Abstract
Stunted growth is an emerging global challenge affecting children under the age of 5 years in low- and middle-income countries. Despite such a high global prevalence of stunting, the mechanism of pathogenesis and the role of associated gut microbiota is poorly understood. The present study was designed to investigate the association of pathogenic strains of E. coli with the residential gut microbiota of stunted growth children. A total of 64 stool sample were collected from children aged ≤ 5 years, and were processed for isolation and molecular characterization of diarrheagenic E. coli. Selected stool samples (n = 39 including three normal controls) were then analysed for microbial community profiling using 16S ribosomal RNA (rRNA) gene sequencing. Furthermore, associations between changes in the microbiota in the presence of different E. coli strains was explored. Pathotyping of the isolated E. coli (n = 64) has shown that 39.68% belonged to one of the five pathotypes of E. coli whilst the remaining ones were non-typeable. Amongst the different pathotypes, EPEC was found to be the most prevalent (52%; n = 13), followed by EAEC (20%; n = 5), EIEC (12%; n = 3), EHEC (8%; n = 2) and ETEC 2 (8%; n = 2). Phylogrouping analysis has shown that majority of the strains belonged to B2 (28.12%). Microbial diversity is shown to be significant and varied when the samples are organized under the recovered phylogroups. Moreover, based on predictive metabolism, the colonization of these strains were found to be significantly associated with energy utilization pathways such as Denovoprine-2 and glyoxylate-by. Differential analysis has shown that Escherichia-Shigella and Enterococcus were altered for the children with stunted growth.
Similar content being viewed by others
Introduction
Globally, approximately 180 million children under 5 years of age are affected by stunted growth, resulting in ~ 3 million child deaths annually and predominantly in low- and middle-income countries1,2,3. Stunted growth may be caused by several factors such as diarrhoea, pneumonia, and other infectious diseases; unhealthy conditions; food insecurity; and inadequate health services and care facilities4. In some cases, it may lead to comorbidity such as impaired cognitive development, decrease life expectancy and an increased risk of chronic diseases later in life. According to the National Nutrition Survey Pakistan (2018)5, four out of ten children under five years of age are stunted, whilst 17.7% suffer from wasting, 28.9% are underweight, and 9.5% are overweight. The most affected districts are in the south of the country located in the provinces of Baluchistan, Khyber Pakhtunkhwa, and Sindh, with more than 50% reported prevalence of stunting6. These results largely match the known socioeconomic status and district-specific multidimensional poverty indices of these regions.
Environmental Paediatric Enteropathy (EEP) is commonly associated with malnourished children and is caused due to prolonged exposure to enteric gut pathogens such as Campylobacter spp, Shigella spp, Enteroinvasive E. coli (EIEC), Shiga toxin-producing enterotoxigenic E. coli (ETEC), and typical enteropathogenic E. coli (tEPEC), norovirus, Cryptosporidium spp. and Giardia spp7,8. Previous studies from Pakistan have shown association of various E. coli pathotypes with the paediatric population showing symptoms of diarrhoea as well as mothers delivering prematurely9,10,11,12. Moreover, E. coli belonging to different phylogroup have also been associated with different clinical outcomes, and among them, phylogroup B2 had been earlier described to be the most prevalent phylogroup. It is associated with infection such as diarrhoea as it carries more virulence genes than any other phylogroup. However, the role of infection caused by E. coli belonging to different pathotypes and phylogroups infections at early age of life and its impact on the growth of children is still unknown.
Previous study from Pakistan highlighted the marked differences amongst rich and poor population with significantly higher prevalence of stunting in the latter13. In order to better understand stunted growth and avoid higher risk of mortality among young children1, it is essential to understand how infections due to different pathotypes and phylogroups of E. coli changes the microflora among stunted children which will in turn provide an insight into developing evidence based intervention approaches. Therefore, the current study investigates the different E. coli pathotypes and phylogroups in low socioeconomic semi-urban/urban children and their associated faecal bacterial taxa.
Material and methods
Ethical approval and consent
Prior written and verbal informed consent was obtained from the accompanying parent or primary caregiver of each child to participate in the study and was approved by the Ethical Review board of Department of Biosciences, CUI, Islamabad Pakistan. All methods were performed in accordance with the relevant guidelines and regulations.
Study design & patients recruitment
The study was carried out in children (aged 1–5 years) living in Islamabad capital territories (ICT), Rawalpindi and Lahore (province-Punjab), Swat (province-KPK) and Mitthi (province-Sindh) all with the low middle income status. Children were defined as stunted if their Height-for-Age Z score (HAZ) is more than two standard deviations below the WHO child growth standards median. Additional information of study design and the criteria for patient recruitment are provided in the supplementary file (Text S1).
Faecal sample collection
A total of 64 stool samples (60 stunted and 4 normal; morning first bowel from children at hospital) were collected using commercially available uBiome microbiome sampling kits (uBiome, USA). These kits follow the protocols outlined by the NIH Human Microbiome Project14. One sterile swab was used to transfer a small amount of faecal material into a vial containing a lysis and stabilization buffer that preserves the DNA for transport at ambient temperatures. The other sterile cotton swab was used to collect the faecal material to be shipped in transport media for culturing. The stool samples from the transport media were then processed by streaking on MacConkey agar (Oxoid) and incubated at 37 °C overnight. Pure cultures were obtained by re-culture of grown colonies for another 18–24 h at the same incubation temperature. E. coli was further confirmed using the standard biochemical tests including catalase, oxidase, and indole tests.
DNA extraction & gene specific PCR for E. coli pathotypes and phylogroup screening
Genomic DNA was extracted by ethanol precipitation technique as described previously13. The extracted DNA was used for polymerase chain reaction (PCR) to screen the isolates for the presence of pathotype using gene specific primers in two-step multiplex PCR. Briefly, nine primer pairs were used, the first set comprised of four primers, including eae, bfp, vt and aggR for EPEC, EAEC, and EHEC, whilst the second set consisted of five primer pairs targeting the genes st, lt, virF, ipaH and daaE as explained previously9. Phylogenetic grouping of E. coli was done using the revised quadruplex PCR method described by Clermont et al. 201315. Each isolate was assigned one of the eight phylogenetic groups (A, B1, B2, C, D, E, F, Clade I) by targeting three marker genes (ArpA, ChuA, YjaA) and a DNA fragment TSPE4.C2 (Supplementary Table 1). Plasmids containing target genes (gift from Oscar G. Gomez-Duarte, International Enteric Vaccines Research Program [IEVRP], University of Iowa Children's Hospital, Iowa City, IA) were used as positive controls and the DNA from E. coli DH5α was used as a negative control.
Microbiome analysis-I6S rRNA sequencing-microbial taxa
For microbiome studies, random 39 stool samples from which E. coli belonging to different phylogroups were processed and these steps were performed in the CLIA-compliant (Clinical Laboratory Improvement Amendments) and CAP-accredited (College of American Pathologists) uBiome laboratory in San Francisco, CA, USA. Samples were lysed using mechanical bead-beating. DNA was extracted and purified by a liquid- handling robot in a class 100 clean room using the method described previously16. The V4 region of 16S rRNA gene was PCR amplified using universal forward and reverse primers (515F: GTGCCAGCMGCCGCGGTAA and 806R: GGACTACHVGGGTWTCTAAT). Primers also contained Illumina tags and barcodes with unique combination of forward and reverse indexes to allow multiplexing. Pooled PCR products were column-purified and selected through microfluidic DNA fractionation based on size17. Real-time qPCR quantified consolidated libraries using the Kapa Bio-Rad iCycler qPCR kit on a BioRad MyiQ prior to sequencing.
Bioinformatics and statistical analysis
Bioinformatics methods were based on Ijaz et al. 2018 with minor modifications18. The VSEARCH v2.3.4 pipeline was used to produce the abundance table by constructing operational taxonomic units (OTUs), a representation of species, as described in http://github.com/torognes/vsearch/wiki/VSEARCH-pipeline). For detailed bioinformatics and statistical analysis, supplementary files are provided in Supplementary_Materials.docx along with meta data information including location and demographic data given in Supplementary_Metadata.xlsx.
Results
Culturing and isolation
E. coli were isolated from all 64 collected stool samples and were initially screened using standard gram staining and biochemical tests including catalase, oxidase and indole tests and all isolates were found to be positive for E. coli.
Recovered E. coli pathotypes
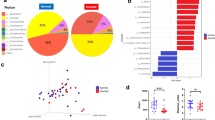
The results suggested that out of a total of 64 samples, 25 (39.68%) were positive for any five out of the six diarrheagenic E. coli pathotypes whereas 39 samples (60.9%) were non-typeable (NT). Within the diarrheagenic pathotypes, the highest ratio of EPEC 13 (52%) was observed, and this was followed by EAEC 5 (20%), EIEC 3 (12%), EHEC 2 (8%) and ETEC 2 (8%) as shown in Fig. 1A,B.
Molecular characterization of E. coli pathotypes and phylogroups. (A) 2% Agarose gel presenting amplified products of multiplex PCR for pathotyping. L represents ladder while lane A, B and C shows the band of eae corresponding to aEPEC while lane D shows the band of eae as well as bfp corresponding to typical EPEC. E Lane presents the band of vt corresponding to EHEC. (B) L represents ladder and lane F shows band for aggR corresponding to EAEC while lane G shows the band of ipaH corresponding to EIEC, and lane I shows band st corresponding to ETEC. (C) 2% Gel image showing Agarose gel presenting amplified products of multiplex PCR for phylotyping combination of four different amplicons, ArpA, chuA, yjaA TSPE4.C2, having 400bps, 288bps, 211bps, 152bps size, representing E. coli different phylogroups isolated from stunted growth children. First lane L shows 100 bp molecular weight ladder, lane A & B shows bands for E. coli clade 1, C lane indicates B2, lane D indicates C phylogroup while lane E presents F phylogroup and F lane shows E phylogroup.
Recovered E. coli phylogroups
Amongst the isolated E. coli strains from 64 samples, 18 of them belonged to B2 (28.12%), 8 belonged to E. coli Clade 1 (12.50%), 8 belonged to A (12.5%), 7 belonged F (10.93%), 6 belonged to E (9.37%), whereas 4 belonged to C (6.25%), and 2 belonged to D (3.12%). 12 of them were non-typeable (18.75%) and none of the isolates belonged to B1 as illustrated in Fig. 1C.
Diversity patterns representative of pathotypes and phylogroups
Microbial diversity of stunted individuals (n = 36) for different pathotypes and phylogroups (both in terms of taxonomy using OTUs and function using MetaCyc pathways recovered from PICRUSt2) was estimated using rarefied richness estimates. There was no significant difference in richness when the stunted samples were organized according to pathotypes. However, when the samples were organised under phylogroups, significant differences in richness were observed during pair-wise comparisons. This affected both taxonomy/composition (between A and B2; A and F; Clade-1 and F) and their recovered metabolic functions (between A and B2; B2 and C; B2 and NT; F and NT; and Clade-1 and NT). Highest richness in terms of taxonomy/composition was observed for phylogroups E and A, whilst phylogroup Clade-1 have shown maximum richness in terms of recovered metabolic functions (Fig. 2).
Beta diversity among different groups were analysed using different dissimilarity indices i.e. Bray–Curtis (for composition) and Hierarchical Meta-storm (HMS; for function). No significant difference in both was observed among pathotypes (Fig. 3A,C). On the other hand, beta diversity varied significantly (PERMANOVA) between different phylogroup for both composition (30% variability explained; p = 0.001) and function (28% variability explained; p = 0.094), respectively (Fig. 3B,D).
Principal coordinate analysis (PCoA) using different several dissimilarity indices [Bray–Curtis distance and hierarchical meta-storm (HMS)] where ellipses were drawn using 95% confidence intervals based on the standard error of ordination points for a given category. Beneath each figure are the R2 values (along with p-values if significant) calculated from PERMANOVA.
To identify the key drivers which result in the changes in beta diversity among the samples, the ‘BVSTEP’ routine was used. The method conserves beta diversity patterns between the samples whilst reducing the feature space, whether it is OTUs or pathways (KEGG KOs and MetaCyc pathways), by following a permutation approach that iterates through subset of features. The method also uses a quality-of-fit criteria, correlation coefficient R, to assess how much beta diversity is conserved between samples when considering a select subset of features. The resulting highly variable OTUs belonged to the genera Prevotella, Faecalibacterium, Streptococcus, Blautia, Erysipelatoclostridium and Megasphaera, and were increased in the EIEC Group. Prevotella, Faecalibacterium, Blautia, Erysipelatoclostridium and Megasphaera were found to be increase in EHEC group as compared to the other groups. Also, Streptococcus and Erysipelatoclostridium were found to be the dominant genera of OTUs in ETEC and EAEC group, respectively (Fig. 4A). In terms of organisation under phylogroups, OTUs belonging to the genera Prevotella and Blautia were more abundant in phylogroup E, with Faecalibacterium, Erysipelatoclostridium and Megasphaera comparatively more abundant in phylogroup A. In phylogroup B2 and F, OTUs of Streptococcus were more abundant as compared to the other groups (Fig. 4B). After applying BVSTEP routine to the PICRUSt2 tables, and irrespective of sample organization used whether pathotypes or phylogroups, we did not find much variation, and only a subset comprising KEGG KOs K00100 and K00854 (encoding for butanol dehydrogenase and Xylulokinase) to have variation between the samples. (Fig. 5A). Similarly, two MetaCyc pathways (DENOVOPURINE2-PWY and GLYOXYLATE-BYPASS) encoding denovopurine-2 and glyoxylate bypass was found to be the main source of variability amongst the samples (Fig. 5B).
Expression of significant OTUs identified from the BVSTEP routine with samples organized under a given convention, whether it is pathotypes (A) or phylogroups (B). The information, and the taxonomy of OTUs from the top subsets is given below the figure with the correlation between Bray–Curtis distances between a particular subset and the full OTU table given in parenthesis.
Expression of significant KEGG Orthologs KOs (A), and MetaCyc pathways (B) identified from the BVSTEP routine with samples organized under a given convention, whether it is pathotypes or phylogroups. For the given data, only one subset was returned with > 0.95 correlation with further details including correlation values given below the figures.
Regression analyses using generalised linear latent variable model (GLLVM) procedure
We ran two different models in GLLVM to associate microbial abundance of top 100 genera with the environmental covariates (Age, Height, and Weight, Gender, pathotypes, phylogroups and Stunting Status), one for samples characterised under pathotypes and one for phylogroups, respectively (Figs. 6, 7, 8 and 9). Here, we have also included the normal samples as a reference (n = 39 with 36 stunted and 3 normal samples). Recovery of beta coefficients on the basis of pathotypes have shown that Anaerostipes and Libanicoccus were negatively associated with height and positively associated with the stunting status. Weight was positively associated with the beneficial flora belonging to Lachnospiracae, Weissella and Clostridium sensu stricto, whilst negative associations were observed for Paraprevotella, Ruminococcus, Lachnospiraceae (Figs. 6 and 7).
Beta-coefficients returned from the GLLVM procedure for covariates considered in this study by considering top 100 most abundant genera incorporating continuous data (Age, Height, and Weight) as well as categorical labelling of samples (Gender, Male, and Pathotypes). Those coefficients which are positively associated with the microbial abundance of a particular species are represented in red colour whilst those that are negatively associated are represented with blue colour, respectively. Where the 95% confidence interval of the beta-coefficients crosses the 0 boundary, i.e., they are insignificant, are represented by grey colour. Since the collation of the OTUs were done at Genus level, all those OTUs that cannot be categorized based on taxonomy are collated under “Unknown” category. The figure only shows beta-coefficients for 48 genera with the remaining ones shown in Fig. 7.
Continuation of Fig. 6 for the remaining beta-coefficients for 52 genera returned from the GLLVM procedure when considering pathotypes in the analysis.
Analysis on the basis of phylogroups have shown that the males showed positive association with normal flora belonging to Actinomyces, Ruminococcaceae, and Lachnospiraceae. Height was positively associated with Tyzzerella and negatively associated with stunting. However, positive association of Eggerthella was observed for stunting. Whilst all the phylogroups have shown negative association with the bacteria belonging to normal flora of human gut, B2 phylogroup has shown a pronounced negative association with majority of normal flora, i.e., Anaerovibrio, Ruminococcus, Paeniclostridium, Lachnospiraceae, Terrisporobacter Baciilus and Prevotella (Figs. 8 and 9).
Beta-coefficients returned from the GLLVM procedure for covariates considered in this study by considering top 100 most abundant genera incorporating continuous data (Age, Height, and Weight) as well as categorical labelling of samples (Gender, Male, and Phylogroups). Those coefficients which are positively associated with the microbial abundance of a particular species are represented in red colour whilst those that are negatively associated are represented with blue colour, respectively. Where the 95% confidence interval of the beta-coefficients crosses the 0 boundary, i.e., they are insignificant, are represented by grey colour. Since the collation of the OTUs were done at Genus level, all those OTUs that cannot be categorized based on taxonomy are collated under “Unknown” category. The figure only shows beta-coefficients for 49 genera with the remaining ones shown in Fig. 9.
Continuation of Fig. 8 for the remaining beta-coefficients for 51 genera returned from the GLLVM procedure when considering phylogroups in the analysis.
Quantifying spatial structuring mechanisms in assemblages of microbial communities of stunted individuals
Lottery model for clade-based assembly were used with the underlying premise being that within a clade, closely related species will exhibit a very strong degree of competition i.e., of all the members in a given clade, the fittest member (or the first member to colonize) will become the most abundant and will dictate the community structure (Fig. 10). These clades were defined taxonomically within each genus. From these clades, lottery ‘winners’ were identified as those species which have > 90% abundance within each clade, with the x-axis representing lottery type clade prevalence as a fraction of the total number of samples, whilst the y-axis represents the diversity if multiple species within a clades are lottery winners. For clades where there is only one lottery winner, the winner diversity values go to zero. The bottom figure (Fig. 10A) shows the winners identified in each clade. The strongest lottery-like groups however have a high diversity and high winner prevalence (Lactobacillus and Catenibacterium) and indicate a more variable and random (stochastic) selection of OTUs as winners within the clade. This may also provide insights into how “stunting microbiome” is shaped. Such winners can be thought of as strong competitors regardless of disease aetiology and are more resilient. Conversely, the lottery-type clades at the bottom always select the same species, suggesting their link to stunting (can be deemed as signature species to stunting). Escherichia-shigella, Enterococcus and Porphyromonas are the prevalence winners and shows a strong link with stunting. The inset (Fig. 10B) is the overall assembly processes breaking down the community assembly mechanisms into the ecological processes using QPE estimates. QPE estimates revealed that the amount of stochasticity (Dispersal limitation, Homogenizing dispersal, and ecological drift explain roughly 55% of community assembly, whilst amongst the deterministic causes (Homogenizing Selection, and Variable Selection), high variable selection implies multiple stable community structures possible in stunted individuals. We then calculated the core microbiome (with 50% minimum prevalence) to identify the signature species associated with stunting. Figure 10C shows that taxa on the right side of the heatmap are not only highly prevalent but also have very high abundances. These taxa include Escherichia–Shigella, Streptococci and Enterococci.
Community structure of samples with “stunted” status. (A) Competitive lottery model for conceptualising community assembly of these samples with genera displaying lottery-like behaviour plotted by prevalence and diversity of the ‘winning’ OTUs; these ‘winning’ OTUs (> 90% abundance for that clade) from each clade are given in the bottom table. The inset (B) gives the assembly processes using QPE estimates. (C) Core microbiome of samples with “stunted” status where we have used a minimum prevalence of 50% of OTUs in these samples as the lower limit.
Other than finding marginal changes in microbiota associated with Gentamicin usage and whether the child was breastfed at birth, we did not find any other observed clinical covariates explaining variability in microbial community structure (Supplementary Table 2).
Discussion
Environmental Paediatric Enteropathy (EEP) is widespread among malnourished children and has been associated with prolonged exposure to enteric gut pathogens including Campylobacter species, Shigella spp, enteroinvasive E. coli, heat-stable toxin-producing enterotoxigenic E. coli, shiga toxin-producing enterotoxigenic E. coli and typical enteropathogenic E. coli, norovirus, Cryptosporidium spp. and Giardia spp leading to stunting at early age19,20,21,22. This study was designed to explore the effect of exposures of different pathogenic strains of E. coli at early age of life and their impact on microbiota in low socioeconomic semi-urban/urban setting.
As majority of the E. coli strains in human gut are part of normal microbiota, infection due to pathogenic strains of E. coli can be determined by the presence of virulence genes. The pathogenic strains can be classified either on the basis of pathotypes or phylogroups9,15. Pathotypes of diarrheagenic E. coli differ from each other in terms of virulence genes and how they interact with eukaryotic cells. Previous studies from Pakistan have shown association of EPEC and EAEC pathotypes with the paediatric population and preterm mothers9,10,11,12,13. In this present study, E. coli were isolated from the gut of asymptomatic children showing stunted growth and were further screened for pathotypes distribution and associated bacterial taxa, and were compared against a normal cohort. The results suggested that a total of 25 (39.06%) were positive for five of diarrheagenic E. coli pathotypes, presenting EPEC 13 (52%) as a dominant pathotype followed by EAEC 5 (20%), EIEC 3(12%), EHEC 2(8%), and ETEC 2(8%), respectively. Our study corroborates with similar study carried out in India, Kenya, Tehran where EPEC was found as the dominant pathotype in children under 5-years of age23,24,25.
Phylogroup characterization of gut E. coli provides insight into the genetic structure of the phylogroups (A, B1, B2, C, D, E, and F) which differ in their phenotypic traits, ecological niche and virulence potential15. In this study, the phylogrouping analysis has shown that majority of the E. coli strains isolated from stunted growth children belonged to Phylogroup B (28.12%). Phylogroup B2 had been earlier described to be the most prevalent phylogroup associated with infection such as diarrhoea, urinary tract infection and colorectal neoplasm, as it carries more virulence genes that any other group26,27,28. Moreover, B2 has the ability to persist for longer periods in infants than other E. coli strains and therefore can influence the microbiota29. Therefore, the association of B2 phylogroup with stunted growth in our study might provide a glimpse of associated illness that might lead to stunting.
The colonization of enteropathogens subsequently results in reduced diversity and abundance of useful gut microbial flora negatively influencing the functions of gut19,30,31,32 by hampering production of important vitamins, antioxidants as well as hindering the complete digestion of food resulting in further loss of 10–15% nutrition as it passes unused through the digestive system33. In this study, we analysed the effect of colonization of different pathotypes and phylogroups of E. coli on the gut microbial diversity, composition and functional makeup in stunted growth children. After identification of pathotypes and phylogroups, a subset of 39 faecal samples were sequenced and analysed for gut microbiome analysis. Both taxonomic and functional diversity were different between the recovered phylogroups of E. coli suggesting that different phylogroups have varying ability to compete for niche and nutrients. The taxonomic richness was found to be lower in samples where Phylogroup B2 and F colonized. However, highest functional richness was found to be in B2 phylogroup which we hypothesize is due to enhance competitive nature of the strains34. This results in alternative pathway to avail nutrient and energy. Diversity patterns were differentially more pronounced when samples were categorized under phylogroups as compared to when they were categorized under pathotypes.
Majority of the key players to vary significantly among samples are strict anaerobes suggesting that colonization of E. coli phylogroups alter the gut niche. In terms of function, Key KEGG orthologs which varied significantly encoded for butanol dehydrogenase and Xylulokinase. Butanol dehydrogenase (K00100) uses butanol as a source of carbon for energy production in nutrient depleted niche and also helps protect against anaerobic stress35. Xylulokinase (K00854) can convert xylose, a pentose, to xylulose 5-phosphate and is also a player in energy production36. Denovoprine-2 and glyoxylate bypass were found to be the two key pathways which varied significantly between the phylogroups. Denovoprine-2 is used in nucleotide biosynthesis process resulting in more energy generation for enhanced colonization and virulence37. Glyoxylate bypass or shunt also varied significantly among is used by microbes when they are in nutrient stress to conserve carbon atoms for gluconeogenesis and promote energy production38. Overall colonization of different phylogroups affect changes in carbohydrate metabolism, and nucleotide synthesis which might be attributed to the competitive nature of the strains.
To further investigate this, GLLVM analysis was carried out by including normal cohort. Colonization of pathotypes were negatively associated with the normal microflora (Paraprevotella, Ruminococcus, Lachnospiraceae) indicating that these strains reduced the beneficial bacteria in the gut which might have led to stunting. Analysis on the basis of phylogroups highlighted height to be positively associated with Tyzzerella and negatively associated with stunting, thus indicating its potential role in normal growth and development39. Among the pathotypes recovered, colonization of B2 phylogroup has shown the most drastic effect on normal microflora as indicated by a negative association with normal flora belonging to genera Anaerovibrio, Ruminococcus, Paeniclostridium, Lachnospiraceae, Terrisporobacter, Baciilus and Prevotella.
Core microbiome analysis for clade-based assembly has shown that Escherichia-shigella, Enterococcus and Porphyromonas are more prevalent and strongly linked with stunting. This is in line with previous studies which have shown that Escherichia-shigella, Streptococcus and Lactobacillus are associated with stunted growth in children39. Our study has shown that colonization of different phylogroups of E. coli (especially B2) significantly changed the intestinal microbiota of stunted growth children. This leads to the conclusion that a disruption in the equilibrium between the bacterial EED aetiology and co-residing bacterial communities might well be linked with subsequent poor growth performance. Furthermore, investigation involving a larger cohort and inclusion of sizeable control group is desirable, as these were the major limitations of the present study.
Conclusions
The present study shows that exposure to pathogenic strains of E. coli drastically changes the gut microbiota of stunted growth children leading to reduction in beneficial microbiota and altered microbial diversity. E coli strains belonging to phylogroup B2 by virtue of their competitive nature had a profound effect on the gut microbial ecology as compared to strains belonging to other phylogroups and pathotypes. Understanding the vulnerable child gut microbiome in relation to the exposome is a route to develop suitable intervention strategies to reduce the burden of EED aetiologies and hence interlinked stunting.
Data availability
The dataset presented in this study is available under ENA repository PRJEB58411 with the meta data provided in the supplementary material.
References
Huey, S. L. & Mehta, S. Stunting: The need for application of advances in technology to understand a complex health problem. EBioMedicine 6, 26–27. https://doi.org/10.1016/j.ebiom.2016.03.013 (2016).
Pennisi, E. The right gut microbes help infants grow. Science 351(6275), 802. https://doi.org/10.1126/science.351.6275.802 (2016).
Gleason, K. M. et al. Stunting is associated with blood lead concentration among Bangladeshi children aged 2–3 years. Environ. Health 15(1), 103. https://doi.org/10.1186/s12940-016-0190-4 (2016).
UNICEF. First Call for Children: World Declaration and Plan of Action from the World Summit for Children. Convention on the Rights of the Child. (UNICEF, 1990).
UNICEF. National Nutrition Survey 2018 Key Findings Report. https://www.unicef.org/pakistan/sites/unicef.org.pakistan/files/2019-07 (UNICEF Pakistan, 2019).
Cesare, D. et al. Geographical and socioeconomic inequalities in women and children’s nutritional status in Pakistan in 2011: An analysis of data from a nationally representative survey. Lancet Glob. Health 3(4), e229–e239. https://doi.org/10.1016/S2214-109X(15)70001-X (2015).
Amour, C. et al. Epidemiology and impact of Campylobacter infection in children in 8 low-resource settings: Results from the MAL-ED study. Clin. Infect. Dis. 63(9), 1171–1179. https://doi.org/10.1093/cid/ciw542 (2016).
Platts-Mills, J. A. et al. Association between enteropathogens and malnutrition in children aged 6–23 mo in Bangladesh: A case–control study. Am. J. Clin. Nutr. 105(5), 1132–1138. https://doi.org/10.3945/ajcn.116.138800 (2017).
Bokhari, H. et al. Escherichia coli pathotypes in Pakistan from consecutive floods in 2010 and 2011. Am. J. Trop. Med. Hyg. 88(3), 519–525. https://doi.org/10.4269/ajtmh.12-0365 (2013).
Younas, M. et al. Characterization of enteropathogenic Escherichia coli of clinical origin from pediatric population in Pakistan. Trans. R. Soc Trop. Med. Hyg. 110(7), 414–420. https://doi.org/10.1093/trstmh/trw047 (2016).
Khalil, U. et al. Phenotypic and genotypic characterization of enteroaggregative Escherichia coli isolates from pediatric population in Pakistan. APMIS 124(10), 872–880. https://doi.org/10.1111/apm.12577 (2016).
Rana, F. et al. Resistance patterns of diversified phylogroups of Escherichia coli associated with mothers having history of preterm births in Pakistan. J. Matern.-Fetal Neonatal Med. 30(1), 68–73. https://doi.org/10.3109/14767058.2016.1163538 (2016).
Mushtaq, S. et al. Phylogenetic diversity of Escherichia coli strains producing NDM-type carbapenemases. J. Antimicrob. Chemother. 66(9), 2002–2005. https://doi.org/10.1093/jac/dkr226 (2011).
McInnes, P., & Cutting, M. Manual of Procedures for Human Microbiome Project: Core Microbiome Sampling Protocol A, HMP Protocol# 07-001, Version Number: 12.0.: Health NIo, ed2010. (2010).
Clermont, O., Christenson, J. K., Denamur, E. & Gordon, D. M. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 5(1), 58–65. https://doi.org/10.1111/1758-2229.12019 (2013) (epub 2012 Dec 24).
Cady, N. C., Stelick, S. & Batt, C. A. Nucleic acid purification using microfabricated silicon structures. Biosens. Bioelectron. 19(1), 59–66. https://doi.org/10.1016/S0956-5663(03)00123-4 (2003).
Minalla, A.R., Dubrow, R., Bousse, L.J. (Eds.) Feasibility of high-resolution oligonucleotide separation on a microchip. in Microfluidics and BioMEMS. https://doi.org/10.1117/12.443044 (International Society for Optics and Photonics, 2001).
Ijaz, U. Z. et al. Comprehensive longitudinal microbiome analysis of the chicken cecum reveals a shift from competitive to environmental drivers and a window of opportunity for Campylobacter. Front. Microbiol. 9, 2452. https://doi.org/10.3389/fmicb.2018.02452 (2018).
Trehan, I. & Manary, M. J. An important chapter in the infection–malnutrition story. Lancet Glob. Health 4(7), e430–e431. https://doi.org/10.1016/S2214-109X(16)30110-3 (2016).
Platts-Mills, J. A. et al. Use of quantitative molecular diagnostic methods to assess the aetiology, burden, and clinical characteristics of diarrhoea in children in low-resource settings: a reanalysis of the MAL-ED cohort study. Lancet Glob. Health 6(12), e1309–e1318. https://doi.org/10.1016/S2214-109X(18)30349-8 (2018).
Petri, W. A. et al. Enteric infections, diarrhea, and their impact on function and development. J. Clin. Invest. 118, 1277–1290. https://doi.org/10.1172/JCI34005 (2008).
Fahim, S. M., Das, S., Gazi, A., Mahfuz, M. & Ahmed, T. Association of intestinal pathogens with faecal markers of environmental enteric dysfunction among slum-dwelling children in the first 2 years of life in Bangladesh. Trop. Med. Int. Health 23(11), 1242–1250. https://doi.org/10.1111/tmi.13141 (2018).
Ajjampur, S. et al. Closing the diarrhoea diagnostic gap in Indian children by the application of molecular techniques. J. Med. Microbiol. 57(11), 1364–1368. https://doi.org/10.1099/jmm.0.2008/003319-0 (2008).
Dallal, M.S., Khorramizadeh, M., & Moez Ardalan, K. Occurrence of enteropathogenic bacteria in children under 5 years with diarrhoea in south Tehran. EMHJ-East. Mediterr. Health J. 12(6), 792–797. https://apps.who.int/iris/handle/10665/117154 (2006).
Pavlinac, P. B. et al. High-risk enteric pathogens associated with HIV-infection and HIV-exposure in Kenyan children with acute diarrhea. AIDS (London, England). 28(15), 2287. https://doi.org/10.1097/QAD.0000000000000396 (2014).
Halaji, M. et al. Phylogenetic group distribution of uropathogenic Escherichia coli and related antimicrobial resistance pattern: A meta-analysis and systematic review. Front. Cell Infect. Microbiol. 12, 790184. https://doi.org/10.3389/fcimb.2022.790184 (2022).
Raisch, J. et al. Colon cancer-associated B2 Escherichia coli colonize gut mucosa and promote cell proliferation. World J. Gastroenterol. 20(21), 6560–6572. https://doi.org/10.3748/wjg.v20.i21.6560 (2014).
Nowrouzian, F. L. et al. Escherichia coli B2 phylogenetic subgroups in the infant gut microbiota: Predominance of uropathogenic lineages in Swedish infants and enteropathogenic lineages in Pakistani infants. Appl. Environ. Microbiol. 85(24), e01681-e1719. https://doi.org/10.1128/AEM.01681-19 (2019).
Sang, W. K., Oundo, V. & Schnabel, D. Prevalence and antibiotic resistance of bacterial pathogens isolated from childhood diarrhoea in four provinces of Kenya. J. Infect. Dev. Ctries. 6(07), 572–578. https://doi.org/10.3855/jidc.2196 (2012).
Sudershan, R. et al. coli pathotypes and their antibiotic resistance in young children with diarrhea in Hyderabad, India. Int. J. Curr. Microbiol. Appl. Sci. 3(9), 647–654 (2014).
Dewey, K. G. & Mayers, D. R. Early child growth: How do nutrition and infection interact?. Matern. Child Nutr. 7, 129–142. https://doi.org/10.1111/j.1740-8709.2011.00357.x (2011).
Gupta, S. S. et al. Metagenome of the gut of a malnourished child. Gut Pathogens 3(1), 7. https://doi.org/10.1186/1757-4749-3-7 (2011).
Iyengar, G. V. & Nair, P. P. Global outlook on nutrition and the environment: Meeting the challenges of the next millennium. Sci. Total Environ. 249(1–3), 331–346. https://doi.org/10.1016/S0048-9697(99)00529-X (2000).
Duan, X., Wang, G., Liu, H., Xue, J. & Zhang, J. Key enzymes in butanol fermentation by a facultative anaerobe Bacillus sp. TSH1. Sheng Wu Gong Cheng Xue Bao (Chinese) 29(5), 620–629 (2013).
Micenková, L. et al. Microcin determinants are associated with B2 phylogroup of human fecal Escherichia coli isolates. Microbiologyopen. 5(3), 490–498. https://doi.org/10.1002/mbo3.345 (2016) (epub 2016 Mar 14).
Lozupone, C. et al. Identifying genomic and metabolic features that can underlie early successional and opportunistic lifestyles of human gut symbionts. Genome Res. 22(10), 1974–1984. https://doi.org/10.1101/gr.138198.112 (2012) (epub 2012 Jun 4).
Goncheva, M. I., Chin, D. & Heinrichs, D. E. Nucleotide biosynthesis: The base of bacterial pathogenesis. Trends Microbiol. 30(8), 793–804. https://doi.org/10.1016/j.tim.2021.12.007 (2022).
Dai, Z. et al. Disinfection exhibits systematic impacts on the drinking water microbiome. Microbiome 8, 42. https://doi.org/10.1186/s40168-020-00813-0 (2020).
Vonaesch, P. et al. Stunted childhood growth is associated with decompartmentalization of the gastrointestinal tract and overgrowth of oropharyngeal taxa. Proc. Natl. Acad. Sci. USA 115(36), E8489–E8498. https://doi.org/10.1073/pnas.1806573115 (2018) (epub 2018 Aug 20).
Acknowledgements
HB was partially supported by uBiome, US. UZI is further supported by a NERC Independent Research Fellowship (NE/L011956/1) and EPSRC (EP/P029329/1 and EP/V030515/1).
Author information
Authors and Affiliations
Contributions
H.B. conceived and designed the study and contributed towards reagents and materials whilst U.Z.I. directed the bioinformatics strategies with support from O.G.. I.Z. and Z.N. performed the experimental work. M.H.H., N.M. and A.K. contributed towards diagnosis and recruitment of stunted patient. Z.N., I.Z., U.Z.I., O.G. and H.B. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Aziz, I., Noreen, Z., Ijaz, U.Z. et al. A prospective study on linking diarrheagenic E. coli with stunted childhood growth in relation to gut microbiome. Sci Rep 13, 6802 (2023). https://doi.org/10.1038/s41598-023-32491-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-32491-x
- Springer Nature Limited