seruca R: Promoter methylation of TGFbeta receptor I and mutation of TGFbeta receptor II are frequent events in MSI sporadic gastric carcinomas. J Pathol. 2003, 200 (1): 32-38. 10.1002/path.1327.
Munoz NM, Upton M, Rojas A, Washington MK, Lin L, Chytil A, Sozmen EG, Madison BB, Pozzi A, Moon RT, et al: Transforming growth factor beta receptor type II inactivation induces the malignant transformation of intestinal neoplasms initiated by Apc mutation. Cancer research. 2006, 66 (20): 9837-9844. 10.1158/0008-5472.CAN-06-0890.
Alazzouzi H, Davalos V, Kokko A, Domingo E, Woerner SM, Wilson AJ, Konrad L, Laiho P, Espin E, Armengol M, et al: Mechanisms of inactivation of the receptor tyrosine kinase EPHB2 in colorectal tumors. Cancer research. 2005, 65 (22): 10170-10173. 10.1158/0008-5472.CAN-05-2580.
Woerner SM, Benner A, Sutter C, Schiller M, Yuan YP, Keller G, Bork P, Doeberitz MK, Gebert JF: Pathogenesis of DNA repair-deficient cancers: a statistical meta-analysis of putative Real Common Target genes. Oncogene. 2003, 22 (15): 2226-2235. 10.1038/sj.onc.1206421.
Korff S, Woerner SM, Yuan YP, Bork P, von Knebel Doeberitz M, Gebert J: Frameshift mutations in coding repeats of protein tyrosine phosphatase genes in colorectal tumors with microsatellite instability. BMC cancer. 2008, 8: 329-10.1186/1471-2407-8-329.
Kim JJ, Baek MJ, Kim L, Kim NG, Lee YC, Song SY, Noh SH, Kim H: Accumulated frameshift mutations at coding nucleotide repeats during the progression of gastric carcinoma with microsatellite instability. Lab Invest. 1999, 79 (9): 1113-1120.
Pre-publication history
The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1471-2407/10/303/prepub
Acknowledgements
We thank Christina Chang, PhD for technical assistance and critical review of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
MY participated in the study design, and carried out PCR experiments, sequencing analysis, interpretation of data, and paper preparation. KH carried out data analysis and interpretation. CRB carried out data analysis, interpretation, and paper preparation. All authors have read and approved of the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Yashiro, M., Hirakawa, K. & Boland, C.R. Mutations in TGFbeta-RII and BAXmediate tumor progression in the later stages of colorectal cancer with microsatellite instability. BMC Cancer 10, 303 (2010). https://doi.org/10.1186/1471-2407-10-303
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2407-10-303