Abstract
Based on recent developments in glioblastoma subtyping, we examined DARPP32 (PPP1R1B), a neuronal marker against STAT5 and STAT3 that are pro-oncogenic in glioblastoma. mRNA ratios of DARPP32, STAT1, STAT3, STAT5A and STAT5B were assessed in routinely diagnosed gliomas s including a series of glioblastomas from patients (n = 67) treated with chemoradiotherapy (temozolomide), out of which 88 % had sequencing validated IDH-negative disease. DARPP32/STAT1 (p = 0.0007), DARPP32/STAT3 (p = 0.0004) and DARPP32/STAT5B (p = 0.0039) ratios were significantly higher in grade II and III as compared to grade IV tumours. The same high ratios were also associated with absence of immunohistochemically assessed AKT/PKB phosphorylation and survivin protein expression. High DARPP32/STAT3, DARPP32/STAT5B, and STAT5B/STAT3 ratios were associated with longer patient progression free (PFS) and overall survival (OS). Upon multivariate analysis, total/subtotal removal of the tumour (HR:0.431; 95%CI:0.241–0.771, Wald p = 0.005), high DARPP32/STAT5B (HR:0.341; 95%CI:0.169–0.690; Wald p = 0.003) and STAT5B/STAT3 mRNA ratios (HR:0.480; 95%CI:0.280–0.824; Wald p = 0.008) were independent favorable parameters for prolonged PFS. Extent of surgery (HR:0.198; 95%CI:0.101–0.390; p < 0.001) and high DARPP32/STAT5A ratios (HR:0.320; 95%CI:0.160–0.638, p = 0.001) were independently predictive for longer OS. The presented approach is applicable for prospective validation and appears promising towards an effective glioblastoma patient stratification in addition to IDH mutations. These data may contribute to understanding the biology of gliomas with respect to their potential neuronal characteristics and justify STAT-inhibiting therapeutic interventions in the same tumour system.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Diffuse gliomas, i.e., tumours with astrocytic, oligodendrocytic, ependymal or mixed cell characteristics, are the most common primary brain tumours in adults and are categorized with increasing grade of malignancy [1]. About 70 % of gliomas are glioblastomas, WHO grade IV, aggressive and currently incurable [2]. Gliomas are presumed to arise from neural stem cells that exist in the brain. The acquisition of IDH1 & 2 mutations in these progenitors is considered as a major event in gliomagenesis [3, 4]. Gliomas developing on an IDH-mutant background are usually of grade II or III with oligodendrocytic or astrocytic or mixed features, and secondary glioblastomas, i.e., tumours with a known favorable outcome. The limited incidence (<5 %) of IDH1-mutant de novo glioblastomas [5] that have a better prognosis than IDH-wild-type ones, presumably reflects a grade II–IV series of events that have gone clinically unnoticed [6]. Hence, all evidence prompts for distinguishing gliomas into IDH-mutant and IDH-wild type disease [6, 7]. Strikingly, IDH-mutant disease is included in the proneural phenotype / subtype [6, 8–10], while it largely overlaps with the methylator phenotype [11], all of which are also associated with a better prognosis of gliomas. However, despite these insights towards comprehending gliomagenesis and although standard treatment (resection, radiotherapy with concomitant and adjuvant temozolomide) appears to benefit patients with glioblastoma [12], the expected median survival is still 12–15 months, while less than 10 % of patients, those with favorable disease characteristics, survive for more than 5 years [13]. Hence, understanding the remaining more than 90 % of glioblastomas and finding alternative treatment options for these patients remains a great challenge.
At the other end of the favorable proneural lies the mesenchymal subtype with worst prognosis [8, 9]. Additional subtypes have been described between these two, such as neural and classical [9], which are backed by specific genomic alterations [14], or proliferative [8], while there is always space for new subtypes to be identified if larger series are examined. As described [9], proneural subtype resembles oligodendrocytes; neural, mainly astrocytes and neurons; classical, murine astrocytes; and, mesenchymal, cultured astroglia characterized by high motility, invasiveness and angiogenic capacity. Main contributors to the acquisition of the mesenchymal phenotype appear to be C/EBPbeta and STAT3 [15]. STAT3 belongs to the Signal Transducers and Activators of Transcription, a family of transcription factors. STAT3 plays key roles in neural stem cell and astrocyte development, and has been found to play dual tumour suppressive and oncogenic roles in gliomas depending on the mutational profile of the tumour (reviewed in [16]). In addition, STAT3 inhibition sensitizes glioblastoma cells to alkylating agents and EGFR inhibitors [17]. Less is known about the role of other usually investigated members of the STAT family in gliomas. Stat5b may be involved in glioblastoma cell growth, cell cycle progression, invasion and migration [18]. Stat1, a potent inhibitor of cell growth and promoter of apoptosis, is ubiquitously expressed in glial tumours, as well as in normal brain tissues [19].
With respect to the subtypes described above it is of interest that neuronal characteristics were represented, except for the neuronal subtype, in the classical and mesenchymal subtypes as well, but not in the proneural one [9]. Herein, we profiled neuronal vs. STAT gene expression in a series of gliomas and associated these profiles with glioblastoma patient outcome. As a neuronal marker we selected a molecule indicative of early neuronal differentiation, PPP1R1B (protein phosphatase 1, regulatory (inhibitor) subunit 1B or dopamine and cAMP-regulated neuronal phosphoprotein 32, DARPP32). DARPP32 is a dual function protein that is capable of inhibiting either protein phosphatase-1 or protein kinase-A, according to the site it is phosphorylated and according to the signal inducing its phosphorylation. DARPP32 is a long known gene and protein [20] that is expressed in most specialized structures in the normal brain [21]. DARPP32 has mainly been studied in the context of central nervous system physiology and pharmacology [22]. Initially regarded as a dopamine signalling mediator, this molecule is currently considered as a signalling hub molecule which may improve the general performance of the neuronal circuits in which it is expressed (reviewed in [23]). DARPP32 may also promote epithelial oncogenesis (Beckler, Moskaluk et al. 2003), while its truncated isoform, t-DARPP, has been associated with resistance to trastuzumab in breast cancer (Belkhiri, Dar et al. 2008). However, full-length DARPP32 antagonizes t-DARPP trastuzumab resistance [24], while in some epithelial cancers DARPP32 may play tumour suppressive (Pimenta, Horta et al. 2007) and migration inhibitory roles [25].
In addition to STAT1, STAT3, STAT5A, STAT5B and DARPP32 mRNA profiles we also examined AKT/PKB phosphorylation and survivin expression in glioma tissues. These markers were chosen because DARPP32 protein is induced through PI3K/AKT activation by the brain-derived neurotrophic factor in the brain [26], while t-DARPP promotes tumour cell growth by activating AKT [27]; and also because STAT3 is essential for survivin expression [28, 29].
Materials and Methods
Tissue Material
Previously diagnosed histologic material from stereotactic biopsies and partial or complete resections of gliomas (formalin-fixed paraffin-embedded tissues [FFPE]) was retrieved from the archives of the Dept of Pathology, Aristotle University of Thessaloniki School of Medicine (n = 135) and from the Tumour Tissue Repository of the Hellenic Cooperative Oncology Group (HeCOG) (n = 70). Out of these 205 tumours, 187 tissue blocks were evaluated as eligible for processing for molecular and immunohistochemical investigations based on adequacy of material, as assessed upon histologic examination. Patient demographic and tumour histologic characteristics are shown in Table 1. With respect to de novo and secondary glioblastomas, the latter were characterized as such (a) in the case of known clinical history of a lower grade tumour, and (b) when areas of grade III or II were present histologically upon examination of all available tissue blocks. The above tumours were used for Tissue MicroArray (TMA) construction (2 × 1.5 mm cores per tumour) and for DNA and RNA extraction. Morphologically non-neoplastic brain (NNBT) tissue was available in eight cases in the same or in separate tissue blocks and was also included in the TMAs.
Patients
Clinical data were available for 67 patients with glioblastoma that had been treated during various time periods (2005–2008) in the Dept of Medical Oncology, Aristotle University of Thessaloniki School of Medicine and in the Dept of Radiation Therapy, both located in Papageorgiou Hospital (54 patients) or in a private clinic (13 patients). As shown in Table 2, the extent of surgical removal of the tumour was known in 65/67 cases. All patients were chemo-naïve. Only one patient with clinically secondary glioblastoma presented in this group; however, histological assessment of all available tissue material showed additional 2 cases with grade II and III areas as described above. Hence, the histologically de novo glioblastomas were 64 out of 67 (95.5 %). Except for two cases where early tumour death occurred, all patients received the same initial treatment with radiotherapy and concomitant temozolomide followed by 6 months adjuvant temozolomide administration (CCRT) [30]. Patients were followed every 3 months with physical examination and brain MRI up to 46 months or until death was reported. Upon relapse, patients (N = 22) received 1st line chemotherapy (temozolomide [N = 7]; bevacizumab and irinotecan [N = 10]; BCNU and irinotecan [N = 3]) with the addition of bevacizumab in one case. Upon further relapse (N = 7), patients received erlotinib, irinotecan and bevacizumab as 2nd line chemotherapy.
Written informed consent had been received by the patients for the use of biologic material and the study was approved by the Bioethics Committee of the School of Medicine, Aristotle University of Thessaloniki under the title: “Molecular and genetic markers associated with response to combined radio/chemotherapy in patients with glioblastoma” (approval #: A13723).
DNA and RNA
from routinely processed formalin-fixed paraffin-embedded tissues were extracted with standard procedures upon enrichment in tumour cells with macrodissection (>50 % tumour cells in the molecular sample in all cases). For DNA extraction the QIAamp mini kit (Qiagen, Hilden, D) was used, while for RNA extraction, overnight paraffin tissue fragment lysis with proteinase K at 56 °C followed by application of the TRIZOL-LS reagent (Life Technologies, Paisley, UK) and reverse transcription with random primers and the Superscript III system (Life Technologies). Commercial reagents were applied according to the instructions of the manufacturers.
Relative Gene Expression
was assessed with RTQ-PCR by using premade Taqman-MGB assays in an ABI7500 system (Applied Biosystems / Life Technologies) equipped with the SDS v1.4 software by using the default conditions. The assays used (NM reference, amplicon size, location) were: for DARPP32, Hs00259967_m1 (NM_001242464.1, NM_032192.3, NM_181505.3, 90 bp, exons 4–5); for STAT1, Hs01014005_m1 (NM_007315.3, NM_139266.2, 67 bp, exons 3–4); for STAT3, Hs01047580_m1 (NM_003150.3, NM_139276.2, NM_213662.1, 87 bp, exons 3–4); for STAT5A, Hs00234181_m1 (NM_003152.3, 63 bp, exons 12–13); and, for STAT5B, Hs00560035_m1 (NM_012448.3, 91 bp, exons 2–3). GUSB (beta-glucuronidase, assay 4333767 F) was selected as the endogenous reference because it does not seem to be represented in pseudogenes and also because it has been identified as one among the best preserved mRNA targets in FFPE tissues [31, 32]. Relative quantification (RQ) analysis was assessed according to the 2^-dCT method that can be applied for small amplicons, whereby dCT = CTtarget—CTGUSB. Samples were tested in 10 ul reactions and were run in duplicates. Duplicate runs were also performed for 32 samples and yielded <1.5 cycles differences in ddCTs (dCTrun1—dCTrun2) per sample and per assay, which would correspond to < ±2-fold change in the evaluation of relative gene expression (inter-run validation). Exclusion criteria for RQ analysis were endogenous reference CT values higher than 36 for each sample; and, dCTs higher than 0.5 per duplicate pair. According to these criteria, in total 145/187 samples (77.5 %) were found eligible for further RQ analysis.
In order to investigate combined effects of DARPP32/STAT gene expression we also assessed RQ ratios for paired gene expression by omitting GUSB. For example, RQ ratio for DARPP32/STAT3 corresponded to 2^-ddCT, whereby ddCT = (CTDARPP32—CTGUSB)—(CTSTAT3—CTGUSB). This approach in fact annihilated reference gene bias for single marker results. That is, changes in single marker DARPP32 and STAT RQ values in one direction might be due to GUSB expression aberrations in the opposite direction; this condition can not be excluded for any endogenous reference, especially in paraffin samples where PCR efficiency unpredictably varies.
Glioma DNA Markers
MGMT promoter methylation, IDH1 and IDH2 mutations (exon 4 for both genes) and EGFR CN (gene copy numbers) were assessed in tumour DNA samples from patients with known follow-up only (Table 2). For MGMT, the SALSA MS-MLPA KIT ME011 (MRC-Holland, Amsterdam, NL) was used, which included probes for three distinct CpG-rich sites in the MGMT enhancer. Normal blood DNA and a reactive lymph node FFPE-DNA sample were used as controls. Products were electrophoresed on an ABI3130 genetic analyzer (Applied Biosystems / Life Technologies). This method gave informative results, i.e., all picks present in the non-digested sample with acceptable method internal controls and reference probe peaks, in 54/67 cases (80.6 %). The results were evaluated according to [33]; relative peak values ≤0.25 were characterized as unmethylated, relative peak values >0.25 as methylated; and, relative peak values >0.75 as completely methylated. The procedure was repeated twice to assess concordance of results (retention in the same category of methylation status).
For mutation analysis, cycle-sequencing with BigDye® Terminators v1.1 (Applied Biosystems) was performed on the ABI3130XL genetic analyzer with the following primers (sequences 5′-3) for IDH1 (nested PCR): forward (F), TCACCAAATGGCACCATACGA, reverse (R) GTTGAGATGGACGCCTATT; Fn, ACCAAATGGCACCATACGA and Rn, GCAAAATCACATTATTGCCAAC (348 bp → 164 bp); for IDH2: F, CCGTCTGGCTGTGTTGTTGCT, R, AGACAAGAGGATGGCTAGGC (326 bp).
Copy number variation (CNV) was assessed with real time PCR (ABI7900HT system) for the EGFR gene with the Hs02822119_cn (exon 10) and the Hs01426560_cn (exon 2) pre-designed TaqMan® Copy Number Assays (Applied Biosystems). The TaqMan® Copy Number Reference Assay RNase P was used as endogenous reference. Reactions were run in quadruplicates in an ABI7900HT system. Three normal blood DNA samples were included in each run as calibrator samples, along with a no-template control (NTC). Results (predicted copy numbers) were obtained automatically with the CopyCaller™ Software v1.0 in comparison to averaged calibrator values upon setting the evaluation threshold at Ct = 32. Average predicted copy numbers higher than 2.5 were considered as amplification.
Immunohistochemistry (IHC)
was performed on 2 μm TMA sections. Phosphorylation of Akt/PKB was investigated with an anti-rabbit monoclonal antibody that detects phosphorylated Ser473 on Akt1 and on corresponding sites on Akt2 and Akt3 proteins (#3787 L, clone 736E11, Cell Signaling Technologies, Beverly, MA). Expression of survivin (BIRC5) was assessed with an anti-mouse monoclonal antibody (#M3624, clone 12C4, DAKO, Glostrup, DK). In both cases, antibodies were diluted 1:50, DAKO Target Retrieval Solution S1699 was used for Akt/PKB and S2367 for survivin epitope unmasking, and anti-rabbit (K4011) and anti-mouse (K4007) Envision systems were used for target visualization. The cut-off for phospho-Akt was set at 5 % of positive tumour cells [34]; nuclear and cytoplasmic staining were separately assessed. With the monoclonal antibody used, survivin expression was identified as intra-nuclear spots in very few glioma cells among all grades, in accordance with observations in a variety of tumours with this antibody [35] but other than reported in former studies using different antibodies [36, 37]. Hence, a cut-off of 1 % was used for survivin positivity. The reported cytoplasmic positivity was not observed. Ki-67 staining (clone MIB-1, DAKO) had also been applied in the diagnostic setting with standard procedures and with 3 % as cut-off between low (II) and high (III and IV) grade tumours [38]
Statistical Analysis
Overall survival (OS) was calculated from initial diagnosis until patient death or last contact. Surviving patients were censored at the date of last contact. Progression free survival (PFS) was defined as the time interval between initial diagnosis and disease progression, which was assessed with clinical examination and with MRI (deaths without verified prior disease progression were also considered as events in the calculation of PFS). Disease progression was considered upon CCRT (initial treatment) only, since treatment in the 1st line was heterogeneous. Log-rank test was used to compare time to event distributions of subgroups. Cox proportional hazards regression models were used to assess the influence of the target genes examined on OS and PFS outcome.
Spearman’s rank correlation was used as a measure of association between all tested targets. The Mann–Whitney test was performed to compare mRNA RQ values with nominal variables. Receiver operating characteristic (ROC) curve analysis was performed to assess the relationship between mRNA expression of genes (corresponding RQ values) and 6-month progression free survival rate (PFS-6 month), which was used as the criterion to assess patient response [39]. For significant results (area under the curve [AUC] >0.6), optimal cut-off values were identified and permitted classification of gene expression into high / low for further analysis. JMP v8.0.2 profiling was used for the identification of associations with histologic grade and with response to initial treatment. All other analyses were performed with SPSS v.15. Association between binary characteristics was tested using Fisher’s exact test. All tests were two sided and the level of significance was set at a = 5 %.
Results
DARPP32 / STAT mRNA Expression in Comparison to Classic Glioma Parameters
Out of the 145 samples with informative RQ data, amplification curves for all five mRNA targets were obtained in 137 cases (DARPP32 was undetectable in 5 and STAT5B in 3 samples). RQ ratios were calculated as described for DARPP32/STAT1, DARPP32/STAT3, DARPP32/STAT5A, and DARPP32/ STAT5B; for STAT5B/STAT1, STAT5B/STAT3, and STAT5B /STAT5A; for STAT3/STAT1, and STAT3/STAT5A; and, for STAT1/STAT5A.
In general, the relative expression of STAT1, a gene located on chromosome 2q32 was higher in comparison to all genes on 17q, i.e., STAT5A and STAT5B on 17q11, DARPP32 on 17q12, and STAT3 on 17q21 (Fig. 1a). The expression of DARPP32 and STAT genes was statistically strongly correlated at the bivariate analysis level (Spearman’s p’s < 0.0001 in all cases). However, strong correlations with r > 0.5 (p < 10−6) were only observed for STAT gene pairs (Fig. 1b). In comparison, all DARPP32/STAT mRNA ratios inter-correlated with r values >0.8, while corresponding correlations between STAT ratios were only occasionally observed and were lower (STAT5B/STAT5A vs. STAT5B/STAT3, r = 0.55; STAT5B/STAT5A vs. STAT3/STAT5A, r = 0.73; STAT3/STAT5A vs. STAT1/STAT5A, r = 0.56). Among the five genes and in comparison to the eight available NNBT samples, only DARPP32 mRNA expression was found significantly decreased in grade II gliomas (mean±SD: 0.84 ± 1.08 in NNBT vs. 0.26 ± 0.25 in grade II, p = 0.0001). STAT gene expression remained constant between NNBT and grade II tumours. However, although the DARPP32 / STAT mRNA ratios were also decreased in grade II as compared to NNBT samples, these did not reach statistical significance. Hence, a possible bias of the endogenous reference (GUSB) could not be excluded for comparisons between non-neoplastic and neoplastic brain tissue. Single marker RQ values and combined marker RQ ratios were not associated with patient age, while STAT5A mRNA was significantly higher in men as compared to women (69 vs. 76, mean±SD: 0.11 ± 0.13 vs. 0.19 ± 0.28, respectively, p = 0.0090). The latter may be attributed to the different regulation of STAT5 regulation in females and in males [40, 41]. No differences in the expression of the genes examined or in their RQ ratios were observed in primary vs. secondary glioblastomas or with respect to the absence vs. presence of oligodendroglial component, although the latter category included a small number of tumours only.
DARPP32 and STAT gene expression descriptives. a The highest relative expression values were obtained for STAT1 and for STAT3, while STAT5A, STAT5B and DARPP32 expression was comparatively low. X-axes: RQ values. b Although the expression of all genes significantly correlated in the bivariate mode, these correlations were stronger for STAT (right panel) than for DARPP32 vs. STAT (left panel) gene pairs. r = Spearman’s bivariate correlation coefficient. In a & b, RQ values were normalized with GUSB
Among the 137 samples with informative RQ data for all targets, 110 corresponded to grade IV, 14 to grade III and 13 to grade II tumours (in total, 27 non-glioblastomas). STAT1 and STAT3 mRNA expression did not differ among glioma grades. By contrast, the remaining genes that were all located on chromosome 17q, i.e. STAT5A, STAT5B and DARPP32, were strongly downregulated in grade IV as compared to grade II tumours (Fig. 2a). The pattern for this change in gene expression was different for STAT5A and STAT5B as compared to DARPP32. DARPP32 mRNA was downregulated with progressive degree of malignancy, while the expression of the two STAT5 genes did not differ in grade II and III tumours. In accordance to single DARPP32 mRNA expression, DARPP32/STAT1, DARPP32/STAT3, STAT5A/STAT3 and STAT5B/STAT3 ratios were strongly decreased in grade IV tumours (Fig. 2b). However, for all expression markers, several outliers were present, particularly in glioblastomas, indicating that some of these tumours expressed high levels of the generally down-regulated DARPP32 and STAT genes. Indeed, profiling of all above RQ values and ratios revealed glioma subgroups with high STAT5B/STAT3 & STAT5B/STAT5A & STAT5B/STAT1 mRNA ratios, high STAT1/STAT3 and STAT1/STAT5A, and high DARPP32/STAT ratios. Further analysis of these profiles showed that high DARPP32/STAT mRNA ratios clustered together in the entire glioma series and were more common in non-glioblastomas (9/27 tumours [33.3 %]) as compared to glioblastomas (12/110 tumours [10.9 %]) (Fig. 2c). By contrast, STAT mRNA ratios did not cluster together in the same setting and their profiles were not associated with glioma grade.
Associations of DARPP32 and STAT gene expression with glioma grade. a Single gene expression normalized with GUSB. Among the five genes examined, DARPP32, STAT5A and STAT5B were significantly down-regulated in grade IV in comparison to grade II and III tumours, while DARPP32 expression was significantly different between grade III and IV tumours as well (Mann–Whitney test). b DARPP32/STAT gene expression ratios (avoiding a possible effect of GUSB) were also strongly related to tumour grade (Mann–Whitney test). However, in line with (a), many outliers were observed in glioblastomas (grade IV tumours). In (c), profiling of all gliomas (blue: low; red: high ratios) revealed non-overlapping subtypes DARPP32/STAT3 & 5 ratios clustered in association with glioma grade (Pearson Chi sq p = 0.0031). Marked cases in the two clusters: non-grade IV cases
MGMT promoter methylation, a marker associated with response to temozolomide, was investigated here only in the group of 67 patients with known follow-up and yielded informative results in 54 samples (Table 2). Single DARPP32, STAT1, STAT3 and STAT5A & B mRNA expression and their ratios were unrelated to MGMT promoter methylation, as assessed with the Mann–Whitney test. No statistics could be performed for IDH1 mutations, since these were found in two tumours only. One of these tumours was a clinically secondary glioblastoma; the second IDH1 mutant tumour was an anaplastic astrocytoma (grade III) with histologic glioblastoma confirmation (grade IV) in a limited area of the available tissue material. Taking into account that IDH status was unknown in six cases due to non-informative sequencing, 59 tumours (88 %) were IDH1 and IDH2 wild type in the cohort with available clinical data. Regarding EGFR CN, the STAT5B/STAT3 profile was marginally inversely associated with higher copy numbers of this gene (Mann Whitney p = 0.058), but no further associations with the remaining single or combined RQ markers were observed.
DARPP32 / STAT mRNA Expression and Activation of the AKT Pathway in Gliomas
Based on TMA core availability, it was possible to evaluate AKT phosphorylation at Ser473 in 128/145 cases with informative RNA data for cytoplasmic positivity and in 133/145 cases for nuclear positivity (in the additional five cases, positive parts of the cores were preserved). Phosphorylated AKT-S473 was present in the cytoplasm of neoplastic cells in 52/128 (40.6 %) cases, while it was localized in the neoplastic nuclei in 24/133 (18.4 %) of the tumours only. Non-cancerous elements, e.g., endothelial cells, were negative. Nuclear and cytoplasmic staining was not necessarily simultaneously present in the same tumour. Nuclear pAKT-S473 positivity was unrelated to glioma grade but cytoplasmic positivity was, since 48 out of the 52 tumours with cytoplasmic pAKT-S473 localization were glioblastomas (p = 0.001).
Among all gliomas, cytoplasmic pAKT-S473 positivity was strongly associated with lower DARPP32 (mean±SD: 0.15 ± 0.19 vs. 0.07 ± 0.10) and STAT5A (mean±SD: 0.19 ± 0.28 vs. 0.10 ± 0.12) expression, as well as with lower DARPP32/STAT1 (mean±SD: 0.08 ± 0.17 vs. 0.03 ± 0.03), DARPP32/STAT3 (mean±SD: 0.13 ± 0.21 vs. 0.06 ± 0.09) and DARPP/STAT5B (mean±SD: 0.40 ± 0.56 vs. 0.16 ± 0.19) ratios (Fig. 3a). When examined in glioblastomas only, cytoplasmic pAKT-S473 was again observed in cases with lower DARPP32 (mean±SD, positive vs. negative pAKT: 0.12 ± 1.7 vs. 0.06 ± 0.09, p = 0.0182) and STAT5A (mean±SD: 0.13 ± 0.12 vs. 0.08 ± 0.07, p = 0.0316).
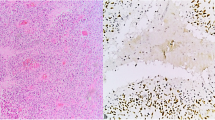
DARPP32 and STAT gene expression, single or in paired ratios, is inversely associated with Akt/PKB phosphorylation (a) and with survivin expression (b). In a, a representative IHC microphotograph (X400) showing a glioblastoma with cytoplasmic localization of phosphorylated Akt/PKB is shown. The Mann–Whitney test was used for all comparisons. Significant inverse associations for the same DARPP32/STAT ratios (shown as Y-axis labels) were observed for both Akt/PKB phosphorylation and survivin. Y axes represent RQ values
DARPP32 / STAT mRNA Expression and the Apoptosis Inhibitor Survivin (BIRC5)
Survivin is a member of the inhibitors of apoptosis protein family [42]; as such, it has often been described as an adverse prognostic factor in several types of cancer, including glioblastomas [43], where it has been associated with proliferation but not with apoptosis [44]. In our series, survivin IHC data were obtained for 116 gliomas, out of which 75 (63.4 %) displayed intra-nuclear spot positivity. As previously reported with different antibodies [36, 37, 43], survivin and Ki-67 positivity were strongly associated (p = 0.001). Survivin positivity was also strongly associated with nuclear AKT-Ser473 phosphorylation (Fisher’s exact p = 0.0028), while a trend was also observed for cytoplasmic pAKT positivity (p = 0.0734).
In line with the above data for AKT-Ser473 phosphorylation, survivin positivity was mostly observed in tumours with low DARPP32/STAT1 (mean±SD, positive vs. negative survivin: 0.10 ± 0.22 vs. 0.04 ± 0.06), low DARPP32/STAT3 (mean±SD: 0.16 ± 0.27 vs. 0.08 ± 0.10), and low DARPP32/STAT5B (mean±SD: 0.52 ± 0.66 vs. 0.22 ± 0.32) (Fig. 3b). In the group of 92 glioblastomas that were evaluable for survivin, positive tumours (n = 61) often expressed low DARPP32/STAT5B (mean±SD, positive vs. negative survivin: 0.42 ± 0.56 vs. 0.18 ± 0.25, p = 0.0127), but higher STAT5B/STAT3 (mean±SD: 0.27 ± 0.14 vs. 0.38 ± 0.29, p = 0.0155).
Relatively High DARPP32 / STAT mRNA Expression is Favorably Associated with Response to Initial Radiotherapy / Temozolomide Treatment and has a Positive Effect on Glioblastoma Patient Survival
Other than expected, MGMT promoter methylation was not associated with response to treatment (PFS-6 month) in this study. PFS-6 mo was further unrelated to patient age, gender and to the extent of surgery. In order to investigate for a possible impact of the observed alterations on the outcome of glioblastoma patients, who were all treated with post-surgery CCRT, we first undertook a profiling screening for DARPP32 and STAT ratios in the group of 67 patients (Fig. 4). Very high DARPP32/STAT ratios clustered together and did not overlap with high STAT5B/STAT5A, STAT5B/STAT3 and STAT1/STAT5A. High DARPP32/STAT ratios were observed in 14/67 patients, 13 out of which responded favorably to CCRT. In comparison, among the 15 cases with low DARPP32/STAT and high STAT/STAT ratios, three were non-responders or experienced early death. Notably, the STAT3/STAT1 ratio did not cluster with the rest of variables in this profile. Based on these observations, we then investigated the impact of single mRNA expression and the corresponding ratios on patient outcome. For this, we classified gene expression into “high” and “low” by using ROC curve analysis with PFS-6 mo as the state variable. Gene expression parameters that failed to yield a significantly (>0.6) large area under the curve (AUC) were not further used for survival analysis. The results of ROC curve analysis and the corresponding cut-off values for gene expression classification are shown in Table 3. Parameters that were thus classified were indeed associated with response to treatment in patients with glioblastoma (Table 4). It appeared that relatively high DARPP32 expression (when calculated vs. GUSB), and relatively high DARPP32 expression ratios with all STAT genes examined (without GUSB interference) were positively associated with absence of relapse in 6 months, as well as with prolonged progression free survival (PFS) (Table 5, Fig. 4). PFS closely reflected treatment response in the cohort examined where all patients had received the same initial treatment. Relatively high STAT5B expression, alone or as STAT5B/STAT3 ratio, was also favorably associated with patient response to CCRT and with prolonged PFS (Fig. 5).
DARPP32 and STAT gene expression ratios were associated with response to initial CCRT treatment, as assessed with PFS-6 mo. Marked rows on the left of the colormap correspond to cases where progression or death was noticed within 6 months (16 out of 67 patients with known follow up). Except for one case, high DARPP32/STAT ratios were observed in patients who responded well to CCRT. In comparison, five non-responders were observed among single or combined high STAT ratios. The two cases marked with black circles correspond to the two IDH1 mutant tumours, the upper one with early progression upon CCRT and the lower one with a favorable outcome. x marks: cases non-informative for IDH1 & 2 sequencing
Effect of DARPP32 and STAT gene expression on glioblastoma patient outcome. a progression-free survival (PFS); b overall survival (OS) Significant gene expression ratios are shown. High—low classification of RQ values according to ROC curve cut-offs; combined vs. STAT3: DARPP32/STAT3 and STAT5B/STAT3; p: log rank test
OS is unequivocally the most solid parameter for assessing patient disease course. However, as shown in Table 1, the administered treatment schemes after the initial course of radiotherapy / temozolomide were not homogeneous and it is possible that these affected patient outcome. Nevertheless, in this group of 67 patients, age and gender were not associated with disease course. In this context, the most important factor affecting patient survival was the extent of surgical removal of the tumours; in cases with total or subtotal excision (n = 48), median OS was 20.52 month (±SD:1.36, 95 % CI: 17.86–23.18) in comparison to cases where tumours were removed by biopsy only (n = 19) and patient OS was 11.6 month (±SD:1.60, 95 % CI: 8.46–14.75, p = 0.002). Out of the immunohistochemical parameters tested, none was associated with patient OS. MGMT promoter methylation was not associated with PFS and showed only a trend for the methylated tumours and prolonged patient survival (p = 0.088).
Relatively high DARPP32 expression as single parameter (normalized vs. GUSB), high DARPP32/STAT3 and DARPP32/STAT5A expression ratios, as well as the combined bivariate profile of DARPP32/STAT3 and STAT5B/STAT3 were strongly associated with longer glioblastoma patient OS (Table 5, Fig. 5).
Last follow up for the clinically secondary IDH1 p.R132H mutant glioblastoma was at 28.9 months from this manifestation with the patient (man, 41 y.o.) alive and well. The IDH1 p.R132G mutant patient (woman, 35 y.o.) with histologically secondary glioblastoma progressed upon CCRT at 4 months and succumbed to disease after 18.6 months from diagnosis. These cases and their DARPP32/STAT profiles are marked in Fig. 4. The third patient (man, 62 y.o.) with histologically secondary IDH1 and IDH2 wild-type glioblastoma succumbed to disease during CCRT at 3.5 months; the tumour had very low DARPP32/STAT3 ratio (RQ 0.002).
Upon multivariate Cox regression analysis adjusted for glioblastoma patient age, extent of surgery, and all mRNA markers presented here, total/subtotal removal of the tumour (HR: 0.431; 95 % CI: 0.241–0.771, Wald p = 0.005), high DARPP32/STAT5B mRNA ratios (HR: 0.341; 95 % CI: 0.169–0.690; Wald p = 0.003) and high STAT5B/STAT3 mRNA ratios (HR: 0.480; 95 % CI: 0.280–0.824; Wald p = 0.008) were independent favorable parameters for prolonged PFS. Patient survival was aggravated with increasing age (HR: 1.028; 95 % CI: 1.003–1.054; p = 0.029), while extent of surgery was independently associated with prolonged survival (HR: 0.198; 95 % CI: 0.101–0.390; p < 0.001). In this setting, high DARPP32/STAT5A mRNA ratios were also independently predictive for longer survival (HR: 0.320; 95 % CI: 0.160–0.638, p = 0.001).
Discussion
The present study shows that transcript ratios of DARPP32, STAT3, STAT5A and STAT5B in glioblastomas are strongly related to patient outcome. The glioblastomas examined here can be considered as de novo in 95 % of the cases and were validated as IDH1 and IDH2 wild-type in 88 % of all patients with available clinical data. In this group of unfavorable disease where patients initially received CCRT / temozolomide, tumour STAT3 mRNA expression lower than STAT5B and DARPP32 was associated with better prognosis upon standard treatment. STAT3 mRNA expression was abundant in glioma tissues and did not differ among glioma grades, which is of no surprise since STAT3 functions in a pro- or anti-tumour mode depending on its molecular environment [15, 16]. The present findings further support STAT3 as an unfavorable prognosticator in glioblastomas [16, 45], and are in line with its mesenchymal-subtype-promoting role, as has been shown with different approaches [9, 15, 46, 47]. Response measured as 6moPFS, and PFS itself, seem to be more reliable endpoints for assessing the markers examined here than OS, which may have been influenced by the subsequent treatment lines administered to about 40 % of the patients. Thus, the fact that STAT3 ratios did not maintain their significance as prognostic for OS may indicate that the favorable effect of relatively low STAT3 is therapy related. STAT3 inhibition was shown to restore temozolomide resistance in glioblastoma cells [48]. Overall, the present and previously published data support STAT3 inhibition for therapeutic purposes in glioblastoma, possibly in combination with other drugs [48, 49].
The Stat5 proteins have been described as downstream targets of EGFRvIII [50] contributing along with STAT3 to the escape of glioblastoma from microglial surveillance [51], without distinguishing between Stat5a and 5b molecules. STAT5A, STAT5B, and STAT3 reside in close neighborhood on chromosome 17q and as shown here, their expression is strongly correlated. Regarding their relative mRNA expression against DARPP32 and STAT3, STAT5A and STAT5B appeared to have different although not contrasting effects on patient outcome in this study, which has been reported for epithelial cancers [52]. Like STAT3, our findings on STAT5B are in accordance with its previously published pro-oncogenic role in glioblastomas [18]. According to the present data, STAT3 effects may depend on STAT5B expression, with high STAT5B/STAT3 ratios being associated with a more indolent glioblastoma course. Relevant information in gliomas is missing but in breast cancer STAT5 and STAT3 mediate opposing effects on several key target genes, with STAT5 exerting a dominant role [53]. The present data on STAT5B and STAT3 may be useful when assessing these molecules for pharmacologic interactions in glioblastomas.
From a different perspective, the most interesting findings in this study concern DARPP32, which, to the best of our knowledge, has not merited attention in brain tumours before but seems worth investigating in this context. It is shown here that DARPP32 is expressed at low levels in gliomas while its expression is further decreased with progressing degree of malignancy. DARPP32 is a key marker of neural specialization marking, for example, differentiation of human embryonic stem cells into specialized neurons [54]. One possible shortcoming regarding our findings is that with the assay used we detected all DARPP32 splice variants; hence, we cannot report on t-DARPP expression. This isoform is pro-oncogenic but nevertheless expressed along with full-length DARPP32, while it is not reported as expressed in the central nervous system [55]. In addition, although it cannot be excluded that DARPP32 expression may derive from entrapped neurons in low grade tumours (for example, in grade II tumours), such an argument can not be justified for glioblastomas that occasionally also express this molecule at levels resembling grade II tumours. Considering that glia has neurotrophic, neuroprotective and synaptogenic function, as well as a major role in neural stem cell differentiation (reviewed in [56]), the observed DARPP32 expression reflecting “neuronal” features in gliomas may indicate some retained degree of differentiation in the neoplastic cells. Overall, our findings with DARPP32 ratios also seem to be in line with previous observations on neuronal-like profiles mainly in the neural, and less so in the classical and mesenchymal subtypes, which correspond to tumours with worsening prognosis in this order [9]. It is currently unknown whether DARPP32 is translated into protein and how it functions in gliomas, in particular with respect to its normal role as an integrator of neurotransmission. In addition, there is a gap in the literature regarding dopamine and monoamine signaling in general in these tumours. However, since dopaminergic compounds inhibit the growth of glioblastoma stem cell enriched cultures [57], the functional aspects of DARPP32 expression in gliomas and the neuronal characteristics of glioblastomas seem worth further pursuing.
A protective role of DARPP32 and an aggravating role of STAT3 and STAT5B in glioblastomas are further supported by the association of the corresponding mRNA ratios with AKT/PKB phosphorylation and survivin expression. Survivin expression is associated with proliferation and poorer prognosis in glioblastomas [43, 44], while it depends on STAT3 [28, 29]. AKT is phosphorylated at Ser473 by the rictor-mTOR complex [58], which may be inhibited experimentally in glioblastoma cells [59], while inhibition of AKT activation results in growth cessation of glioma cells [60]. These IHC markers did not have an effect on patient survival in the present series. It should be noticed that survivin is a protein with very short half-life [61], while the performance of the antibody used for phospho-AKT largely depends on time-to-fixation [62]. Hence, IHC results for these markers may not reflect the actual condition of these proteins in tumours, in spite that every care was taken for prompt tissue fixation upon surgical removal Nevertheless, high DARPP32/STAT1, DARPP32/STAT3 and DARPP32/STAT5B ratios were associated with the absence of phosphorylated AKT and survivin in the tumours examined, indicating that DARPP32 negatively and STAT positively interfere with the expression of these pro-oncogenic protein markers.
The value of the present findings is that they potentially represent markers for the assessment of IDH-wild-type glioblastomas with unfavorable prognosis. Clearly, these results need prospective validation in larger multi-center sample and patient cohorts. In terms of methodology, the approach used here allows for the evaluation of glioma-relevant gene expression ratios without the involvement of a reference gene, which was used only for template evaluation. In a sense, this approach may be described as a clustering pre-stage, since it allows for gene expression ratios to be examined in duets. From a different aspect, mRNA ratios may be described as yin-yang: since molecules act in concert, their ratios may be more important than individual up- or down-regulation, which can not be determined by this approach. DNA / RNA extraction from routine FFPE diagnostic material is increasingly applied in pathology labs, while RT-qPCR is a very robust method for obtaining quantitative results. One of the problems with such results is the assessment of cut-offs. With the ROC curve cut-offs applied here, DARPP32 and STAT5B transcripts should be roughly higher than STAT3 transcripts by 2 % and 25 %, respectively, for the classification of favorable vs. unfavorable ratios. Although these results are unavoidably fitted to the cohort, as would be the case with any single continuous marker (including clusters that derive from such assessments), these analogies are easier to keep when standardizing the method.
Given the heterogeneity of gliomas and the devastating course of glioblastomas, glioblastoma subtyping is necessary for a more efficient assessment of patients for the application of individualized treatment [63]. With all subtyping efforts so far, which may be replicated retrospectively but seem to suffer from inherent errors for wide application [64], the best as yet distinction of glioblastomas appears to be IDH positive and negative disease [6, 7]. This study provides an applicable approach for the patient stratification in addition to IDH mutations. The DARPP32/STAT ratios presented here seem worth for further testing as prognostic and potentially as markers predictive for response to standard, as well as to new drug combinations for glioblastoma treatment. The neuronal aspect of glioma differentiation, in particular glioblastoma, as evidenced with DARPP32 expression, merits further elucidation for understanding the behaviour of these tumours.
References
WHO (2007) WHO Classification of Tumors of the Central Nervous System. International Agency for Research on Cancer, City
Wen PY, Kesari S (2008) Malignant gliomas in adults. N Engl J Med 359:492–507
Riemenschneider MJ, Jeuken JW, Wesseling P, Reifenberger G (2010) Molecular diagnostics of gliomas: state of the art. Acta Neuropathol 120:567–584
Yan H, Bigner DD, Velculescu V, Parsons DW (2009) Mutant metabolic enzymes are at the origin of gliomas. Cancer Res 69:9157–9159
Ohgaki H, Kleihues P (2011) Genetic profile of astrocytic and oligodendroglial gliomas. Brain Tumor Pathol 28:177–183
Lai A, Kharbanda S, Pope WB et al (2011) Evidence for sequenced molecular evolution of IDH1 mutant glioblastoma from a distinct cell of origin. J Clin Oncol 29:4482–4490
Hartmann C, Hentschel B, Wick W et al (2011) Patients with IDH1 wild type anaplastic astrocytomas exhibit worse prognosis than IDH1-mutated glioblastomas, and IDH1 mutation status accounts for the unfavorable prognostic effect of higher age: implications for classification of gliomas. Acta Neuropathol 120:707–718
Phillips HS, Kharbanda S, Chen R et al (2006) Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 9:157–173
Verhaak RG, Hoadley KA, Purdom E et al (2010) Integrated Genomic Analysis Identifies Clinically Relevant Subtypes of Glioblastoma Characterized by Abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17:98–110
Brennan C, Momota H, Hambardzumyan D et al (2009) Glioblastoma subclasses can be defined by activity among signal transduction pathways and associated genomic alterations. PLoS One 4:e7752
Noushmehr H, Weisenberger DJ, Diefes K et al (2010) Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 17:510–522
Stupp R, Tonn JC, Brada M, Pentheroudakis G (2010) High-grade malignant glioma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 21(Suppl 5):v190–v193
Stupp R, Hegi ME, Mason WP et al (2009) Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol 10:459–466
CGARN (2008) Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455:1061–1068
Carro MS, Lim WK, Alvarez MJ et al (2010) The transcriptional network for mesenchymal transformation of brain tumours. Nature 463:318–325
de la Iglesia N, Puram SV, Bonni A (2009) STAT3 regulation of glioblastoma pathogenesis. Curr Mol Med 9:580–590
Anderson JC, McFarland BC, Gladson CL (2008) New molecular targets in angiogenic vessels of glioblastoma tumours. Expert Rev Mol Med 10:e23
Liang QC, Xiong H, Zhao ZW et al (2009) Inhibition of transcription factor STAT5b suppresses proliferation, induces G1 cell cycle arrest and reduces tumor cell invasion in human glioblastoma multiforme cells. Cancer Lett 273:164–171
Haybaeck J, Obrist P, Schindler CU, Spizzo G, Doppler W (2007) STAT-1 expression in human glioblastoma and peritumoral tissue. Anticancer Res 27:3829–3835
Hemmings HC Jr, Greengard P, Tung HY, Cohen P (1984) DARPP-32, a dopamine-regulated neuronal phosphoprotein, is a potent inhibitor of protein phosphatase-1. Nature 310:503–505
Brene S, Lindefors N, Ehrlich M et al (1994) Expression of mRNAs encoding ARPP-16/19, ARPP-21, and DARPP-32 in human brain tissue. J Neurosci 14:985–998
Svenningsson P, Nishi A, Fisone G, Girault JA, Nairn AC, Greengard P (2004) DARPP-32: an integrator of neurotransmission. Annu Rev Pharmacol Toxicol 44:269–296
Yger M, Girault JA (2011) DARPP-32, Jack of All Trades… Master of Which? Frontiers in behavioral neuroscience 5:56
Gu L, Waliany S, Kane SE (2009) Darpp-32 and its truncated variant t-Darpp have antagonistic effects on breast cancer cell growth and herceptin resistance. PLoS One 4:e6220
Hansen C, Howlin J, Tengholm A et al (2009) Wnt-5a-induced phosphorylation of DARPP-32 inhibits breast cancer cell migration in a CREB-dependent manner. J Biol Chem 284:27533–27543
Bogush A, Pedrini S, Pelta-Heller J et al (2007) AKT and CDK5/p35 mediate brain-derived neurotrophic factor induction of DARPP-32 in medium size spiny neurons in vitro. J Biol Chem 282:7352–7359
Belkhiri A, Dar AA, Zaika A, Kelley M, El-Rifai W (2008) t-Darpp promotes cancer cell survival by up-regulation of Bcl2 through Akt-dependent mechanism. Cancer Res 68:395–403
Chen F, Xu Y, Luo Y et al (2010) Down-regulation of Stat3 decreases invasion activity and induces apoptosis of human glioma cells. J Mol Neurosci 40:353–359
Konnikova L, Kotecki M, Kruger MM, Cochran BH (2003) Knockdown of STAT3 expression by RNAi induces apoptosis in astrocytoma cells. BMC Cancer 3:23
Stupp R, Mason WP, van den Bent MJ et al (2005) Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 352:987–996
Zhang F, Wang ZM, Liu HY et al (2011) Application of RT-PCR in formalin-fixed and paraffin-embedded lung cancer tissues. Acta Pharmacol Sinica 31:111–117
Sanchez-Navarro I, Gamez-Pozo A, Gonzalez-Baron M et al (2010) Comparison of gene expression profiling by reverse transcription quantitative PCR between fresh frozen and formalin-fixed, paraffin-embedded breast cancer tissues. Biotechniques 48:389–397
Jeuken JW, Cornelissen SJ, Vriezen M et al (2007) MS-MLPA: an attractive alternative laboratory assay for robust, reliable, and semiquantitative detection of MGMT promoter hypermethylation in gliomas. Lab Invest 87:1055–1065
Mizoguchi M, Betensky RA, Batchelor TT, Bernay DC, Louis DN, Nutt CL (2006) Activation of STAT3, MAPK, and AKT in malignant astrocytic gliomas: correlation with EGFR status, tumor grade, and survival. J Neuropathol Exp Neurol 65:1181–1188
Spaulding B, Pan D, Ghadersohi A et al (2006) Characterization of the 12C4 survivin monoclonal antibody and insight into the expression of survivin in human adult tissues. Histopathology 49:622–633
Uematsu M, Ohsawa I, Aokage T et al (2005) Prognostic significance of the immunohistochemical index of survivin in glioma: a comparative study with the MIB-1 index. J Neurooncol 72:231–238
Preusser M, Gelpi E, Matej R et al (2005) No prognostic impact of survivin expression in glioblastoma. Acta Neuropathol 109:534–538
Johannessen AL, Torp SH (2006) The clinical value of Ki-67/MIB-1 labeling index in human astrocytomas. Pathol Oncol Res 12:143–147
Stupp R, Roila F (2009) Malignant glioma: ESMO clinical recommendations for diagnosis, treatment and follow-up. Ann Oncol 20(Suppl 4):126–128
Feltus FA, Groner B, Melner MH (1999) Stat5-mediated regulation of the human type II 3beta-hydroxysteroid dehydrogenase/delta5-delta4 isomerase gene: activation by prolactin. Mol Endocrinol 13:1084–1093
Waxman DJ (2000) Growth hormone pulse-activated STAT5 signalling: a unique regulatory mechanism governing sexual dimorphism of liver gene expression. Novartis Found Symp 227:61–74, discussion 75–81
Viglietto G, Motti ML, Bruni P et al (2002) Cytoplasmic relocalization and inhibition of the cyclin-dependent kinase inhibitor p27(Kip1) by PKB/Akt-mediated phosphorylation in breast cancer. Nat Med 8:1136–1144
Shirai K, Suzuki Y, Oka K et al (2009) Nuclear survivin expression predicts poorer prognosis in glioblastoma. J Neurooncol 91:353–358
Mellai M, Caldera V, Patrucco A, Annovazzi L, Schiffer D (2008) Survivin expression in glioblastomas correlates with proliferation, but not with apoptosis. Anticancer Res 28:109–118
Brantley EC, Benveniste EN (2008) Signal transducer and activator of transcription-3: a molecular hub for signaling pathways in gliomas. Mol Cancer Res 6:675–684
Bhat KP, Salazar KL, Balasubramaniyan V et al (2011) The transcriptional coactivator TAZ regulates mesenchymal differentiation in malignant glioma. Genes Dev 25:2594–2609
Cooper LA, Gutman DA, Chisolm C et al (2012) The tumor microenvironment strongly impacts master transcriptional regulators and gene expression class of glioblastoma. Am J Pathol 180:2108–2119
Kohsaka S, Wang L, Yachi K et al. (2012) STAT3 inhibition overcomes temozolomide resistance in glioblastoma by downregulating MGMT expression. Mol Cancer Ther
Lo HW, Cao X, Zhu H, Ali-Osman F (2008) Constitutively activated STAT3 frequently coexpresses with epidermal growth factor receptor in high-grade gliomas and targeting STAT3 sensitizes them to Iressa and alkylators. Clin Cancer Res 14:6042–6054
Chumbalkar V, Latha K, Hwang Y et al (2011) Analysis of phosphotyrosine signaling in glioblastoma identifies STAT5 as a novel downstream target of DeltaEGFR. J Proteome Res 10:1343–1352
Kostianovsky AM, Maier LM, Anderson RC, Bruce JN, Anderson DE (2008) Astrocytic regulation of human monocytic/microglial activation. J Immunol 181:5425–5432
Tan SH, Nevalainen MT (2008) Signal transducer and activator of transcription 5A/B in prostate and breast cancers. Endocr Relat Cancer 15:367–390
Walker SR, Nelson EA, Zou L et al (2009) Reciprocal effects of STAT5 and STAT3 in breast cancer. Mol Cancer Res 7:966–976
Aubry L, Bugi A, Lefort N, Rousseau F, Peschanski M, Perrier AL (2008) Striatal progenitors derived from human ES cells mature into DARPP32 neurons in vitro and in quinolinic acid-lesioned rats. Proc Natl Acad Sci U S A 105:16707–16712
Beckler A, Moskaluk CA, Zaika A et al (2003) Overexpression of the 32-kilodalton dopamine and cyclic adenosine 3′,5′-monophosphate-regulated phosphoprotein in common adenocarcinomas. Cancer 98:1547–1551
He F, Sun YE (2007) Glial cells more than support cells? Int J Biochem Cell Biol 39:661–665
Visnyei K, Onodera H, Damoiseaux R et al (2011) A molecular screening approach to identify and characterize inhibitors of glioblastoma stem cells. Mol Cancer Ther 10:1818–1828
Sarbassov DD, Ali SM, Sabatini DM (2005) Growing roles for the mTOR pathway. Curr Opin Cell Biol 17:596–603
Fan QW, Weiss WA (2012) Inhibition of PI3K-Akt-mTOR signaling in glioblastoma by mTORC1/2 inhibitors. Methods Mol Biol 821:349–359
Koul D, Shen R, Bergh S et al (2006) Inhibition of Akt survival pathway by a small-molecule inhibitor in human glioblastoma. Mol Cancer Ther 5:637–644
Zhao J, Tenev T, Martins LM, Downward J, Lemoine NR (2000) The ubiquitin-proteasome pathway regulates survivin degradation in a cell cycle-dependent manner. J Cell Sci 113(Pt 23):4363–4371
Espina V, Edmiston KH, Heiby M et al (2008) A portrait of tissue phosphoprotein stability in the clinical tissue procurement process. Mol Cell Proteomics 7:1998–2018
Weller M, Stupp R, Hegi M, Wick W (2012) Individualized targeted therapy for glioblastoma: fact or fiction? Cancer J 18:40–44
Marko NF, Quackenbush J, Weil RJ (2011) Why is there a lack of consensus on molecular subgroups of glioblastoma? Understanding the nature of biological and statistical variability in glioblastoma expression data. PLoS One 6:e20826
Acknowledgments
The authors wish to thank Dr. Athanassios B.-T. Fassas, MD, Private Medical Center “Agios Loukas” for providing patient material, Mrs. Emily Daskalaki for excellent technical assistance and Mrs. Anastasia Eleftheraki for support with statistics. Work supported by PENED 03/583, a research grant from the General Secretariat for Research and Technology (Ministry of Development, Athens, Greece); supported in part by a Hellenic Cooperative Oncology Group research grant HE R_17/09.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Televantou, D., Karkavelas, G., Hytiroglou, P. et al. DARPP32, STAT5 and STAT3 mRNA Expression Ratios in Glioblastomas are Associated with Patient Outcome. Pathol. Oncol. Res. 19, 329–343 (2013). https://doi.org/10.1007/s12253-012-9588-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12253-012-9588-7