Abstract
Background
Acinetobacter species are recognised as important nosocomial pathogens that have become a major cause of invasive opportunistic infections in hospitalised patients. Their clinical significance is largely due to the rapid development of antimicrobial resistance among strains. The development of antibiotic resistance among bacterial strains occurs frequently by the acquisition of resistance genes by gene transfer systems such as bacterial plasmids.
Method
Multi-antibiotic resistant Acinetobacter nosocomialis strain 178 was isolated from a hospital in Melbourne, Australia. This strain was screened for the presence of plasmids. The two plasmids isolated were sequenced and annotated.
Results
Two plasmids isolated from a single clinical Acinetobacter nosocomialis strain were sequenced. One plasmid, designated pRAY*-v3, appears to have evolved via the same lineage as the pRAY plasmid isolated from an Acinetobacter baumannii in South Africa. The other plasmid, designated pAB49-v1, appears to be an evolutionary descendent from a cryptic plasmid isolated from an A. baumannii almost 20 years ago. Both of the plasmid sequences here share a high level of sequence similarity with their ancestors, however differences are noted.
Conclusion
The isolation of these plasmid-lineages across different decades and continents suggests their global dissemination.
Similar content being viewed by others
Background
The Acinetobacter calcoaceticus-baumannii (ACB) complex [1] consists of four closely related genospecies, namely A. calcoaceticus (genomic sp. 1), A. baumannii (genomic sp. 2) [2], A. nosocomialis (former genomic sp. TU13) and A. pittii (former genomic sp. 3) [3], along with two additional unnamed genomic species [4]. These species, particularly A. baumannii, A. nosocomialis and A. pittii, are recognised as important nosocomial pathogens that have become a major cause of invasive opportunistic infections among critically ill hospitalised patients [5, 6]. Their clinical significance is largely implicated by the rapid development of multi-antimicrobial resistance among these species, making hospital-acquired infection difficult to treat [5, 6]. The development of antibiotic resistance among bacterial strains occurs frequently by the acquisition of resistance genes by gene transfer systems, such as bacterial plasmids, transposable elements and integrons, where plasmids serve as the vehicles for resistance gene capture and their subsequent dissemination throughout the microbial population [7]. Bacterial plasmids are commonly observed among members of the Acinetobacter genus where they are suggested to play an important role in the adaptive evolution of these organisms [7–9]. Considering this suggested role for plasmids, coupled with their implication in serious antibiotic resistance issues in hospitals, makes further study into the detail of their functions of paramount importance.
We here report the isolation and characterization of two plasmids isolated from a single clinical strain of A. nosocomialis obtained from a Melbourne hospital.
Methods
The bacterial strain Acinetobacter sp. strain 178 was provided to us from a local Melbourne (Australia) hospital and is the subject of this study. The source of the strain is unknown and de-identified. The Escherichia coli K-12 derivative used for transformation of cloned DNA fragments was DH5α (Δlac169 endA1 recA1 Res- Mod+ Nalr) [10]. The plasmid used for cloning was pBluescript KS+. Bacteria were grown on Luria-Bertani (LB) medium and incubated at 30 °C for Acinetobacter strains and 37 °C for E. coli. To select E. coli strains with conferred resistance (or A. nosocomialis strains, as indicated) antimicrobials were added to LB at the following final concentrations (μ g mL-1): ampicillin (Ap) 50 and 100; chloramphenicol 32; tetracycline 80; kanamycin 12.5; rifampicin 2; gentamycin 10; streptomycin 10.
To determine the identity of the clinical isolate str. 178, the 16S rRNA gene and 16S-23S Internal Transcribed Spacer (ITS) region were PCR amplified using the primers 6R (GGGTTYCCCCRTTCRGAAAT) [11] and 27 F (GAG TTT GAT CMT GGC TCA G) [12]. Sequencing was performed by Australian Genome Research Facility (AGRF) (Brisbane, Australia) using the primers 530 F (GTG CCA GCM GCC GCG G), 519R (GWA TTA CCG CGG CKG CTG), 1114 F (GCA ACG AGC GCA ACC C), 907R(CCG TCA ATT CMT TTR AGT TT) [12], 1512 F (GTCGTAACAAGGTAGCCGTA) and 6R (GGGTTYCCCCRTTCRGAAAT) [11]. Phylogenetic placement was assessed with the alignment and construction of a phylogenetic tree of related sequences [13]. The ITS region was also sequenced to complement 16S rRNA gene analysis, as the 16S rRNA gene is reportedly unable to provide phylogenetic resolution between species of the ACB complex [14].
Plasmids were isolated, digested with restriction endonucleases Hin dIII and the fragments were cloned into pBluescript KS+. The fragments were sequenced and assembled, and primer walking was used to close any remaining gaps. The fragments were sequenced using pUC/M13 forward and reverse primers [15]. Cloning, plasmid DNA isolation and other manipulations were performed as described previously [16]. DNA sequence analysis was performed using the Geneious Pro 4.0.4 and the BlastN algorithm at National Centre Biotechnology Information (NCBI).
PCR amplification employed novel primers, BG1 (CGATTTATGTCAGTTCGCGG) and BG2 (CTCTGAGTTGGCTGACAATG), to amplify a region within pAB49 and its relatives using the conditions described in Petrovski & Stanisich, [16].
The nucleotide sequence of the 16S rRNA gene and ITS region has been deposited in GenBank under accession number KC257034. The plasmid sequences for pRAY*-v3 and pAB49-v1 have been deposited in GenBank under the accession numbers KC417494 and KC417495 respectively.
Results and discussion
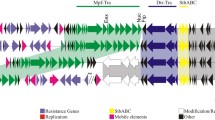
A. nosocomialis strain 178 was selected from 41 clinical Acinetobacter strains collected from a local Melbourne (Australia) hospital. This strain was of particular interest because of its relatively broad antibiotic resistance profile, growing in the presence of gentamicin, streptomycin, chloramphenicol and kanamycin. Gel electrophoresis of plasmid DNA isolated from strain 178 indicated the presence of two small plasmids ~6 kb and ~2.2 kb in size. The plasmids were sequenced revealing they were 6,078 bp and 2,200 bp in size, correlating well with the estimated sizes. Both plasmids had a GC mol% content of 39%, which is similar to the genome of their host species [17], and are here named pRAY*-v3 and pAB49-v1.
The compiled sequence of the 6,078 bp plasmid is almost identical to the sequence of the plasmid pRAY from Acinetobacter sp. SUN, which was isolated in the 1990s in South Africa [18]. More recently plasmids similar to pRAY with small variations have been isolated from A. baumannii strains in New South Wales (Australia) and named pRAY*, pRAY*-v1 and pRAY*-v2 [19, 20]. We therefore here name this plasmid pRAY*-v3. pRAY*-v3 is most closely related to pRAY*-v1 with only a single nucleotide substitution at position 1656 from a C > G. The other two Australian isolates pRAY* and pRAY*-v2 share 99% identity with pRAY*-v3. Comparison of pRAY*-v3 with both pRAY* and pRAY*-v2 reveals that an additional 55 to 60 bp substitution occurs within the mobA gene (Figure 1a). pRAY*-v2 also contains an insertion of a putative IS-element [20].
Our annotation of the pRAY*-v3 plasmid reveals five putative ORFs designated as aadB, orf1, orf2, mobA and mobC (Figure 1a). The aadB gene encodes an aminoglycoside resistance protein, enabling the host to have a competitive advantage at survival in the presence of such antimicrobials. However, this gene is generally located within the attI site of integrons and appears to be a gene cassette. Despite this finding, no integrase gene is present in pRAY*-v3 so the integration of this gene cassette probably occurred when an integrase was provided in trans from another DNA molecule [21]. The DNA sequence upstream of the aadB gene must also encode a promoter region, as gene cassettes traditionally solely rely on the Pc promoter, located within an integrase, for their expression [21]. Two putative genes of unknown function are present downstream of the aadB gene, here named orf1 and orf2. Following these two genes are two additional genes, mobA and mobC, thought to encode putative mobilisation proteins. The presence of such proteins suggests that the pRAY plasmids could be mobilisable and the presence of an IS-element in pRAY*-v2 suggests that these pRAY plasmids could potentially act as vehicles for the dissemination of transposable elements in Acinetobacter species, particularly the ACB complex.
Comparisons of pRAY*-v3 with its close relatives reveals most mutations have occurred within the mobA gene, suggesting that this gene is either not essential for plasmid replication or maintenance or that these mutations have no affect on the function of the encoded protein. This plasmid and its derivatives have been reportedly detected in various locations around the world that include South Africa, Australia, Europe and the USA [20] suggesting stable maintenance within Acinetobacter strains. This is the first reported isolation of a pRAY lineage plasmid from A. nosocomialis, having previously only been detected in A. baumannii strains, suggesting that the pRAY-lineage plasmids are at least more widely distributed among members of the ACB complex.
The DNA sequence of the smaller plasmid (2,200 bp), isolated from A. nosocomialis str. 178, was compared to other DNA sequences in GenBank and appears to be 78% similar to the first 1850 bp of the DNA sequence of plasmid pAB49 (Acc No L77992), deposited in the database in 1996 but is as yet unpublished. The remaining 350 bp of the sequence are novel. We therefore here named this plasmid pAB49-v1.
The identification of pAB49-v1 15 years after the sequence submission of pAB49, without any apparent selection pressure, is interesting and suggests that perhaps these pAB49 type plasmids are stably maintained in some Acinetobacter strains. The plasmid described here encodes two putative genes presumably involved in plasmid replication (Figure 1b).
To determine the distribution of the plasmid among related clinical ACB complex strains, we designed primers that would amplify a 1 kb region of its sequence and conducted PCR on 41 Acinetobacter isolates from a local Melbourne hospital. Only the 178 strain gave a PCR product, indicating the presence of the pAB49-v1.
We cloned the entire pAB49-v1 into pBluescript KS + (see materials and methods). This newly created plasmid contained the origin of vegetation of the E. coli plasmid pBluescript KS + and that of the A. nosocomialis plasmid pAB49-v1. Furthermore the plasmid now had a selectable marker, namely ampicillin. To determine its host range we attempted to transform this plasmid into ampicillin sensitive (at 50 μg/ml) Acinetobacter baumannii. The plasmid was unable to replicate in the type strain suggesting a narrow host range potentially restricted to A. nosocomialis. Despite these findings, the detection of the plasmid in two separate continents 15 years apart indicates that this small plasmid may have disseminated between Acinetobacter strains throughout the world.
Conclusion
In conclusion we isolated two plasmids from a clinical A. nosocomialis strain from Melbourne Australia. Ancestors of the two plasmids have been previously isolated and sequenced from different continents in Acinetobacter strains. Plasmid pRAY has a selective advantage to its host, providing a mechanism for resistance to aminoglycosides, whereas pAB49 does not. The fact that such plasmids can be isolated many years later, in geographically distinct locations, suggest they are strongly maintained within their hosts, despite the smaller plasmid having no evident selective advantage. The work presented here suggests that these two plasmids may be widely disseminated among members of the ACB complex throughout the world and thus could play an important role in their ecology.
References
Gerner-Smidt P, Tjernberg I, Ursing J: Reliability of phenotypic tests for identification of Acinetobacter species. J Clin Microbiol. 1991, 29: 277-282.
Bouvet PJM, Grimont PAD: Taxonomy of the genus Acinetobacter with the recognition of Acinetobacter baumannii sp.nov., Acinetobacter junii sp. nov. and emended descriptions of Acinetobacter calcoaceticus and Acinetobacter lwoffii. Int J Syst Bacteriol. 1986, 36: 228-240. 10.1099/00207713-36-2-228.
Nemec A, Krizova L, Maixnerova M, van der Reijden TJ, Deschaght P, Passet V, Vaneechoutte M, Brisse S, Dijkshoorn L: Genotypic and phenotypic characterization of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex with the proposal of Acinetobacter pittii sp. nov. (formerly Acinetobacter genomic species 3) and Acinetobacter nosocomialis sp. nov. (formerly Acinetobacter genomic species 13TU). Res Microbiol. 2011, 162: 393-404. 10.1016/j.resmic.2011.02.006.
Gerner-Smidt P, Tjernberg I: Acinetobacter in Denmark: II. Molecular studies of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex. APMIS. 1993, 101: 826-832. 10.1111/j.1699-0463.1993.tb00187.x.
Lee NY, Lee HC, Ko NY, Chang CM, Shih HI, Wu CJ, Ko WC: Clinical and economic impact of multidrug resistance in nosocomial Acinetobacter baumannii bacterium. Infect Contol Hosp Epidemiol. 2007, 28: 713-719. 10.1086/517954.
Lee K, Yong D, Jeong SH, Chong Y: Multidrug-resistant Acinetobacter spp.: increasingly problematic nosocomial pathogens. Yonsei Med J. 2011, 52: 879-891. 10.3349/ymj.2011.52.6.879.
Bennett PM: Plasmid encoded antibiotic resistance: acquisition and transfer of antibiotic resistance genes in bacteria. Br J Pharmacol. 2008, 153: S347-S357. 10.1038/sj.bjp.0707569.
Alekshun MN, Levy SB: Molecular mechanisms of antibacterial multidrug resistance. Cell. 2007, 128: 1037-1050. 10.1016/j.cell.2007.03.004.
Van Looveren M, Goossens H, ARPAC Steering Group: Antimicrobial resistance of Acinetobacter spp. In Europe. Clin Microbiol Infect. 2004, 10: 684-704. 10.1111/j.1469-0691.2004.00942.x.
Hanahan D: Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983, 166: 557-580. 10.1016/S0022-2836(83)80284-8.
Relman DA: Universal bacterial 16S rDNA amplification and sequencing. Diagnostic molecular microbiology. Edited by: Persing DH, Smith TF, Tenover FC, White TJ. 1993, Washington, D.C: American Society for Microbiology, 489-495.
Lane D: 16S/23S rRNA sequencing. Modern Microbial Methods: Nucleic Acid Techniques in Bacterial Systematics. 1991, England: John Wiley & Sons, 115-175.
Maslunka C, Gifford B, Tucci J, Gurtler V, Seviour RJ: Insertions and deletions (Indels) in the rrn 16S-23S rRNA gene internal transcribed spacer region (ITS) compromise the typing and identification of strains within Acinetobacter calcoaceticus-baumannii (Acb) complex and closely related members. Plos One. 2014, 9: e105390-10.1371/journal.pone.0105390.
Keum KC, Yoo SM, Lee SY, Chang KH, Yoo NC, Yoo WM, Kim JM, Choi JY, Kim JS, Lee G: DNA microarray based detection of nosocomial pathogenic Pseudomonas aeruginosa and Acinetobacter baumannii. Mol Cell Probes. 2006, 20: 42-50. 10.1016/j.mcp.2005.09.001.
Sambrook J, Fritsch EF, Maniatis T: Molecular cloning: a laboratory manual. 1989, Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press, 2
Petrovski S, Stanisich VA: Tn502 and Tn512 are res site hunters that provide evidence of resolvase-independent transposition to random sites. J Bacteriol. 2010, 192: 1865-1874. 10.1128/JB.01322-09.
Vallenet D, Nordmann P, Barbe V, Poirel L, Mangenot S, Bataille E, Dossat C, Gas S, Kreimeyer A, Lenoble P, Oztas S, Poulain J, Segurens B, Robert C, Abergel C, Claverie JM, Raoult D, Medigue C, Weissenbach J, Cruveiller S: Comparative analysis of Acinetobacters: three genomes for three lifestyles. PLoS One. 2008, 19: e1805-
Segal H, Elisha BG: Characterization of the Acinetobacter plasmid, pRAY, and the identification of regulatory sequences upstream of an aadB gene cassette on this plasmid. Plasmid. 1999, 42: 60-66. 10.1006/plas.1999.1403.
Nigro SJ, Post V, Hall HM: Aminoglycoside resistance in multiply antibiotic-resistant Acinetobacter baumannii belonging to global clone 2 from Australian hospitals. J Antimicrob Chemother. 2011, 66: 1504-1509. 10.1093/jac/dkr163.
Hamidian M, Nigro SJ, Hall RM: Variants of the gentamicin and tobramycin resistance plasmid pRAY are widely distributed in Acinetobacter. J Antimicrob Chemother. 2012, 67: 2833-2836. 10.1093/jac/dks318.
Segal H, Francia MV, Lobo JM, Elisha G: Reconstruction of an active integron recombination site after integration of a gene cassette at a secondary site. Antimicrob Agents Chemother. 1999, 43: 2538-2541.
Acknowledgements
Bianca Gifford was supported by an Australian Postgraduate Award.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
BG carried out the experiments, analysed the data and drafted the manuscript. JT funded the research and drafted the manuscript. SJM assisted in data analysis and the drafting of the manuscript. SP conceived and designed the experiments, interpreted the data, and drafted the manuscript. All authors have read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Gifford, B., Tucci, J., McIlroy, S.J. et al. Isolation and characterization of two plasmids in a clinical Acinetobacter nosocomialis strain. BMC Res Notes 7, 732 (2014). https://doi.org/10.1186/1756-0500-7-732
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1756-0500-7-732