Abstract
Background
The proteome is an important resource for exploring potential diagnostic and therapeutic targets for cancer. This study aimed to investigate the causal associations between plasma proteins and prostate cancer (PCa), and to explore the downstream phenotypes that plasma proteins may influence and potential upstream intervening factors.
Methods
Proteome-wide Mendelian randomization was used to investigate the causal effects of plasma proteins on PCa. Colocalization analysis examined the common causal variants between plasma proteins and PCa. Summary-statistics-based Mendelian Randomization (SMR) analyses identified associations between the expression of protein-coding genes and PCa. Phenome-wide association study was performed to explore the effect of target proteins on downstream phenotypes. Finally, a systematic Mendelian randomization analysis between lifestyle factors and plasma proteins was performed to assess upstream intervening factors for plasma proteins.
Results
The findings revealed a positive genetic association between the predicted plasma levels of nine proteins and an elevated risk of PCa, while four proteins exhibited an inverse association with PCa risk. SMR analyses revealed ZG16B, PEX14 in blood and ZG16B, NAPG in prostate tissue were potential drug targets for PCa. The genetic association of PEX14 with PCa was further supported by colocalization analysis. Further Phenome-wide association study showed possible side effects of ZG16B, PEX14 and NAPG as drug targets. 10 plasma proteins (RBP7, TPST1, NFASC, LAYN, HDGF, SERPIMA5, DLL4, EFNA3, LIMA1, and CCL27) could be modulated by lifestyle-related factors.
Conclusion
This study explores the genetic associations between plasma proteins and PCa, provides evidence that plasma proteins serve as potential drug targets and enhances the understanding of the molecular etiology, prevention and treatment of PCa.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Prostate cancer (PCa) is one of the most common malignant tumors in men and the global burden of this disease is rising. It is estimated that the number of new cases of PCa will reach approximately 1.7 million worldwide in 2030 [1]. Epidemiologic studies have shown that age, family history, and genetic susceptibility are significant risk factors of PCa [2]. The incidence of PCa is highest among men of African descent, followed by men of European and Asian ancestry, underscoring the important role of genetics in the development of PCa [3]. Lifestyle modifications, such as smoking cessation, exercise and weight control, may reduce the risk of PCa, but few modifiable risk behavioral factors have been identified [4].
Proteins are important molecules that perform biological functions. Plasma proteins secreted by tissues or organs can serve as predictive targets for many diseases or symptoms in humans [5]. Previous epidemiologic studies have identified a number of circulating proteins that are associated with cancer risk [6,7,8]. However, most of these studies have been based on observational designs or have focused on only a few proteins. Currently, Mendelian randomization (MR) analysis has emerged as a novel tool for re-evaluating drugs and discovering new therapeutic targets [9]. With large-scale proteomics studies, more than 18,000 protein quantitative trait loci (pQTL) for more than 4,000 proteins have been identified [10]. This has provided a valuable source of data for exploring associations between circulating proteins and disease using MR. MR uses these pQTLs as instrumental variables to explore potential causal relationships between exposures and outcomes and to screen for biomarkers that can be used as drug targets [11]. Compared with observational studies, MR helps to mitigate the effects of confounding factors, thereby enhancing the reliability and accuracy of causality assessment [12]. In addition, with the application of phenotype-wide association studies (PheWAS), the prediction of target-associated adverse effects can be widely investigated [13].
Plasma proteins play crucial roles in a variety of cellular biological processes such as signaling, transport, growth, repair and immune defense. Dysregulation of plasma proteins is strongly associated with many diseases and is an important source of targets for drug development [14]. Therefore, we performed a MR analysis of plasma proteomics to explore its causal association with PCa.
2 Methods
2.1 Study design
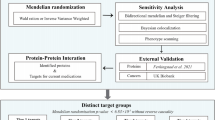
The flowchart of study design is shown in Fig. 1. We first performed a two-sample MR analysis using 4907 plasma proteins from the deCODE genetics dataset with the PCa GWAS dataset from the PRACTICAL consortium to obtain 143 positive plasma proteins that were causally associated with PCa. Similar analysis of the UK Biobank GWAS dataset for PCa yielded 126 positive plasma proteins. Thirteen common plasma proteins were obtained by taking intersections. These 13 proteins were then analyzed for colocalization and drug target summary-statistics-based Mendelian Randomization (SMR). Subsequently, we performed downstream PheWAS study of three plasma proteins to identify possible side effects of target drugs. Finally, a systematic MR analysis was performed on 26 lifestyle-related factors with 13 plasma proteins to determine which lifestyle factors might serve as upstream intervening factors for the target proteins.
2.2 Study population and data sources
Summary statistics of genetic associations for plasma proteins were obtained from a large-scale proteomics study (deCODE genetics) that included 35,559 Icelandic participants measured using the SOMAscan, which provides comprehensive pQTLs localization for 4907 plasma proteins.10 Summary-level data of PCa and lifestyle-related factors were obtained from publicly available datasets of genome-wide association studies (GWASs). Details for related GWASs are described in Supplementary Table 1). All participants were of European ancestry and provided informed consent. Considering that our study was based on publicly available summary statistics, no further ethical review was required.
2.3 Statistical analysis
2.3.1 Proteome-wide Mendelian randomization analysis
We performed two-sample MR analyses with plasma proteins as the exposure and PCa as the outcome. The screening criteria for pQTLs as instrumental variables were as follows: First, SNPs (cis-pQTLs) within ± 1 mb around the gene region. Second, the genome-wide significance threshold for SNPs highly associated with plasma proteins was P < 5 × 10−8. Finally, in order to alleviate the effect of linkage disequilibrium (LD) on the results, we set a threshold of 0.001 for the linkage disequilibrium parameter (r2) and a genetic distance of 10,000 kb. The strength of instrumental variables was measured using the F-statistic, with an F-statistic of less than 10 considered a weak instrumental variable [15]. Detailed information on the instrumental variables is provided in Supplementary Table 2.
Proteome-wide MR analysis was performed using the R package “TwoSampleMR”. When only one SNP was available for a specific protein, we applied the Wald ratio method. If two or more SNPs were available, we applied the inverse variance weighting (IVW) method. A P value below 0.05 was considered statistically significant.
2.3.2 Colocalization analysis
We performed Bayesian colocalization analysis using the coloc package to detect SNPs within ± 1 MB around the gene region (cis-pQTLs) with positive MR results, and further analyzed whether the identified plasma proteins shared common causal genetic variants with PCa [16]. The colocalization analysis involves five hypotheses: H0 indicates that the selected SNP within the locus is unrelated to both protein A and disease B; H1 suggests that the SNP within the selected locus is associated with protein A but not with disease B; H2 implies that the SNP within the selected locus is related to disease B but not to protein A; H3 states that the SNP within the selected locus is associated with either protein A or disease B, but the two are independent SNPs; H4 signifies that the SNP within the selected locus is concurrently associated with both protein A and disease B, and is a shared SNP. In view of the tissue-specificity of the blood and prostate, we performed the analysis in blood (eQTLgen and GTEx V8 Blood) and prostate tissue (GTEx V8 Prostate) for colocalization, respectively.
2.3.3 SMR analysis
We applied the SMR approach to test for associations between PCa and expression levels of plasma protein-coding genes using GWAS data of PCa from PRACTICAL and pooled data from eQTL, and subsequently validated this using the same methodology on data from UK Biobank. eQTL pooled data includes blood and prostate data from eQTLgen and GTEx. eQTLgen summary statistics include blood gene expression genetic data from 31,684 individuals of 37 datasets [17]. GTEx summary statistics include gene expression genetic data from 670 blood samples and 221 prostate tissue samples [18]. A Psmr value below 0.05 was considered statistically significant.
2.3.4 Phenome-wide association study
PheWAS, a method used to explore the relationship between SNPs or specific phenotypes and a broad range of phenotypes, is particularly valuable for investigating potential side effects associated with drug targets [19, 20]. PheWAS can identify diseases or traits that are positively or negatively correlated with the target protein, which can be used as indicators of potential side effects or complications when developing agonists or inhibitors on this target for appropriate clinical observation. We chose three plasma proteins (ZG16B, PEX14 and NAPG) that had a Psmr < 0.05 in the SMR analyses as exposures, obtained data of 2803 phenotypes from the FinnGen database. Screening criteria for instrumental variables remained consistent with those described previously. A P value below 0.05 was considered statistically significant.
2.3.5 Modulation of PCa-related plasma proteins by lifestyle-related factors
A total of 26 lifestyle-related factors (Supplementary Table 1) were used to assess associations with PCa-related proteins. The analytical methods for the MR were consistent with the description provided in the proteome-wide MR analysis. All statistical analyses were performed in R Software 4.1.0.
3 Results
3.1 Proteome-wide MR identified 13 PCa-related plasma proteins
With strict adherence to the instrumental variable screening criteria of this study, 1605 (PRACTICAL) and 1603 (UK Biobank) plasma proteins were included in the MR analysis, respectively. MR analysis results based on IVW or Wald ratio method showed that 143 plasma proteins (PRACTICAL) and 126 plasma proteins (UK Biobank) were associated with PCa (Fig. 2). We took the intersection of the two lists of positive proteins and ended up with 13 common plasma proteins. 9 proteins were positively associated with PCa (EFNA3, LIMA1, HDGF, PEX14, DLL4, NAPG, ZG16B, TPST1, and NFASC) and 4 proteins were negatively associated with PCa (SERPINA5, RBP7. CCL27, and LAYN). Detailed results of MR analysis are shown in Fig. 3 and Supplementary Tables 3, 4. Detailed results of heterogeneity and horizontal pleiotropy analyses are shown in Supplementary Tables 5–8.
3.2 Results of colocalization analysis
We performed colocalization analysis of the genes encoding each of the 13 proteins obtained in the previous step in blood and prostate tissues, respectively. The results showed that PEX14 (PPH3 + PPH4 = 0.99/0.98, eQTLgen/GTEx Blood), RBP7 (PPH3 + PPH4 = 0.96/0.99, eQTLgen/GTEx Blood) and NFASC (PPH3 + PPH4 = 0.98, GTEx Prostate) were respectively Higher likelihood of sharing genetic variants with PCa in blood and prostate tissue (PPH3 + PPH4 > 0.8). Detailed results of colocalization analysis are shown in Fig. 4 and Supplementary Table 9.
Heatmap of colocalization analysis of 13 plasma proteins and prostate cancer in eQTLgen, GTEx Blood and Prostate. H3: SNP within the selected locus is associated with either protein or disease, but the two are independent SNPs; H4: SNP within the selected locus is concurrently associated with both protein and disease, and is a shared SNP
3.3 Results of drug target SMR analysis
We performed SMR analysis of the expression levels of genes encoding plasma proteins in blood and prostate tissues with GWAS data (PRACTICAL) for PCa and validated them against GWAS data from UK Biobank. The results showed that the expression levels of ZG16B and PEX14 in blood were positively correlated with PCa. The expression levels of ZG16B and NAPG in prostate tissues were positively correlated with PCa. Detailed results of SMR analysis are shown in Fig. 5A–F and Supplementary Table 10.
SMR analysis of the expression levels of genes encoding plasma proteins in blood tissue (eQTLgen) with prostate cancer in two GWAS datasets. A–C ZG16B, PEX14 and NAPG (PRACTICAL). D–F. ZG16B, PEX14 and NAPG (UK Biobank). G Dotplot of result of PheWAS analysis of associations between ZG16B, PEX14 and NAPG and other disease outcomes.
3.4 Phenome-wide association study of three plasma proteins associated with PCa
We applied Phenome-wide association study to assess the potential impact of the three plasma proteins associated with prostate cancer in the SMR analysis on other phenotypes. Among 2803 phenotypes screened from the Finnish database, we observed a significant causal association (P < 0.05) between plasma ZG16B and 78 phenotypes, in which ZG16B level were positively associated with the risk of diseases such as corneal degeneration, thyroid disorders, and chronic bronchitis, and negatively associated with the risk of disorders of mineral metabolism, cerebrovascular disorders, and intervertebral disc infections. There was a significant causal relationship (P < 0.05) between plasma PEX14 and 88 phenotypes, in which PEX14 levels were positively associated with the risk of diseases such as disorders of porphyrin and bilirubin metabolism, liver disease, and esophagitis, and negatively associated with the risk of diseases such as benign tumors of the rectum or anal canal, autism, and urticaria nodosa. There was a significant causal relationship (P < 0.05) between plasma NAPG and 121 phenotypes, in which NAPG levels were positively associated with the risk of diseases such as personality or behavioral disorders, vascular occlusive disease, and pulmonary cysts, whereas they were negatively associated with the risk of diseases such as parathyroid disorders, liver disease, and ulcerative colitis. Detailed data are shown in Fig. 5G and Supplementary Table 11.
3.5 Effects of 26 life-related factors on PCa-related proteins
Among 26 common lifestyle-related factors we selected, the frequency of alcohol intake was positively correlated with TPST1 and SERPINA5; BMI was positively correlated with RBP7, TPST1, NFASC, LAYN, HDGF, SERPINA5, DLL4, LIMA1, and CCL27; body weight was positively correlated with RBP7, LAYN, TPST1, HDGF SERPINA5, DLL4, NFASC, and LIMA1; hip circumference was positively associated with RBP7, LAYN, and HDGF; Cannabis use was negatively associated with SERPINA5; fat intake was negatively associated with EFNA3; hypertension was negatively associated with EFNA3; aspirin use was positively associated with LAYN and NFASC; and type 2 diabetes was negatively associated with EFNA3 (Pfdr < 0.05). Detailed results are shown in Fig. 6 and Supplementary Table 12.
4 Discussion
PCa is the second most common cause of cancer-related deaths in men worldwide [21]. A deficiency in the lack of specificity of serum prostate-specific antigen, the most widely used test, has been shown to lead to over-diagnosis and over-treatment of PCa [22]. Prostate gland is located deep in the pelvis, which makes it difficult to have a direct contact with other organs. The circulatory system is the primary object that comes into contact with and potentially affects the prostate, and blood samples are relatively easy to obtain and less invasive to the patient. Therefore, investigating the association between plasma proteins and PCa and discovering protein targets in the blood has positive and practical clinical implications for the prediction and treatment of PCa.
In this study, the plasma proteins positively associated with PCa were EFNA3, LIMA1, HDGF, PEX14, DLL4, NAPG, ZG16B, TPST1 and NFASC. EFNA3 (Ephrin-A3) belongs to the family of membrane-bound ligands of hepatic collagens. Many members of the hepatic collagen family are aberrantly expressed in cancer cells, often predicting more aggressive tumors, higher likelihood of metastasis, and poorer prognosis [23]. Overexpression of EFNA3 accelerates self-renewal of hepatocellular carcinoma cells, and enhances proliferation and migration [24]. Gastric cancer patients with high expression of EFNA3 have a worse overall and disease-free survival, the mechanisms of which may involve immune cell infiltration and immune checkpoint regulation [25]. LIMA1 (LIM Domain And Actin Binding 1) plays an important role in regulating actin cytoskeleton dynamics, and its defective expression may lead to dysregulation of cytoskeleton dynamics, altered cell motility, and inter-cellular adhesion breaks, thus promoting tumor proliferation, invasion, and migration [26]. LIMA1 up-regulation leads to prostate cancer cell invasion and reduced extracellular matrix adhesion [27, 28]. These studies emphasize the importance of endogenous LIMA1 in regulating PCa cells growth and invasiveness. However, tumor cells still require sufficient exogenous LIMA1 protein to shape their own cytoskeleton. In addition, whether plasma LIMA1 protein inhibits LIMA1 expression levels in tumor cells remains to be further investigated. HDGF (Hepatoma Derived Growth Factor) up-regulation is present in many types of tumors and positively correlates with clinicopathological features [29]. Mechanisms by which HDGF regulates tumor progression include PI3K/AKT and ERK signaling pathway activation29, epithelial-mesenchymal transition promotion [30] and vascular endothelial growth factor induction [31]. HDGF plays an important role in prostate cancer cell growth, apoptosis, and invasion, which may be mediated by the AKT and NF-κB pathways [32]. PEX14 (Peroxisomal Biogenesis Factor 14), a cellular biosynthesis factor involved in cellular redox homeostasis and lipid metabolism, is a peroxisome [33]. Its aberrant regulation causes tumor angiogenesis and extracellular matrix degradation [34]. PEX14 is up-regulation in aggressive PCa with loss-of-function mutations in TP53, suggesting the importance of peroxisomes in advanced PCa [35]. The results of colocalization and SMR analyses in our study support that PEX14 expression levels are positively correlated with PCa. According to the results of PheWAS analysis, the use of PEX14 inhibitors may be beneficial in ameliorating disorders of porphyrin and bilirubin metabolism, tear overflow, and liver-related disorders, but may increase the risk of diseases such as esophagitis and benign tumors of the rectum and anal canal. DLL4 (Delta Like Canonical Notch Ligand 4) plays an important role in tumor angiogenesis [36]. Knockdown of DLL4 impedes Notch-1 signaling pathway activation and inhibits the self-renewal and invasive ability of gastric cancer stem cells as well as resistance to 5-FU chemotherapy [37]. NAPG (NSF Attachment Protein Gamma) encodes a gamma-soluble NSF attachment protein that mediates platelet cytokinesis and controls membrane fusion events. Mutations in NAPG may lead to insufficient platelet spreading, triggering autosomal dominant angiodysplasia syndrome [38]. Our study found that high expression of NAPG in prostate tissues increases the risk of PCa, but there is no other relevant literature to support this view. Interestingly, a paper describing genetic variation and adaptive phenotypes in high-altitude populations triggered our thinking. NAPG may be involved in oxygen sensing, metabolism, and vascular homeostasis, and is associated with physiological adaptations in low-pressure hypoxic environments [39]. In PheWAS analysis, plasma NAPG was positively associated with vascular occlusion, HER2-negative breast cancer, and nonorganic psychiatric disorders, and negatively associated with hyperparathyroidism, colitis, and retinal chorioretinitis. The biological function of NAPG in PCa remains to be further explored. ZG16B (Zymogen granule protein 16B) as a growth factor overexpressed in pancreatic cancer, inhibits NF-κB signaling thereby promoting tumor proliferation and enabling immune escape [40]. ZG16B also enhances angiogenesis through activation of CXCR4 and FAK, increases vascular permeability, and facilitates pancreatic cancer progression and metastasis [41]. In colorectal cancer, up-regulation of ZG16B, through modulation of the Wnt/β-catenin pathway enhances immunosuppressive activity in the tumor microenvironment and enhances tumor cell migration and invasion, leading to a poor prognosis [42]. In addition, ZG16B is similarly highly expressed in cervical, oral squamous cell, and ovarian cancers [43,44,45], which is consistent with our findings in PCa. Of interest, ZG16B upregulation seems to be associated with a better prognosis in PCa patients [46]. This phenomenon suggests that the biological mechanisms of ZG16B may change with the progression of PCa. The results of the PheWAS analysis suggest that the use of ZG16B inhibitors may ameliorate diseases such as corneal degeneration, chronic bronchitis, and thyrotoxicosis, but may increase risk of neurologic abnormalities, mineral metabolism disorders and cerebrovascular disease. Therefore, the development of drugs targeting ZG16B still requires more comprehensive and careful evaluation. TPST1 (Tyrosylprotein Sulfotransferase 1) is overexpressed in breast cancer [47], oral squamous cell carcinoma [48] and nasopharyngeal carcinoma [49]. In bladder cancer, elevated expression of TPST1 is associated with high pathologic stage and poor survival, and correlates with the immune profile and responsiveness to immunotherapy in bladder cancer. characteristics and responsiveness to immunotherapy [50]. NFASC (Neurofascin), a member of the immunoglobulin superfamily of adhesion molecules, may play a role in the regulation of the cytoskeleton and migration of cancer cells [51]. NFASC and its potential regulatory variants suggest that long-range chromatin interactions may be an etiological factor in PCa [52].
The plasma proteins that were negatively associated with prostate cancer risk in this study were SERPINA5, RBP7, CCL27, and LAYN. SERPINA5 (Serpin Family A Member 5), a serine protease inhibitor, belongs to the serine protease inhibitor superfamily. Previous reports have shown that SERPINA5 is lowly expressed in a range of cancers, including renal, breast, thyroid, colorectal, prostate, and ovarian cancers [53,54,55,56,57,58]. SERPINA5 exerts its anticancer properties through anti-angiogenesis and inhibits tumor metastasis [59]. RBP7 (Retinol Binding Protein 7), a member of the cellular retinol-binding protein family, is involved in immune regulation and can be used to predict the prognosis of patients with uroepithelial carcinoma of the bladder [60]. High expression of RBP7 correlates with tumor invasion and epithelial mesenchymal transition, which predicts a poor prognosis for patients with colorectal carcinoma [61]. CCL27 (C-C Motif Chemokine Ligand 27), which is mainly produced by keratinocytes, is critical in directing immune cells to epithelial and mucosal tissues. Down-regulation of CCL27 in melanoma, basal cell carcinoma, colon cancer, and breast cancer allows tumor cells to achieve immune escape, which in turn leads to tumor progression [62]. LAYN (Layilin), which is located on chromosome 11 and shares homology with C-type lectins, also acts as a hyaluronic acid surface receptor, which plays an important role in cell adhesion, motility, and migration [63]. In most tumors, LAYN up-regulation is associated with poor prognosis, but LAYN expression levels are low in PCa. This phenomenon reflects the differential expression of LAYN among tumors as well as different biological functions [64]. A similar situation can be seen in melanoma, where LAYN is highly expressed on CD8T cells, which promotes integrin-mediated cell adhesion and thus enhances anti-tumor immunity [65].
The strengths of the current study are as follows. First, this is a systematic and extensive study exploring the causal relationship between plasma proteins and PCa, which helps to provide a comprehensive perspective to understand the etiologic role of circulating proteins in PCa. Second, this study employed a variety of analytical methods based on MR and validated them in multiple datasets of different dimensions to identify the association between plasma proteins and PCa in blood and prostate tissues respectively, to improve the accuracy and robustness of the analysis. Third, we performed an extensive evaluation of potential downstream associated phenotypes and upstream intervening factors for PCa-related plasma proteins. However, this study also has limitations. First, all GWAS participants are from Europe, which can greatly avoid the interference of geographical differences, but also limits the general applicability of the analysis results. Second, the expression levels or predictive values of the target genes we identified may fluctuate in different types of PCa or different stages of PCa progression. Therefore, future studies are best conducted in specific types or stages of prostate cancer, and experimental studies are needed to confirm these findings.
5 Conclusion
In conclusion, by applying an MR approach to a broad range of plasma proteins we identified causal associations between 13 plasma proteins and PCa, and prioritized the potential intervention protein targets by drugs or lifestyle changes, which provided new insights into the etiology, prevention and treatment of PCa.
Data availability
All the summary-level GWAS and eQTL in this study are publicly available for download by qualified researchers as shown in Supplementary Table 1. All data generated in this study can be obtained from the Supplementary Tables.
References
Ferlay J, Shin H, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Intl J Cancer. 2010;127(12):2893–917. https://doi.org/10.1002/ijc.25516.
Bergengren O, Pekala KR, Matsoukas K, et al. 2022 update on prostate cancer epidemiology and risk factors—a systematic review. Eur Urol. 2023;84(2):191–206. https://doi.org/10.1016/j.eururo.2023.04.021.
Eeles R, Goh C, Castro E, et al. The genetic epidemiology of prostate cancer and its clinical implications. Nat Rev Urol. 2014;11(1):18–31. https://doi.org/10.1038/nrurol.2013.266.
Cuzick J, Thorat MA, Andriole G, et al. Prevention and early detection of prostate cancer. Lancet Oncol. 2014;15(11):e484–92. https://doi.org/10.1016/S1470-2045(14)70211-6.
Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteom. 2003;2(1):50. https://doi.org/10.1074/mcp.A300001-MCP200.
Xu JY, Zhang C, Wang X, et al. Integrative proteomic characterization of human lung adenocarcinoma. Cell. 2020;182(1):245–e26117. https://doi.org/10.1016/j.cell.2020.05.043.
Cairat M, Rinaldi S, Navionis AS, et al. Circulating inflammatory biomarkers, adipokines and breast cancer risk—a case-control study nested within the EPIC cohort. BMC Med. 2022;20(1):118. https://doi.org/10.1186/s12916-022-02319-y.
Mukama T, Fortner RT, Katzke V, et al. Prospective evaluation of 92 serum protein biomarkers for early detection of ovarian cancer. Br J Cancer. 2022;126(9):1301–9. https://doi.org/10.1038/s41416-021-01697-z.
Storm CS, Kia DA, Almramhi MM, et al. Finding genetically-supported drug targets for Parkinson’s disease using mendelian randomization of the druggable genome. Nat Commun. 2021;12(1):7342. https://doi.org/10.1038/s41467-021-26280-1.
Ferkingstad E, Sulem P, Atlason BA, et al. Large-scale integration of the plasma proteome with genetics and disease. Nat Genet. 2021;53(12):1712–21. https://doi.org/10.1038/s41588-021-00978-w.
Mälarstig A, Grassmann F, Dahl L, et al. Evaluation of circulating plasma proteins in breast cancer using mendelian randomisation. Nat Commun. 2023;14(1):7680. https://doi.org/10.1038/s41467-023-43485-8.
Walker VM, Zheng J, Gaunt TR, Smith GD. Phenotypic causal inference using genome-wide association study data: mendelian randomization and beyond. Annu Rev Biomed Data Sci. 2022;5(1):1–17. https://doi.org/10.1146/annurev-biodatasci-122120-024910.
Jerome RN, Joly MM, Kennedy N, et al. Leveraging human genetics to identify safety signals prior to drug marketing approval and clinical use. Drug Saf. 2020;43(6):567–82. https://doi.org/10.1007/s40264-020-00915-6.
Sun BB, Maranville JC, Peters JE, et al. Genomic atlas of the human plasma proteome. Nature. 2018;558(7708):73–9. https://doi.org/10.1038/s41586-018-0175-2.
Burgess S, Thompson SG. Bias in causal estimates from mendelian randomization studies with weak instruments. Stat Med. 2011;30(11):1312–23. https://doi.org/10.1002/sim.4197.
Giambartolomei C, Vukcevic D, Schadt EE, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014. https://doi.org/10.1371/journal.pgen.1004383.
Võsa U, Claringbould A, Westra HJ, et al. Large-scale cis- and trans-eQTL analyses identify thousands of genetic loci and polygenic scores that regulate blood gene expression. Nat Genet. 2021;53(9):1300–10. https://doi.org/10.1038/s41588-021-00913-z.
GTEx Consortium. The GTEx consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369(6509):1318–30. https://doi.org/10.1126/science.aaz1776.
Julian TH, Girach Z, Sanderson E, et al. Causal factors in primary open angle glaucoma: a phenome-wide mendelian randomisation study. Sci Rep. 2023;13(1):9984. https://doi.org/10.1038/s41598-023-37144-7.
Cai YX, Wu YQ, Liu J, et al. Proteome-wide analysis reveals potential therapeutic targets for colorectal cancer: a two-sample mendelian randomization study. BMC Cancer. 2023;23(1):1188. https://doi.org/10.1186/s12885-023-11669-6.
Lorenc T, Klimczyk K, Michalczewska I, Słomka M, Kubiak-Tomaszewska G, Olejarz W. Exosomes in prostate Cancer diagnosis, prognosis and therapy. IJMS. 2020;21(6):2118. https://doi.org/10.3390/ijms21062118.
Teng M, Zhou S, Cai C, Lupien M, He HH. Pioneer of prostate cancer: past, present and the future of FOXA1. Protein Cell. 2021;12(1):29–38. https://doi.org/10.1007/s13238-020-00786-8.
Kou CTJ, Kandpal RP. Differential expression patterns of eph receptors and ephrin ligands in human cancers. Biomed Res Int. 2018;2018:1–23. https://doi.org/10.1155/2018/7390104.
Husain A, Chiu YT, Sze KMF, et al. Ephrin-A3/EphA2 axis regulates cellular metabolic plasticity to enhance cancer stemness in hypoxic hepatocellular carcinoma. J Hepatol. 2022;77(2):383–96. https://doi.org/10.1016/j.jhep.2022.02.018.
Zheng P, Liu X, Li H, et al. EFNA3 is a prognostic biomarker correlated with Immune Cell Infiltration and Immune checkpoints in gastric Cancer. Front Genet. 2022;12:796592. https://doi.org/10.3389/fgene.2021.796592.
Wu D. Epithelial protein lost in neoplasm (EPLIN): beyond a tumor suppressor. Genes Dis. 2017;4(2):100–7. https://doi.org/10.1016/j.gendis.2017.03.002.
Sanders AJ, Martin TA, Ye L, Mason MD, Jiang WG. EPLIN is a negative Regulator of prostate Cancer Growth and Invasion. J Urol. 2011;186(1):295–301. https://doi.org/10.1016/j.juro.2011.03.038.
Collins RJ, Morgan LD, Owen S, Ruge F, Jiang WG, Sanders AJ. Mechanistic insights of epithelial protein lost in neoplasm in prostate cancer metastasis. Intl J Cancer. 2018;143(10):2537–50. https://doi.org/10.1002/ijc.31786.
Bao C, Wang J, Ma W, Wang X, Cheng Y. HDGF: a Novel Jack-of-All-trades in Cancer. Future Oncol. 2014;10(16):2675–85. https://doi.org/10.2217/fon.14.194.
Chen S, Kung M, Hu T, et al. Hepatoma-derived growth factor regulates breast cancer cell invasion by modulating epithelial–mesenchymal transition. J Pathol. 2012;228(2):158–69. https://doi.org/10.1002/path.3988.
Okuda Y, Nakamura H, Yoshida K, et al. Hepatoma-derived growth factor induces tumorigenesis in vivo through both direct angiogenic activity and induction of vascular endothelial growth factor. Cancer Sci. 2003;94(12):1034–41. https://doi.org/10.1111/j.1349-7006.2003.tb01397.x.
Shetty A, Dasari S, Banerjee S, et al. Hepatoma-derived growth factor: a survival-related protein in prostate oncogenesis and a potential target for vitamin K2. Urologic Oncology: Seminars Original Investigations. 2016;34(11):483. https://doi.org/10.1016/j.urolonc.2016.05.027.
Feng Y, Rhie SK, Huo D, et al. Characterizing genetic susceptibility to breast Cancer in women of African ancestry. Cancer Epidemiol Biomarkers Prev. 2017;26(7):1016–26. https://doi.org/10.1158/1055-9965.EPI-16-0567.
Kim JA. Peroxisome metabolism in Cancer. Cells. 2020;9(7):1692. https://doi.org/10.3390/cells9071692.
Zhang Y, Song X, Yu B, et al. TP53 loss-of‐function causes vulnerability to autophagy inhibition in aggressive prostate cancer. Int J Urol. 2022;29(9):1085–94. https://doi.org/10.1111/iju.15021.
Fasoulakis Z, Koutras A, Ntounis T, et al. The prognostic role and significance of Dll4 and toll-like receptors in Cancer Development. Cancers. 2022;14(7):1649. https://doi.org/10.3390/cancers14071649.
Liu Z, Fan F, Wang A, Zheng S, Lu Y. Dll4-Notch signaling in regulation of tumor angiogenesis. J Cancer Res Clin Oncol. 2014;140(4):525–36. https://doi.org/10.1007/s00432-013-1534-x.
Xu Y, Zhang YB, Liang LJ, et al. NAPG mutation in family members with hereditary hemorrhagic telangiectasia in China. BMC Pulm Med. 2021;21(1):197. https://doi.org/10.1186/s12890-021-01524-4.
Sharma S, Koshy R, Kumar R, et al. Hypobaric hypoxia drives selection of altitude-associated adaptative alleles in the himalayan population. Sci Total Environ. 2024;913:169605. https://doi.org/10.1016/j.scitotenv.2023.169605.
Park HD, Lee Y, Oh YK, et al. Pancreatic adenocarcinoma upregulated factor promotes metastasis by regulating TLR/CXCR4 activation. Oncogene. 2011;30(2):201–11. https://doi.org/10.1038/onc.2010.401.
Kim SJ, Lee Y, Kim NY, et al. Pancreatic adenocarcinoma upregulated factor, a novel endothelial activator, promotes angiogenesis and vascular permeability. Oncogene. 2013;32(31):3638–47. https://doi.org/10.1038/onc.2012.366.
Escudero-Paniagua B, Bartolomé RA, Rodríguez S, et al. PAUF/ZG16B promotes colorectal cancer progression through alterations of the mitotic functions and the Wnt/β-catenin pathway. Carcinog Published Online May. 2019;17:bgz093. https://doi.org/10.1093/carcin/bgz093.
Zhang GH, Chen MM, Kai JY, et al. Molecular profiling of mucinous epithelial ovarian cancer by weighted gene co-expression network analysis. Gene. 2019;709:56–64. https://doi.org/10.1016/j.gene.2019.05.034.
Choi CH, Chung JY, Park HS, et al. Pancreatic adenocarcinoma up-regulated factor expression is associated with disease-specific survival in cervical cancer patients. Hum Pathol. 2015;46(6):884–93. https://doi.org/10.1016/j.humpath.2015.02.016.
Sasahira T, Kurihara M, Nishiguchi Y, Nakashima C, Kirita T, Kuniyasu H. Pancreatic adenocarcinoma up-regulated factor has oncogenic functions in oral squamous cell carcinoma. Histopathology. 2017;70(4):539–48. https://doi.org/10.1111/his.13097.
Zhang T, Wang Y, Dong Y, et al. Identification of Novel diagnostic biomarkers in prostate adenocarcinoma based on the stromal-immune score and analysis of the WGCNA and ceRNA network. Disease Markers. 2022;2022:1–10. https://doi.org/10.1155/2022/1909196.
Zhao H, Langerød A, Ji Y, et al. Different gene expression patterns in Invasive Lobular and Ductal carcinomas of the breast. MBoC. 2004;15(6):2523–36. https://doi.org/10.1091/mbc.e03-11-0786.
Toruner GA, Ulger C, Alkan M, et al. Association between gene expression profile and tumor invasion in oral squamous cell carcinoma. Cancer Genet Cytogenet. 2004;154(1):27–35. https://doi.org/10.1016/j.cancergencyto.2004.01.026.
Xu J, Deng X, Tang M, et al. Tyrosylprotein sulfotransferase-1 and tyrosine sulfation of chemokine receptor 4 are induced by epstein-barr virus encoded latent membrane protein 1 and associated with the metastatic potential of human nasopharyngeal carcinoma. PLoS ONE. 2013;8(3):e56114 https://doi.org/10.1371/journal.pone.0056114.
Zhang Y, Lin Y, Lv D, et al. Identification and validation of a novel signature for prediction the prognosis and immunotherapy benefit in bladder cancer. PeerJ. 2022;10:e12843. https://doi.org/10.7717/peerj.12843.
Samulin Erdem J, Arnoldussen YJ, Skaug V, Haugen A, Zienolddiny S. Copy number variation, increased gene expression, and molecular mechanisms of neurofascin in lung cancer. Mol Carcinog. 2017;56(9):2076–85. https://doi.org/10.1002/mc.22664.
Du M, Tillmans L, Gao J, et al. Chromatin interactions and candidate genes at ten prostate cancer risk loci. Sci Rep. 2016;6(1):23202. https://doi.org/10.1038/srep23202.
Asanuma K, Yoshikawa T, Hayashi T, et al. Protein C inhibitor inhibits breast cancer cell growth, metastasis and angiogenesis independently of its protease inhibitory activity. Intl J Cancer. 2007;121(5):955–65. https://doi.org/10.1002/ijc.22773.
Bijsmans ITGW, Smits KM, De Graeff P, et al. Loss of SerpinA5 protein expression is associated with advanced-stage serous ovarian tumors. Mod Pathol. 2011;24(3):463–70. https://doi.org/10.1038/modpathol.2010.214.
Wakita T, Hayashi T, Nishioka J, et al. Regulation of carcinoma cell invasion by protein C inhibitor whose expression is decreased in renal cell carcinoma. Intl J Cancer. 2004;108(4):516–23. https://doi.org/10.1002/ijc.11594.
Zhang T, Yuan K, Wang Y, et al. Identification of candidate biomarkers and prognostic analysis in Colorectal Cancer Liver metastases. Front Oncol. 2021;11:652354. https://doi.org/10.3389/fonc.2021.652354.
Brenner AV, Neta G, Sturgis EM, et al. Common single nucleotide polymorphisms in genes related to immune function and risk of papillary thyroid cancer. PLoS ONE. 2013;8(3):e57243. https://doi.org/10.1371/journal.pone.0057243.
Cao Y, Becker C, Lundwall Å, et al. Expression of protein C inhibitor (PCI) in benign and malignant prostatic tissues. Prostate. 2003;57(3):196–204. https://doi.org/10.1002/pros.10296.
Jing Y, Jia D, Wong CM, et al. SERPINA5 inhibits tumor cell migration by modulating the fibronectin-integrin β1 signaling pathway in hepatocellular carcinoma. Mol Oncol. 2014;8(2):366–77. https://doi.org/10.1016/j.molonc.2013.12.003.
Jin K, Qiu S, Jin D, et al. Development of prognostic signature based on immune-related genes in muscle-invasive bladder cancer: bioinformatics analysis of TCGA database. Aging. 2021;13(2):1859–71. https://doi.org/10.18632/aging.103787.
Elmasry M, Brandl L, Engel J, Jung A, Kirchner T, Horst D. RBP7 is a clinically prognostic biomarker and linked to tumor invasion and EMT in colon cancer. J Cancer. 2019;10(20):4883–91. https://doi.org/10.7150/jca.35180.
Lin HY, Sun SM, Lu XF, et al. CCR10 activation stimulates the invasion and migration of breast cancer cells through the ERK1/2/MMP-7 signaling pathway. Int Immunopharmacol. 2017;51:124–30. https://doi.org/10.1016/j.intimp.2017.07.018.
Bono P, Rubin K, Higgins JMG, Hynes RO. Layilin, a novel integral membrane protein, is a hyaluronan receptor. MBoC. 2001;12(4):891–900. https://doi.org/10.1091/mbc.12.4.891.
Pan Jhua, Zhou H, Cooper L, et al. LAYN is a prognostic biomarker and correlated with Immune infiltrates in gastric and Colon cancers. Front Immunol. 2019;10:6. https://doi.org/10.3389/fimmu.2019.00006.
Mahuron KM, Moreau JM, Glasgow JE, et al. Layilin augments integrin activation to promote antitumor immunity. J Exp Med. 2020;217(9):e20192080. https://doi.org/10.1084/jem.20192080.
Acknowledgements
The authors thank all the participants from proteomic studies, prostate cancer genome-wide association studies, and FinnGen cohort, the institutions and their staff for providing data.
Funding
This study was supported by the National Natural Science Foundation of China (No.82160148); Gansu Province Science and Technology Department Youth Science Fund Project (No.23JRRA1640); Gansu Province Department of Education College Teachers’ Innovation Fund Project (No.2023B-032); 2023 Natural Science Central Universities Basic Scientific Research Business Fee Interdisciplinary Innovation Team Construction Project (No.lzujbky-2023-ct06); Gansu Province Science and Technology Department Joint Research Fund General Project (No.23JRRA1509), and the Second Hospital of Lanzhou University, Cuiying Science and Technology Innovation Plan Project, Applied Basic Research - Youth Fund Project (No.CY2022-QN-A09).
Author information
Authors and Affiliations
Contributions
Conceptualization, Zhilong Dong and Long Cheng; methodology, Long Cheng and Zeming Qiu; data curation, Xuewu Wu; writing-original draft preparation, Long Cheng; writing-review and editing, Zhilong Dong.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All the summary-level GWAS and eQTL data used in the analyses are publicly available, and therefore ethical approval was not imperative for this study. Ethical approval for the GWASs can be found in the corresponding GWAS publications cited in the manuscript.
Consent for publication
All authors have read and agreed to the published version of the manuscript.
Competing interests
The authors declare no potential Competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Cheng, L., Qiu, Z., Wu, X. et al. Evaluation of circulating plasma proteins in prostate cancer using mendelian randomization. Discov Onc 15, 453 (2024). https://doi.org/10.1007/s12672-024-01331-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12672-024-01331-3