Abstract
Background
Despite numerous advances in the identification of risk factors for the development of severe coronavirus disease 2019 (COVID-19), factors that promote recovery from COVID-19 remain unknown. Natural killer (NK) cells provide innate immune defense against viral infections and are known to be activated during moderate and severe COVID-19. Killer immunoglobulin-like receptors (KIR) mediate NK cell cytotoxicity through recognition of an altered MHC-I expression on infected target cells. However, the influence of KIR genotype on outcome of patients with COVID-19 has not been investigated so far. We retrospectively analyzed the outcome associations of NK cell count and KIR genotype of patients with COVID-19 related severe ARDS treated on our tertiary intensive care unit (ICU) between February and June 2020 and validated our findings in an independent validation cohort of patients with moderate COVID-19 admitted to our tertiary medical center.
Results
Median age of all patients in the discovery cohort (n = 16) was 61 years (range 50–71 years). All patients received invasive mechanical ventilation; 11 patients (68%) required extracorporeal membrane oxygenation (ECMO). Patients who recovered from COVID-19 had significantly higher median NK cell counts during the whole observational period compared to patients who died (121 cells/µL, range 16–602 cells/µL vs 81 cells/µL, range 6–227 cells/µL, p-value = 0.01). KIR2DS5 positivity was significantly associated with shorter time to recovery (21.6 ± 2.8 days vs. 44.6 ± 2.2 days, p-value = 0.01). KIR2DS5 positivity was significantly associated with freedom from transfer to ICU (0% vs 9%, p-value = 0.04) in the validation cohort which consisted of 65 patients with moderate COVID-19.
Conclusion
NK cells and KIR genotype might have an impact on recovery from COVID-19.
Similar content being viewed by others
Background
A novel coronavirus (SARS-Cov-2) causing coronavirus disease 2019 (COVID-19) was first identified in December 2019 in Wuhan, China, upon a series of pneumonias of unknown cause [1]. Since then, SARS-CoV-2 spread worldwide, reaching pandemic levels [2]. The clinical presentation of COVID-19 covers a broad spectrum, ranging from asymptomatic upper respiratory infections to severe acute respiratory distress syndrome (ARDS) [3]. Genome-wide association analyses in a large case–control study including healthy volunteers as a control group identified certain gene clusters to be associated with patients suffering from COVID-19 and requiring oxygen supplementation [4]. Yet, associations of individual genetic imprints with outcome in patients already suffering from moderate or severe COVID-19 are still missing.
Natural killer (NK) cells play a central role in the immune response against viral infections [5,6,7,8]. In patients with COVID-19, especially in those with severe symptoms, lower NK cell counts were consistently reported [9]. In contrast to peripheral blood, the human lung is enriched in NK cells during the course of COVID-19 [10]. Moreover, there has been evidence for an exhausted NK cell phenotype in COVID-19 [11]. Further characterization analyses identified distinct NK cell immunotypes to be associated with COVID-19 severity and an increase in adaptive NK cells in SARS-CoV-2-infected patients with severe disease [12]. Taking it all together, these data underline why NK cells might influence the course of COVID-19.
NK cells express a highly polymorphic group of killer immunoglobulin-like receptors (KIR), capable of binding specific human leukocyte antigens (HLA) in order to recognize an altered MHC-I expression, mostly HLA-C antigens on virus-infected target cells (missing-self principle) [13, 14]. Evidence linking specific KIR genotypes and susceptibility to viral infections has been broadly published [15, 16]. However, the influence of KIR genotype on the course of COVID-19 has not been investigated so far. Therefore, we aimed to investigate the associations of KIR genotypes and clinical course of COVID-19-related severe ARDS in patients treated on our tertiary intensive care unit (ICU) between February and June 2020 and validated our findings in an independent validation cohort of patients with moderate COVID-19 admitted to our tertiary medical center.
Methods
Patient characteristics and study design
In this retrospective study, we included all patients with confirmed severe COVID-19 who were treated on our ICU between February and June 2020 (discovery cohort) and all patients with moderate COVID-19 admitted to our tertiary medical center (validation cohort) of whom written informed consent by themselves or their next of kin was obtained. In the same time period, a total of 40 patients with COVID-19 were treated on our intensive care unit.
Severe COVID-19 was defined as polymerase chain reaction (PCR)-confirmed SARS-COV2 infection and severe ARDS as defined by the Berlin definition with a PaO2/FiO2 ratio of less than 100 mmHg (Horowitz Index (P/F ratio) < 100 mmHg) and bilateral pulmonal infiltration on radiological imaging [17]. Moderate COVID-19 was defined as PCR-confirmed SARS-COV2 infection and evidence of lower respiratory disease during clinical assessment or imaging and who have an oxygen saturation (SpO2) ≥ 94% on room air at sea level.
This study and post hoc analysis were part of the COORSAAR register study and approved by the local ethics committee (Ethikkommission der Ärztekammer des Saarlandes, Ethics approval number 62/20). The study was performed in accordance with the rules of the Declaration of Helsinki.
Study endpoints
Study endpoint of this retrospective study for the discovery cohort was recovery from COVID-19 at day 28 after ICU admission. Patients were considered recovered if they were free from oxygen supplementation regardless of their hospitalization status.
Study endpoint for the validation cohort was freedom from transfer to ICU.
KIR genotyping
We extracted genomic deoxyribonucleic acid (DNA) from whole blood samples obtained from patients using the Qiagen column-based method (QIAmp DNA Blood Mini Kid, Qiagen, Chatsworth, CA). We performed KIR genotyping by real-time polymerase chain reaction (RT-PCR) (BioRad CFX Connect Real-Time System) as previously described [18]. In brief, PCR reaction mixtures contained KIR-specific primers, internal control primers (GALC), patient DNA and SYBR Green I Master Mix (Applied Biosystems, Foster City, CA). Presence of the specific KIR was confirmed by analyzing individual melting temperatures. Seventeen KIR genes were analyzed in this study. KIR haplotype was discriminated according to existing nomenclature [19].
Natural killer cells and laboratory parameters
Measurements of leucocytes were performed using a Sysmex XN-L TM automated hematology analyzer. Immune characterization of lymphocytes was performed by flow cytometry using a BD FACSCanto™ II cell analyzer. Flow cytometry was performed according to the “Guidelines for the use of flow cytometry and cell sorting in immunological studies” [20]. NK cell subpopulation was characterized using a Beckman–Coulter Navios TM Analyzer. Total counts of lymphocytes and NK cell subpopulation were therefore calculated using a dual-platform system. Tests were performed twice weekly until transfer to normal care unit or hospital discharge due to clinical improvement or death. Cellular parameters included lymphocytic differentiation into NK cells (CD56+, CD16+). Laboratory parameters included complete blood cell count, CRP and IL-6 and were also obtained at least twice weekly until transfer from ICU to normal care unit due to clinical improvement or death.
Statistical analysis
Patient baseline characteristics were summarized descriptively according to data type. One-sample Kolmogorov–Smirnov test and one-sample Chi-square test were used to assess the distribution of data. Differences between groups were assessed with Chi-square and independent median test. All statistical tests were two-sided with a significance level of 5%. Adjustment for multiple comparison was performed in all presented data. Kaplan–Meier analysis was used to assess the differences in outcome between the groups. Log-rank pooled over strata was used as a comparing method. All data were analyzed using SPSS v25.0 (IBM, Ehningen, Germany).
Results
Severe COVID-19 discovery cohort
In the discovery cohort, we identified 16 patients with COVID-19-related severe ARDS and written informed consent obtained from themselves or their next of kin, treated on our intensive care unit between February and June 2020. Patient baseline (day 1 of ICU admission) characteristics are shown in Table 1. Median age of all patients was 61 years (range 50–71 years). All analyzed patients were Caucasian males. Main comorbidities were arterial hypertension (8/16, 50%) and diabetes mellitus type 2 (2/16, 12.5%). Pre-existing diseases were equally distributed between outcome groups (Table 1).
Patient outcome
Median time of stay on ICU ward was 21.5 days (range 11–47 days) (Table 2). All patients received invasive mechanical ventilation and 11 patients (68%) required extracorporeal membrane oxygenation (ECMO). Regarding clinical outcome, 6 (37.5%) patients died, and 10 (62.5%) patients fully recovered. The cause of death was COVID-19 related in all patients. Complications during ICU stay included pneumothorax in 3 (18%) patients, catheter-related sepsis in 2 (12%), acute mesenteric ischemia with compartment syndrome in 1 (6%) patient and aortic dissection in 1 (6%) patient. Median time to recovery from COVID-19 was 40.5 days (range 11–47 days). Median time to death was 21 days (range 15–39 days). Recovery at day 28 after admission was reached in 4/16 (25%) patients. Clinical characteristics, laboratory parameters, duration of ECMO therapy and Horowitz Index (P/F-ratio) were similar in patients who recovered at 28 days and those who did not (Additional file 1: Table S1).
Laboratory parameters
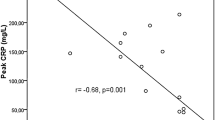
Baseline laboratory parameters are summarized in Table 1 and Additional file 1: Table S1. Median leukocyte count on admission was 9940 cells/µL (range 6390–31,500 cells/µL). Median natural-killer cell count on ICU admission was 58 cells/µL (range 6–236 cells/µL). Median baseline leukocyte count was comparable between outcome groups (recovered: 12,020 cells/µL, range 6390–31.500 cells/µL vs. dead: 9105 cells/µL, range 8000–14,210 cells/µL, p-value = 0.6). On admission, NK cell count was similar in patients who recovered and those who died (recovered: 72.5 cells/µL vs. 25 cells/µL, p-value = 0.11). However, patients who recovered showed significantly higher median NK cells counts during the whole observational period compared to patients who died (121 cells/µL, range 16–602 cells/µL vs. 81 cells/µL, range 6–227 cells/µL, p-value = 0.01) (Table 2 and Fig. 1). Furthermore, NK cell counts correlated negatively with disease severity (Pearson correlation − 0.31, p-value = 0.001) and C-reactive protein (CRP) levels (Pearson correlation − 0.31, p-value = 0.001). Patients who died of COVID-19 had significantly higher median CRP levels during whole observational period compared to patient who recovered (Table 2). IL-6 showed no significant differences at baseline between the two outcome groups (Table 1). However, during the whole observational period, patients who died had significantly higher IL-6 levels compared to patients who recovered (p-value = 0.007) (Table 2).
KIR genotype and recovery
KIR genotype distribution across all patients is presented in Additional file 1: Table S2. The frequency of different KIR genes of all patients suffering from COVID-19-related severe ARDS was comparable to existing data on KIR genotype distribution upon the general German Caucasian population (Additional file 1: Table S2). In 16 patients suffering from COVID-19-related severe ARDS, we did not observe a significant difference in single KIR and KIR haplotype distribution between patients who died and those who recovered (Additional file 1: Table S3). Hence, only KIR2DS5 was significantly associated with time to recovery by day 28 (Fig. 2 and Additional file 1: Figure S1). In particular, KIR2DS5-positive patients demonstrated shorter time to recovery compared to KIR2DS5-negative patients, respectively (mean 21.6 ± 2.8 days vs. mean 44.6 ± 2.2 days, p-value = 0.01). By day 28 after admission, 60% of KIR2DS5-positive patients had recovered compared to only 9% of KIR2DS5-negative patients (p-value = 0.01).
There was no significant difference between KIR2DS5-positive and negative patients regarding clinical and laboratory baseline characteristics (Additional file 1: Table S4). Furthermore, the rate of complications in KIR2DS5-positive and negative patients was comparable (Additional file 1: Table S4).
Moderate COVID-19 validation cohort
To validate our findings from the discovery cohort, we determined the KIR2DS5 status in 65 Caucasian patients with moderate COVID-19 and written informed consent from themselves or their next of kin admitted to our tertiary medical center during the same time period. KIR2DS5 positivity was observed in 20 of 65 patients (30%). Median age was 68 years (IQR: 22 years) (Additional file 1: Table S5). Males represented 57% of all patients. 23 (35%) patients required oxygen supplementation. KIR2DS5-positive patients required less oxygen supplementation than KIR2DS5-negative patients, but without reaching statistical significance (22% vs. 46%, p-value = 0.08). Only 1 (5%) KIR2DS5-positive patient died of COVID-19, compared to 6 (14%) KIR2DS5-negative patients (p-value = 0.33). None of the KIR2DS5-positive patients required transfer to intensive care unit (ICU), compared to 9 (22%) KIR2DS5-negative patients (p-value = 0.04) (Additional file 1: Figure S2).
Discussion
In this retrospective, single-center study, we analyzed the associations of KIR genotype with recovery of patients with COVID-19-associated severe ARDS and validated our findings in patients with moderate COVID-19. We identified KIR2DS5 positivity to be associated with improved clinical course in these patients.
All of our analyzed patients from the discovery cohort of patients with severe COVID-19 were males, which is in line with previous reports of COVID-19 showing consistently a male predominance regarding infection rates [21, 22]. Additionally, the comorbidity rates were comparable with previously published data [23]. Cumulative data from 18 studies, including 2984 COVID-19 patients, reported increased white blood cell and neutrophil counts and decreased lymphocyte, platelet and eosinophil counts in patients with severe forms of respiratory distress, emphasizing the cellular involvement in the course of COVID-19 [24]. Regarding NK cell counts, our results are consistent with previously published literature, showing reduced NK cell numbers in patients who died compared to those who survived COVID-19 [22]. Additionally, higher CRP and IL-6 levels were associated with adverse outcome, which is in line with previously published studies [25,26,27].
Our data show a comparable distribution between KIR genotype in patients with severe COVID-19 and the general German Caucasian population, suggesting, that the individual KIR status does not correlate with disease severity. Nevertheless, we found, that KIR2DS5 positivity was significantly associated with a superior course of disease in patients already suffering from moderate or severe COVID-19.
Although the ligand of KIR2DS5 remains unknown, activating KIR2DS5 was shown to have a protective role in different diseases, including acute rejection reaction of kidney grafts, malaria, human immunodeficiency virus infection and Hepatitis C-induced hepatocellular carcinoma [28]. The activation of KIR2DS5 triggers both, NK cell cytotoxicity and interferon γ (IFN‐γ) release [29]. This enhanced activation could explain the observed shorter mean time to recovery, since an exhaustion of NK cells was previously shown to be associated with inferior outcomes in patients suffering from COVID-19 patients [30]. Moreover, reduced expression of IFN-γ by NK cells in patients with severe disease as compared to patients with moderate disease was observed in a previous immunological characterization analysis of 21 patients with COVID-19 [31].
However, our study does have some limitations. First, it is a single-center retrospective study with a small sample size of patients even though it was performed during very challenging clinical circumstances of the pandemic-imposed limitations. Second, IFN‐γ levels of patients participating in this study were not available. Nevertheless, our results provide a first look into the possible role of KIR in the context of COVID-19 and suggest KIR2DS5 to promote disease recovery from moderate or severe COVID-19.
Although the number of patients included in our analysis does not allow for valid conclusions, association of both, stimulating KIR2DS5 and higher NK cell counts with recovery in patients with severe COVID-19 and validation of our findings in patients with moderate COVID-19 warrant further analyses regarding the impact of KIR genotype on the course of COVID-19.
Conclusions
NK cell count and KIR genotype might influence the course COVID-19. Stimulating KIR2DS5 was associated with superior outcome in moderate or severe COVID-19.
Availability of data and materials
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Code availability
Not applicable.
Abbreviations
- ARDS:
-
Severe acute respiratory distress syndrome
- COVID-19:
-
Coronavirus disease 2019
- CRP:
-
C-reactive protein
- DNA:
-
Deoxyribonucleic acid
- ECMO:
-
Extracorporeal membrane oxygenation
- HLA:
-
Human leukocyte antigen
- ICU:
-
Intensive care unit
- IFN‐γ:
-
Interferon γ
- IL-6:
-
Interleukin-6
- IMC:
-
Intermediate care ward
- KIR:
-
Killer-immunoglobulin-like receptor
- NK:
-
Natural killer
- PCR:
-
Polymerase chain reaction
- RT-PCR:
-
Real-time polymerase chain reaction
- SARS-Cov-2:
-
Severe acute respiratory syndrome coronavirus type 2
References
Yang X, Yu Y, Xu J et al (2020) Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. https://doi.org/10.1016/S2213-2600(20)30079-5
Petersen E, Koopmans M, Go U et al (2020) Comparing SARS-CoV-2 with SARS-CoV and influenza pandemics. Lancet Infect Dis 20:e238–e244
Kannan S, Shaik Syed Ali P, Sheeza A, Hemalatha K (2020) COVID-19 (Novel Coronavirus 2019)—recent trends. Eur Rev Med Pharmacol Sci. https://doi.org/10.26355/eurrev_202002_20378
(2020) Genomewide association study of severe Covid-19 with respiratory failure. N Engl J Med. https://doi.org/10.1056/nejmoa2020283
Björkström NK, Lindgren T, Stoltz M et al (2011) Rapid expansion and long-term persistence of elevated NK cell numbers in humans infected with hantavirus. J Exp Med. https://doi.org/10.1084/jem.20100762
Zimmer CL, Cornillet M, Solà-Riera C et al (2019) NK cells are activated and primed for skin-homing during acute dengue virus infection in humans. Nat Commun. https://doi.org/10.1038/s41467-019-11878-3
Blom K, Braun M, Pakalniene J et al (2016) NK cell responses to human tick-borne encephalitis virus infection. J Immunol. https://doi.org/10.4049/jimmunol.1600950
Scharenberg M, Vangeti S, Kekäläinen E et al (2019) Influenza A virus infection induces hyperresponsiveness in human lung tissue-resident and peripheral blood NK cells. Front Immunol. https://doi.org/10.3389/fimmu.2019.01116
Wen W, Su W, Tang H et al (2020) Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell Discov. https://doi.org/10.1038/s41421-020-0168-9
Zhou Z, Ren L, Zhang L et al (2020) Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe. https://doi.org/10.1016/j.chom.2020.04.017
Wilk AJ, Rustagi A, Zhao NQ et al (2020) A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat Med. https://doi.org/10.1038/s41591-020-0944-y
Maucourant C, Filipovic I, Ponzetta A et al (2020) Natural killer cell immunotypes related to COVID-19 disease severity. Sci Immunol. https://doi.org/10.1126/SCIIMMUNOL.ABD6832
Ljunggren HG, Kärre K (1990) In search of the “missing self”: MHC molecules and NK cell recognition. Immunol Today. https://doi.org/10.1016/0167-5699(90)90097-S
Winter CC, Long EO (1997) A single amino acid in the p58 killer cell inhibitory receptor controls the ability of natural killer cells to discriminate between the two groups of HLA-C allotypes. J Immunol 158:4026–4028
Zwolińska K, Błachowicz O, Tomczyk T et al (2016) The effects of killer cell immunoglobulin-like receptor (KIR) genes on susceptibility to HIV-1 infection in the Polish population. Immunogenetics. https://doi.org/10.1007/s00251-016-0906-1
Aranda-Romo S, Garcia-Sepulveda CA, Comas-García A et al (2012) Killer-cell immunoglobulin-like receptors (KIR) in severe A (H1N1) 2009 influenza infections. Immunogenetics. https://doi.org/10.1007/s00251-012-0623-3
Ranieri VM, Rubenfeld GD, Thompson BT et al (2012) Acute respiratory distress syndrome: the Berlin definition. JAMA J Am Med Assoc. https://doi.org/10.1001/jama.2012.5669
Alves LGT, Rajalingam R, Canavez F (2009) A novel real-time PCR method for KIR genotyping. Tissue Antigens. https://doi.org/10.1111/j.1399-0039.2008.01184.x
Marsh SGE, Parham P, Dupont B et al (2003) Killer-cell immunoglobulin-like receptor (KIR) nomenclature report, 2002. Tissue Antigens 62:79–86
Cossarizza A, Chang HD, Radbruch A et al (2017) Guidelines for the use of flow cytometry and cell sorting in immunological studies. Eur J Immunol. https://doi.org/10.1002/eji.201646632
Bhatraju PK, Ghassemieh BJ, Nichols M et al (2020) Covid-19 in critically ill patients in the seattle region—case series. N Engl J Med. https://doi.org/10.1056/nejmoa2004500
Guan W, Ni Z, Hu Y et al (2020) Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. https://doi.org/10.1056/nejmoa2002032
Tian W, Jiang W, Yao J et al (2020) Predictors of mortality in hospitalized COVID-19 patients: a systematic review and meta-analysis. J Med Virol 92:1875–1883
Henry BM, De Oliveira MHS, Benoit S et al (2020) Hematologic, biochemical and immune biomarker abnormalities associated with severe illness and mortality in coronavirus disease 2019 (COVID-19): a meta-analysis. Clin Chem Lab Med 58:1021–1028
Herold T, Jurinovic V, Arnreich C et al (2020) Elevated levels of IL-6 and CRP predict the need for mechanical ventilation in COVID-19. J Allergy Clin Immunol. https://doi.org/10.1016/j.jaci.2020.05.008
Liu F, Li L, Da XuM et al (2020) Prognostic value of interleukin-6, C-reactive protein, and procalcitonin in patients with COVID-19. J Clin Virol. https://doi.org/10.1016/j.jcv.2020.104370
Han H, Ma Q, Li C et al (2020) Profiling serum cytokines in COVID-19 patients reveals IL-6 and IL-10 are disease severity predictors. Emerg Microbes Infect. https://doi.org/10.1080/22221751.2020.1770129
Chavan VR, Ahir S, Ansari Z et al (2016) Diversity in KIR gene repertoire in HIV-1 exposed infected and uninfected infants: a study from India. J Med Virol. https://doi.org/10.1002/jmv.24348
Della Chiesa M, Romeo E, Falco M et al (2008) Evidence that the KIR2DS5 gene codes for a surface receptor triggering natural killer cell function. Eur J Immunol. https://doi.org/10.1002/eji.200838434
Zheng M, Gao Y, Wang G et al (2020) Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell Mol Immunol 17:533–535
Chen G, Wu D, Guo W et al (2020) Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest. https://doi.org/10.1172/JCI137244
Acknowledgements
The authors are grateful to all patients and their relatives as well as to the colleagues from the Department of Pneumology, Saarland University Hospital, Germany, for taking care of the patients and providing the data for this analysis. This analysis is part of the CoRSAAR project.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
VL, MB, CM, AB, SM, ER, LT, FN, IK, KC, RB, SS, GD, PL, DKM and TR contributed to collection, review, and/or analysis of the data; VL and TR performed statistical analysis of the data; VL, TR and DKM wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study and post hoc analysis were part of the COORSAAR register study and approved by the local ethics committee (Ethikkommission der Ärztekammer des Saarlandes, Ethics approval number 62/20). The study was performed in accordance with the rules of the Declaration of Helsinki. Informed consent was obtained from all individual participants included in the study.
Consent for publication
Consent to publication was obtained from all individual participants included in the study.
Competing interests
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1
: Table S1: Clinical characteristics and laboratory parameters based on recovery by day 28 (discovery cohort). Table S2: KIR prevalence in 16 patients with COVID-19 related severe ARDS compared to the general German Caucasian population. Table S3: KIR genes and Haplotype regarding clinical outcome. Figure S1. Kaplan Meier Estimates on Recovery regarding KIR genotype. Cumulative estimates on recovery from COVID-19 related severe ARDS showing no statistically significant differences regarding specific KIR genotype. Table S4. Clinical characteristics and laboratory parameters regarding KIR2DS5 status. Table S5. Baseline characteristics of 65 patients with moderate COVID-19 (validation cohort) according to KIR2DS5 status. Figure S2. Kaplan Meier Estimates on transfer to Intensive Care Unit (ICU) of 65 patients with moderate COVID-19 (validation cohort) according to KIR2DS5 Status. Cumulative estimates on time to transfer to intensive care unit (ICU) showing statistical significant difference regarding KIR2DS5 status.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lesan, V., Bewarder, M., Metz, C. et al. Killer immunoglobulin-like receptor 2DS5 is associated with recovery from coronavirus disease 2019. ICMx 9, 45 (2021). https://doi.org/10.1186/s40635-021-00409-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40635-021-00409-4