Abstract
BH3-mimetic drugs are an anti-cancer therapy that can induce apoptosis in malignant cells by directly binding and inhibiting pro-survival proteins of the BCL-2 family. The BH3-mimetic drug venetoclax, which targets BCL-2, has been approved for the treatment of chronic lymphocytic leukaemia and acute myeloid leukaemia by regulatory authorities worldwide. However, while most patients initially respond well, resistance and relapse while on this drug is an emerging and critical issue in the clinic. Though some studies have begun uncovering the factors involved in resistance to BCL-2-targeting BH3-mimetic drugs, little focus has been applied to pre-emptively tackle resistance for the next generation of BH3-mimetic drugs targeting MCL-1, which are now in clinical trials for diverse blood cancers. Therefore, using pre-clinical mouse and human models of aggressive lymphoma, we sought to predict factors likely to contribute to the development of resistance in patients receiving MCL-1-targeting BH3-mimetic drugs. First, we performed multiple whole genome CRISPR/Cas9 KO screens and identified that loss of the pro-apoptotic effector protein BAX, but not its close relative BAK, could confer resistance to MCL-1-targeting BH3-mimetic drugs in both short-term and long-term treatment regimens, even in lymphoma cells lacking the tumour suppressor TRP53. Furthermore, we found that mouse Eµ-Myc lymphoma cells selected for loss of BAX, as well as upregulation of the untargeted pro-survival BCL-2 family proteins BCL-XL and A1, when made naturally resistant to MCL-1 inhibitors by culturing them in increasing doses of drug over time, a situation mimicking the clinical application of these drugs. Finally, we identified therapeutic approaches which could overcome these two methods of resistance: the use of chemotherapeutic drugs or combined BH3-mimetic treatment, respectively. Collectively, these results uncover some key factors likely to cause resistance to MCL-1 inhibition in the clinic and suggest rational therapeutic strategies to overcome resistance that should be investigated further.
Similar content being viewed by others
Introduction
The BCL-2 family-regulated (a.k.a. intrinsic, mitochondrial) pathway to apoptosis is critical for the removal of damaged or unwanted cells, and the killing of cells in response to many anti-cancer agents [1, 2]. Following a death stimulus, the levels of BH3-only proteins (e.g., BIM, NOXA, PUMA, BMF) are increased and these critical inducers of apoptosis bind via their BH3 domain into the hydrophobic pocket of the pro-survival BCL-2 family proteins (BCL-2, MCL-1, BCL-XL, BCL-W, A1/BFL-1). Some BH3-only proteins (e.g., PUMA, BIM) have also been reported to activate the pro-apoptotic effector proteins BAX and BAK directly [1, 2]. Together these events allow activated BAX and BAK to oligomerise and form pores in the mitochondrial outer membrane. Release of apoptogenic factors from the mitochondria leads to the activation of caspases and, ultimately, dismantling of the cell [3].
Direct activation of the intrinsic apoptotic pathway using drugs termed BH3-mimetics has shown success for therapy of certain blood cancers [4, 5]. These drugs mimic the interaction of the BH3 domain from the pro-apoptotic BH3-only proteins with the hydrophobic groove of the pro-survival BCL-2 proteins, thereby allowing activated BAX and BAK to induce apoptosis [5, 6]. The most clinically advanced BH3-mimetic drug is the BCL-2-specific inhibitor venetoclax, which is approved by the FDA and many other international regulatory authorities for therapy of chronic lymphocytic leukemia (CLL) and acute myeloid leukemia (AML) [7,8,9]. BH3-mimetic drugs targeting related pro-survival BCL-2 proteins have also been developed for cancer therapy. A wealth of evidence supports the notion that MCL-1 inhibitors would be efficacious for a broad range of haematological malignancies, including for aggressive MYC-driven lymphomas, such as Burkitt Lymphomas (BL), which are known to be highly dependent on MCL-1 for continued survival and proliferation [10,11,12,13,14,15,16]. Such MYC-driven lymphomas can be modelled pre-clinically using the Eµ-Myc transgenic mouse model, which recapitulates the immunoglobulin/c-MYC chromosomal translocation characteristic of BL, and results in pre-B/B cell lymphomas with almost 100% incidence [17]. When these mouse lymphoma cells are transplanted into recipient mice, loss of even one allele of Mcl-1 can invoke tumour regression, demonstrating their high dependence on MCL-1 for sustained survival and tumour expansion [14]. There is also evidence that MCL-1-targeting BH3-mimetics could be effective for certain solid cancers when used alongside inhibitors of oncogenic kinases [18,19,20]. The clinical progression of MCL-1-targeting BH3-mimetics has been limited by on-target toxicity, since MCL-1 is required for the survival of many healthy cell types, including cardiomyocytes [5]. Nevertheless, evidence suggests a therapeutic window for MCL-1 inhibitors can be achieved, where tumour cells are killed but healthy cells are spared [20]. As such, six MCL-1 inhibitors have entered into stage 1 clinical trials for relapsed/refractory lymphomas, AML and multiple myeloma [5].
Recent evidence from clinical trials with venetoclax in CLL and AML patients has shown that drug resistance can emerge over time despite initial complete responses [21,22,23]. Therefore, understanding the underlying mechanisms of resistance to BH3-mimetic drugs is critical to improve therapeutic outcomes in patients by developing rational drug combination therapies. In previous studies using cell lines and patient samples derived from diverse blood cancers, we identified loss of the tumour suppressor TP53 as a resistance factor to single agent therapy using either BCL-2-specific or MCL-1-specific BH3-mimetic drugs [24]. This was consistent with the analysis of CLL and AML patients that relapsed on venetoclax therapy, as well as with other published studies seeking to identify resistance factors to BCL-2 inhibitors [22, 23, 25,26,27,28,29]. Whilst performing CRISPR/Cas9 whole genome knockout (KO) screens to identify factors that confer resistance to MCL-1-targeting BH3-mimetics in Eµ-Myc lymphoma cells, we additionally identified loss of the apoptotic effector BAX as a top hit [24]. Here we delve deeper into this result, showing that loss of BAX is also a hit when such screens are performed in Trp53-deficient Eµ-Myc lymphoma cells, and additionally arises as the key mediator of resistance in longer-term CRISPR/Cas9 gene KO screens using suboptimal doses of MCL-1 inhibitors. Loss of BAX expression is also a prominent feature in Eµ-Myc lymphoma cells selected to be BH3-mimetic drug resistant through culturing for extended periods in increasing doses of these agents. These data reveal that BAX, but not BAK, is the key mediator of optimal MCL-1 inhibitor-induced killing of malignant cells. Importantly, such BAX-deficient lymphoma cells, unlike TRP53-deficient lymphoma cells, can still be potently killed by standard chemotherapeutic drugs, indicating a potential therapeutic strategy to overcome this mode of resistance to BH3-mimetic drugs.
Results
Whole genome CRISPR/Cas9 gene knockout screens consistently identify loss of BAX as a resistance factor to MCL-1-targeting BH3-mimetic drugs in Eµ-Myc mouse lymphoma cells
We previously identified loss of TRP53 as a factor which could confer resistance to MCL-1-targeting BH3-mimetic drugs in short-term CRISPR/Cas9 KO screens performed in Eµ-Myc mouse lymphoma cells [24]. To explore this further, we generated a Trp53 KO Eµ-Myc lymphoma cell line (AH15A Trp53 KO) using CRISPR/Cas9 gene editing, and validated loss of TRP53 by western blotting (Fig. 1A). Despite being the top hit identified in these previous screens, we found that loss of TRP53 provided only modest resistance to MCL-1 inhibitors during short-term killing assays (24 h) (Fig. 1B). Therefore, as many human cancers have TP53 mutations, we sought to identify additional factors that might enable TRP53-deficient lymphoma cells to resist high doses of BH3-mimetics. To this end, we performed a whole genome CRISPR/Cas9 KO screen in Trp53 KO cells using the mouse YUSA sgRNA library [30] (Fig. 1C). We treated library-infected, Cas9-expressing cells with a high dose of the MCL-1-targeting BH3-mimetic S63845 (IC99) for a short timeframe (24 h) and found that sgRNAs targeting the pro-apoptotic BCL-2 family gene Bax were significantly enriched by drug treatment (Fig. 1D). No other significant hits were identified from this screen, suggesting that loss of TRP53 and BAX are the most potent resistance factors for MCL-1 inhibitors.
A Western blot to validate CRISPR/Cas9-mediated KO of Trp53 in AH15A Eµ-Myc mouse lymphoma cells. Cells were treated with the TRP53-activating drug Nutlin-3a for 24 h to enable visualisation of stabilised wildtype TRP53 protein in Trp53 wildtype cells. Probing for β-ACTIN served as a loading control. B Dose–response curves for control AH15A Eµ-Myc lymphoma cells (containing Cas9 and a non-targeting control sgRNA) and isogenic Trp53 KO cells treated with the MCL-1 inhibitor S63845 for 24 h. Live cells were identified as Annexin V/PI double negative by flow cytometry. Trp53 KO cells showed a ~2-fold increase in IC50 for S63845 treatment. C Schematic of CRISPR/Cas9 screen performed in the AH15A Trp53 KO mouse Eµ-Myc lymphoma cell line. Cells were treated with DMSO (vehicle, control) or 1 µM of S63845 (~IC99 dose) for 24 h and surviving cells were collected for next-generation sequencing. D MAGeCK analysis to identify top enriched sgRNAs in S63845-resistant cell populations. Significant hits (FDR < 0.05) are indicated with black text. Loss of Bax was identified as the top, and only significant, hit that promoted drug resistance in the AH15A Trp53 KO Eµ-Myc lymphoma cell line.
We next aimed to identify factors that conferred resistance to suboptimal doses of S63845 over a longer timeframe, as this may be more representative of drug resistance that arises in patients (Fig. 2A). To this end, two independent, YUSA library-infected, Cas9-expressing Eµ-Myc lymphoma cell lines with wildtype Trp53 expression were treated with suboptimal doses (<IC50) of S63845 for 2 weeks. Significant enrichment of sgRNAs targeting Bax were identified (Fig. 2B). As in the previous screen performed in Trp53 KO cells, sgRNAs targeting the related pro-apoptotic gene Bak were not enriched after drug treatment. This indicates that loss of BAX, but not loss of BAK, confers resistance to MCL-1 inhibitors in these cells.
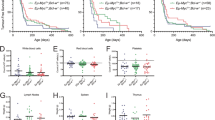
A Schematic of CRISPR/Cas9 screen performed in Eµ-Myc lymphoma cells with wildtype Trp53. Cells were treated with DMSO (vehicle, control) or 100 nM of S63845 (~IC50 dose) for 2 weeks and surviving cells were collected for next-generation sequencing. B MAGeCK analysis to identify top enriched sgRNAs in S63845-resistant cell populations. Significant hits (FDR < 0.05) are indicated with black text. Loss of Bax was identified as the top hit in the two independent Eµ-Myc lymphoma cell lines used. C Western blots to validate CRISPR/Cas9-mediated loss of BAK, BAX or both BAK and BAX in three independent Eµ-Myc lymphoma cell lines. Probing for HSP70 served as a loading control. D Dose–response curves for control Eµ-Myc lymphoma cells (containing Cas9 and a non-targeting control sgRNA) and isogenic Bak as well as Bax single KO or Bak/Bax double KO cells treated with the MCL-1 inhibitor S63845 for 24 h. Live cells were identified as Annexin V/PI double negative by flow cytometry. E Isogenic WT, Bak KO, Bax KO or Bak/Bax double KO AF47A lymphoma cells treated with the cytotoxic drugs doxorubicin, vincristine, cisplatin, etoposide or ionomycin. All data are presented as mean ± SD for 3 independent experiments. One-way ANOVA was used to measure statistical significance (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns = not significant).
To confirm this result, we used CRISPR/Cas9 to generate isogenic Eµ-Myc lymphoma cell lines that lack either BAK, BAX, or both of these effectors of apoptosis, as confirmed by western blotting (Fig. 2C). When treated with S63845, Bax KO lymphoma cells showed a 10-fold increase in IC50 compared to cells containing non-targeting (control) sgRNAs (control cells; Fig. 2D). In contrast, loss of BAK did not confer protection from S63845. Interestingly, loss of both BAK and BAX profoundly protected cells from even high doses of S63845, beyond that of BAX loss alone. This reveals that whilst BAX is the primary mediator of the apoptotic response in Eµ-Myc lymphoma cells following exposure to MCL-1-targeting BH3-mimetic drugs, BAK does play an important complementary role in this apoptotic process.
Eµ-Myc lymphoma cells lacking BAX expression can still be killed by cytotoxic drugs
One plausible explanation for our finding that loss of BAX but not loss of BAK confers resistance to MCL-1 inhibitors is that Eµ-Myc lymphoma cells depend only on BAX to undergo cell death. To determine whether loss of BAX conferred resistance selectively to S63845, or more generally to all cytotoxic drugs that induce apoptosis, isogenic Bak KO, Bax KO and Bak/Bax double KO cell lines from three independent Eµ-Myc lymphomas were generated using CRISPR/Cas9 gene editing and treated with standard-of-care agents that are part of the R-CHOP regimen, namely doxorubicin and vincristine [31]. In addition, we tested the sensitivity to cisplatin, which is used as salvage therapy for lymphoma patients [32] and also to the DNA-damaging agent etoposide as well as the calcium ionophore ionomycin (Fig. 2E and Supplementary Fig. 1). In all cases, loss of BAX conferred no substantial protection against these cytotoxic drugs compared to control cells, showing that loss of BAX is not a general resistance factor in these Eµ-Myc lymphoma cells, but confers specific resistance to S63845. In fact, the Bak KO Eµ-Myc lymphoma cells seemed more resistant to some of the standard-of-care drugs than the Bax KO lymphoma cells. The Bak/Bax double KO Eµ-Myc lymphoma cells were markedly resistant to all agents (Fig. 2E and Supplementary Fig. 1), confirming that these drugs kill these cells by inducing apoptosis.
We confirmed these results by crossing Eµ-Myc transgenic mice to mice that had been genetically engineered to have Bax or Bak gene KO. Cell lines were established from independent lymphomas that arose in three sick Eµ-Myc/Bak KO and three sick Eµ-Myc/Bax KO mice (Fig. 3A). Western blotting for TRP53 in these cell lines revealed that one Eµ-Myc/Bax KO cell line (171) likely possessed a Trp53 mutation, as indicated by stabilisation of TRP53 protein (Fig. 3A). Similar to the results in the CRISPR/Cas9-edited cells, Eµ-Myc/Bax KO cells were >10-fold more resistant to S63845 than Eµ-Myc/Bak KO cells (Fig. 3B). Neither lack of BAK nor lack of BAX protected these cells from etoposide or ionomycin (Fig. 3C, D). Together, these results confirm that conventional chemotherapies are still effective at killing BAX KO lymphoma cells that are resistant to S63845.
A Cell lines were derived from lymphomas arising in Eµ-Myc/Bak KO and Eµ-Myc/Bax KO mice. Western blotting was used to validate the loss of BAK or BAX protein in these cell lines, respectively. Probing for HSP70 was used as a loading control. B Dose–response curves for Eµ-Myc/Bak KO and Eµ-Myc/Bax KO lymphoma cell lines treated with the MCL-1 inhibitor S63845 for 24 h. Live cells were identified as Annexin V/PI double negative by flow cytometry. Three independent cell lines were tested for each genotype. C, D Survival of Eµ-Myc/Bak KO and Eµ-Myc/Bax KO lymphoma cell lines treated with the cytotoxic drugs etoposide (C) or ionomycin (D). Loss of BAX provided no protection from these drugs. One Eµ-Myc/Bax KO cell line (171) showed resistance to etoposide treatment, likely due to the presence of an additional mutation in Trp53. All data are presented as mean ± SD for 3 independent experiments.
Generation of MCL-1 inhibitor-resistant cells in culture selects for loss of BAX protein expression
The second method we employed to identify factors that confer resistance to MCL-1 inhibitors was through the generation of naturally resistant cell populations. We cultured Eµ-Myc lymphoma cell lines in increasing doses of S63845 over time, until the cells were resistant to doses of >1 µM (10× the IC50 of S63845 in parental cells; Fig. 4A). Two independent culturing experiments were undertaken, resulting in a total of four drug-resistant derivatives for each independent Eµ-Myc cell line.
To generate MCL-1 inhibitor-resistant cells, three independent Eµ-Myc lymphoma cell lines were cultured in increasing amounts of S63845 over time, until they were resistant to 1 µM S63845 (10× IC50). Cell lines were then either collected for analysis (resistant cell lines R1.1 and R2.1) or continued to be cultured until they were resistant to 5 µM S63845 (resistant cell lines R1.2 and R2.2). Two independent culturing campaigns were undertaken. A Cell viability assay following treatment of parental and drug-resistant cell lines with 1 µM S63845 for 24 h. Live cells were identified as Annexin V/PI double negative by flow cytometry. B Western blotting for TRP53 as well as for pro-apoptotic and pro-survival BCL-2 family proteins in parental and drug-resistant cell lines. Probing for HSP70 was used as a loading control. * indicates non-specific bands. C Parental and drug-resistant cell lines derived from three independent Eµ-Myc lymphoma cell lines were treated with the chemotherapeutic drug etoposide for 24 h. Live cells were identified as Annexin V/PI double negative by flow cytometry. D Treatment of Eµ-Myc lymphoma cell lines exhibiting upregulated pro-survival proteins BCL-XL or A1 (AH15A R1.2, R2.2) with the BH3-mimetic drugs S63845 (MCL-1 inhibitor), A-1331852 (BCL-XL inhibitor) and venetoclax/ABT-199 (BCL-2 inhibitor), either alone or in the indicated combinations for 24 h. Live cells were identified as Annexin V/PI double negative by flow cytometry. In (D), significance is measured versus the DMSO-treated sample for each cell line. All data are presented as mean ± SD for 2–3 independent experiments. One-way ANOVA was used to measure statistical significance (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns = not significant).
To explore the underlying molecular changes that were conferring resistance to S63845 in these cells, western blotting was performed for TRP53, as well as both pro- and anti-apoptotic BCL-2 family proteins (Fig. 4B). In resistant variants derived from two of the three Eµ-Myc lymphoma cell lines (AF47A and 560), the most common abnormality identified was loss of BAX protein expression. Resistant cells derived from one cell line (AH15A) did not exhibit loss of BAX, and in fact one of the derived S63845-resistant variants (R2.1) actually showed increased BAX protein expression compared to the parental cell line. Instead, the AH15A drug-resistant variants showed increased expression of the pro-survival BCL-2 family members BCL-XL and A1 that are not targeted by S63845. We found that the AF47A and 560-derived S63845-resistant cell lines that had lost expression of BAX protein could still be killed by the TRP53-activating drug Nutlin-3a, indicating that they retained TRP53 function (Supplementary Fig. 2). Importantly, these resistant cell lines also retained sensitivity to the chemotherapeutic drug etoposide (Fig. 4C), similar to the Bax KO Eµ-Myc lymphoma cells we examined (Figs. 2 and 3). In contrast, etoposide could not effectively kill the AH15A-derived S63845-resistant cell lines that had increased expression of BCL-XL and A1. However, these lymphoma cells could be killed by combined targeting of MCL-1 and BCL-XL using S63845 and the BCL-XL-specific BH3-mimetic drug A-1331852 (Fig. 4D). These results suggest that BCL-XL is the primary mediator of resistance in these cells over A1, but the lack of an A1-targeting BH3-mimetic precludes a firm conclusion in this regard. The data do indicate that co-targeting of compensatory pro-survival proteins, which are upregulated in response to prolonged BH3-mimetic drug treatment, could be an effective therapeutic strategy.
Splicing mutations in the Bax gene were identified in some MCL-1 inhibitor-resistant Eµ-Myc lymphoma cells
To investigate the underlying mechanisms that may have caused the loss of BAX protein expression in the S63845-resistant cells, next-generation sequencing of the Bax exons and proximal promoter was undertaken. This region of the Bax promoter contains important transcription factor binding sites, including for the tumour suppressor protein TRP53 [33]. Two of the four cell lines (AF47A R1.2 and 560 R1.2) examined were found to possess a G>A mutation of base pair 504 downstream of the transcriptional start site (Fig. 5A). This mutation disrupts the final base of the first mRNA intron consensus sequence splicing site of the canonical Bax transcript, Bax-201, which would likely result in this intron being retained in the mRNA. As we did not detect an elongated BAX protein in these cells by western blotting (Fig. 4B), it is likely that the mRNA encoding this mutant BAX protein is degraded, resulting in a BAX KO phenotype.
A Next-generation sequencing of the Bax coding regions revealed a 504G>A substitution downstream of the transcriptional start site in two drug-resistant Eµ-Myc lymphoma cell lines which lack BAX protein expression (AF47A R1.2 and 560 R1.2). This mutation disrupts the mRNA splicing acceptor site and would likely result in the retention of intron 1 in the consensus mouse Bax transcript Bax-201 (ENSEMBL ENSMUST00000033093.10). Gene sequencing coverage is indicated by green bars. B qRT-PCR of parental and drug-resistant Eµ-Myc lymphoma cells treated with DMSO (vehicle, negative control) or the TRP53-activating drug Nutlin-3a for 24 h, of which Bax is a transcriptional target. Data are shown normalised to the housekeeping gene Hmbs, and relative to the parental Eµ-Myc lymphoma cell line treated with DMSO. C qRT-PCR of parental and drug-resistant Eµ-Myc lymphoma cells treated with DMSO or the hypomethylating agent 5’azacytidine (inhibitor of DNMT1) for 24 h. Data are shown normalised to the housekeeping gene Hmbs, and relative to the parental cell line treated with DMSO. D Parental and drug-resistant Eµ-Myc lymphoma cell lines exhibiting loss of BAX protein expression were treated with 5’azacytidine for 24 h, alone or in combination with S63845. Live cells were identified as Annexin V/PI double negative by flow cytometry. All flow cytometry data are presented as mean ± SD for 3 independent experiments. One-way ANOVA was used to determine statistical significance (**p < 0.01; ****p < 0.0001, ns = not significant). All qRT-PCR data are presented as mean ± SD for 2 independent experiments.
For the other resistant Eµ-Myc lymphoma cell lines that had lost BAX expression, we examined whether BAX may be dysregulated at the pre-transcriptional or post-transcriptional levels by performing quantitative reverse-transcriptase PCR (qRT-PCR) for Bax mRNA following treatment of the cells with the TRP53-activating drug Nutlin-3a (Fig. 5B). Bax is a known target gene of TRP53 and therefore, as expected, we observed an increase in Bax transcript levels in the parental Eµ-Myc lymphoma cell lines treated with Nutlin-3a compared to those treated with DMSO (vehicle, negative control). In the S63845 drug-resistant cell lines with Bax splicing mutations (AF47A R1.2 and 560 R1.2), as well as in AF47A R2.2, we saw upregulation of Bax mRNA following treatment with Nutlin-3a. However, in the 560 R2.2 lymphoma cells, no induction of Bax mRNA was observed after Nutlin-3a treatment, indicating that transcription of this gene can no longer be induced by TRP53. As a control, we confirmed the impact of Nutlin-3a treatment, which was able to induce expression of the TRP53 target gene Puma in all parental and S63845-resistant cell lines tested, as expected (Supplementary Fig. 3A).
To examine whether the Bax gene might be silenced at the epigenetic level in the 560 R2.2 lymphoma cells in which Bax transcripts could not be detected, we treated these cells with the hypomethylating agent 5’azacytidine, which inhibits DNMT1, and performed qRT-PCR to look for the restoration of Bax mRNA expression levels (Fig. 5C). However, while treatment with 5’azacytidine could induce expression of the known DNMT1-regulated gene Noxa in all lymphoma cell lines tested (Supplementary Fig. 3B), no substantial induction of Bax mRNA was observed in the 560 R2.2 cells. Therefore, it is possible that Bax expression is being prevented by a transcriptional repressor protein or a microRNA in these cells, or by a genetic mutation that we were unable to identify. Interestingly, while 5’azacytidine treatment could increase Bax transcript levels in the other drug-resistant BAX KO cell lines tested, it could not restore sensitivity of any of these drug-resistant lymphoma cells to S63845, confirming that a functional BAX protein was not being made in these cells (Fig. 5D).
Unbiased CRISPR/Cas9 whole genome knockout screens in a human Burkitt lymphoma cell line show that loss of BAX is a major resistance factor for MCL-1-targeting BH3-mimetic drugs
To examine whether our results in the Eµ-Myc mouse model could be applied to human cancer cells, we performed a whole genome CRISPR/Cas9 KO screen in the human Burkitt lymphoma cell line BL2 that has wildtype TP53 and is highly sensitive to MCL-1 inhibition [24]. We used the human GeCKO v2 sgRNA library [34], and treated library-infected, Cas9-expressing BL2 cells with either DMSO (vehicle control) or a high dose (IC99) of S63845 for 24 h (Fig. 6A). As in our previous screens in the Eµ-Myc mouse lymphoma cell lines, loss of BAX was identified as the top hit conferring resistance to S63845, whereas sgRNAs targeting BAK were not enriched (Fig. 6B).
A Schematic of CRISPR/Cas9 KO screen performed in the human Burkitt lymphoma cell line BL2. Cells were treated with DMSO (vehicle, negative control) or 100 nM of S63845 (~IC99 dose) for 24 h and surviving cells were sorted by flow cytometry and collected for next-generation sequencing. B MAGeCK analysis to identify top enriched sgRNAs in drug-resistant cell populations. Significant hits (FDR < 0.05) are indicated with black text. Loss of BAX was identified as the top factor conferring resistance to S63845 in BL2 cells. C Western blot analysis to validate CRISPR/Cas9-mediated loss of BAK or BAX in BL2 cells. Probing for HSP70 was used as a loading control. D Dose–response curves for control BL2 cells (containing Cas9 and a non-targeting control sgRNA) and isogenic BAK KO or BAX KO BL2 lymphoma cells treated with the MCL-1 inhibitor S63845 for 24 h. Live cells were identified as Annexin V/PI double negative by flow cytometry. E Isogenic WT, BAK KO or BAX KO BL2 cells were treated with etoposide, doxorubicin, vincristine or cisplatin for 24 h and cell survival was determined as described above. In (E), significance is measured versus the DMSO-treated sample for each cell line. One-way ANOVA was used to determine statistical significance (****p < 0.0001). All data are presented as mean ± SD for 3 independent experiments.
To validate these results, we used CRISPR/Cas9 to generate isogenic BL2 lymphoma cell lines with BAK or BAX KO and confirmed the absence of these proteins by western blotting (Fig. 6C). Treatment of these cell lines with S63845 revealed that the BAX KO cells were highly resistant, whereas the BAK KO cells showed sensitivity similar to the parental cells (Fig. 6D). Of note, single agent treatment with vincristine, doxorubicin, cisplatin and etoposide (all relevant to the treatment of human lymphoma patients [31, 32]) could still kill the BAX KO BL2 cells (Fig. 6E), as was observed for the Eµ-Myc BAX KO lymphoma cells. This suggests that human MYC-driven lymphomas, with loss of BAX function that arise in response to BH3-mimetic drug treatment, may still respond to conventional chemotherapeutic agents as salvage therapy.
Discussion
Resistance to BH3-mimetic drugs is a critical and emerging issue which threatens to limit the benefit of this class of anti-cancer therapy. While genetic changes underlying resistance to the BCL-2 inhibitor venetoclax are beginning to be uncovered [5, 23, 26, 35,36,37,38,39], less attention has been focused on resistance to MCL-1 inhibitors, which have entered clinical trials for diverse haematological malignancies [5]. To this end, we sought to identify factors that could confer resistance to MCL-1 inhibitors using murine and human models of highly aggressive, MYC-driven lymphoma.
Our previous work identified TP53 as a critical mediator of the response to MCL-1 and BCL-2 inhibitors in diverse blood cancer cell lines and patient samples [24]. Therefore, we first sought to identify any additional factors which might enable cells lacking TRP53 to escape BH3-mimetic-mediated cell death. Using whole genome CRISPR/Cas9 KO screens, we identified loss of BAX as the top factor conferring resistance of both Trp53 wildtype and Trp53-deficient mouse Eµ-Myc lymphoma cell lines to BH3-mimetics targeting MCL-1. Similarly, in a CRISPR/Cas9 KO screen performed in human BL2 cells, loss of BAX was also the top hit. Validation experiments revealed that the IC50 for the MCL-1 inhibitor S63845 in cells lacking BAX was >10-fold higher compared to control cells. Consistent with our results, BAX has been identified as a top hit conferring resistance of AML cells to venetoclax in other CRISPR/Cas9 KO screens [25, 26]. Furthermore, AML patient samples exhibiting loss of BAX expression [40], and AML cell lines with CRISPR/Cas9-mediated BAX KO [41] were both found to be resistant to venetoclax. Importantly, in our study, we found that human BL2 and mouse Eµ-Myc lymphomas lacking BAX expression could still be killed by both standard-of-care drugs and salvage therapies for lymphoma, indicating that tumours which acquire BAX mutations following BH3-mimetic drug treatment may still respond to chemotherapy. These results suggest that the upregulation of pro-apoptotic BH3-only proteins by chemotherapy drugs may act to counter the loss of pro-apoptotic BAX, allowing apoptosis to proceed. While our data are compelling in this regard, further research will be needed to bring these findings to the clinic.
We next wanted to identify resistance-conferring factors that would be likely to occur spontaneously in patients treated with MCL-1 inhibitors. Therefore, we generated Eµ-Myc lymphoma cell lines that are resistant to S63845 by culturing them in increasing doses of this drug over time. We identified two major mechanisms driving resistance: loss of BAX expression, and increased expression of pro-survival proteins other than MCL-1. The most prominent was the loss of BAX expression. In two of these cell lines lacking BAX, we identified a splicing mutation likely to result in the preservation of intron 1 in the canonical and most highly expressed mouse Bax transcript, Bax-201. In a third case, we identified a complete lack of any Bax transcription, which could not be restored by treatment with the hypomethylating agent 5’azacytidine, suggesting that this is not due to epigenetic silencing.
The selection via diverse mechanisms to disable BAX expression in these cells illustrates its importance in facilitating cell death in the face of MCL-1 inhibition. Despite these mutations, all Bax KO Eμ-Myc lymphoma cell lines remained sensitive to killing by diverse chemotherapeutic drugs. Our findings are consistent with recent studies which found that loss of BAX could confer resistance to both venetoclax and S63845 in AMLs [42] and a panel of non-Hodgkin B cell lymphoma cell lines [43]. In addition, in this latter study, low levels of BAX mRNA were identified in samples from 3 CLL patients treated with venetoclax, and in CLL cells from another patient it was shown that BAX mutations were enriched during venetoclax treatment. Our work complements these findings and reveals that loss of BAX is the most potent resistance factor for MCL-1 inhibition in lymphoma, and also that such resistance can be overcome by treatment with standard-of-care chemotherapeutic drugs. Interestingly, selection for loss of BAX has also been reported in non-malignant myeloid cells of patients undergoing venetoclax therapy for CLL [36].
A major question that arises from our research, and that of the others discussed above, is why the loss of BAX, and not the loss of BAK, is the major mediator of specific resistance to MCL-1 targeting BH3-mimetic drugs. BAX and BAK both function as effectors of apoptosis that, when activated, oligomerise and form pores in the outer mitochondrial membrane. BAX and BAK have largely overlapping functions, best demonstrated by the observation that single KO mice exhibit only minor abnormalities, whereas Bax/Bak double KO mice have severe (usually fatal) developmental abnormalities [44, 45]. However, BAX and BAK proteins are found in distinct sub-cellular localisations [46,47,48]. In addition, they show differential binding to the BCL-2 pro-survival proteins [49,50,51]. From our earlier studies we do know that both BAX and BAK are activated to some degree after BH3-mimetic drug treatment [24]. Therefore, one hypothesis that could explain our findings is that when a BH3-mimetic drug binds to MCL-1, another pro-survival protein such as BCL-XL preferentially binds to free BAK over BAX, leaving BAX to be the primary mediator of cell death in this context. Another possibility is that the MCL-1 targeting BH3-mimetic S63845 preferentially inhibits BAX over BAK from binding to MCL-1. These different scenarios are difficult to measure because of the different states in which BAX and BAK can exist within cells, including (inert) monomers, homodimers, higher-order oligomers, as well as in complexes with different pro-survival proteins. The future generation of antibodies that can distinguish between these states, as well as affinity measurements for the full-length proteins in a mitochondrial context, is essential to interrogate this with greater precision.
The second major mechanism driving resistance we identified in our S63845-resistant cells was upregulation of the pro-survival proteins BCL-XL and A1. Many studies have reported the over-expression of pro-survival BCL-2 proteins as resistance factors for venetoclax therapy, including in patient samples and in in vitro whole genome CRISPR/Cas9 screens [35, 37, 39, 52, 53]. In our study, Eµ-Myc lymphoma cells that had achieved resistance to S63845 through upregulation of BCL-XL and A1 also showed resistance to etoposide treatment due to the over-expression of these pro-survival proteins preventing apoptosis. However, the lymphoma cells with upregulation of BCL-XL/A1 could be re-sensitised to apoptosis by co-inhibition of BCL-XL alongside MCL-1, illustrating that inhibition of compensatory, non-targeted pro-survival BCL-2 proteins is an effective therapeutic strategy to kill malignant cells that have become resistant to a particular BH3-mimetic drug. However, it must be noted that the combination of MCL-1 and BCL-XL targeting BH3-mimetics is predicted to be toxic to healthy cells [54]. Therefore, these therapeutic strategies are likely to only be possible in patients when such drugs can be targeted to the cancer cells directly through, for example, antibody conjugation.
Understanding the factors which can guard against cell killing by BH3-mimetic drugs is the first step in enabling the long-term success of these new anti-cancer therapies. In this study, we have identified factors (loss of BAX; gain of BCL-XL and/or A1) that confer resistance to the new class of BH3-mimetics targeting MCL-1, drugs which are currently being evaluated in clinical trials for diverse blood cancers. Furthermore, we identified therapeutic strategies to overcome resistance based on an understanding of underlying gene expression changes, including combining MCL-1 targeting BH3-mimetic drugs with conventional chemotherapeutic drugs (e.g., etoposide, vincristine, doxorubicin or cisplatin) or with BH3-mimetics that target other pro-survival proteins. This work illustrates how mechanistic studies can inform new therapeutic strategies to overcome resistance and therefore enable durable responses to BH3-mimetic drugs in the clinic.
Materials and methods
Cell lines
Mouse Eµ-Myc lymphoma-derived cell lines AH15A, AF47A and 560 were cultured in high glucose Dulbecco’s modified Eagle’s medium (DMEM) containing 10% heat-inactivated foetal bovine serum (FBS; Sigma #F9423), 100 µM asparagine (Sigma #A4284), 50 µM β-mercaptoethanol (Sigma #M3148), 100 U/mL penicillin and 100 µg/mL streptomycin (Gibco #15140122). Eµ-Myc lymphoma cells were maintained in a humidified incubator at 37 °C with 10% CO2. The human Burkitt lymphoma cell line BL2 was a kind gift from Prof Alan Rickinson, The University of Birmingham, UK. BL2 cells were authenticated by STR profiling at the Australian Genome Research Facility and maintained in RPMI-1640 medium containing 10% heat-inactivated FBS, 1 mM sodium pyruvate (Gibco #11360070), 2 mM L-glutamine (Gibco #25030081), 50 μM α-thioglycerol (Sigma #M-6145), 100 U/mL penicillin and 100 µg/mL streptomycin in a humidified incubator at 37 °C with 5% CO2. HEK293T cells were cultured in DMEM containing 10% heat-inactivated FBS, 100 U/mL penicillin and 100 µg/mL streptomycin, and maintained in a humified incubator at 37 °C with 10% CO2. Mycoplasma screening of all cell lines used was conducted regularly (MycoAlert kit; Lonza #LT07-118).
CRISPR/Cas9 whole genome knockout screening
Eµ-Myc and BL2 cell lines were stably transduced with lentiviruses carrying FuCas9-Cherry (Addgene Plasmid #70182) using established methods [15]. Lentiviral supernatant was used to transduce 1 × 105 Eµ-Myc or BL2 cells by centrifugation at 2200 rpm for 2 h at 32 °C in the presence of polybrene (made in house, final concentration 8 µg/mL). After 72 h, mCherry-positive cells were sorted on a FACSAria Fusion flow cytometer (BD Biosciences). Three to six replicates of 3 × 105 Cas9-expressing lymphoma cells were then transduced with the whole genome CRISPR knockout libraries, either the YUSA library for mouse Eµ-Myc lymphoma cells (87,897 sgRNAs targeting 19,150 protein-coding genes) [30] or the GeCKO v2 library for human BL2 cells (123,411 sgRNAs targeting 19,050 protein-coding genes) [34]. Cells were expanded for 2–3 days and then each replicate was split into two flasks of 7 × 106 cells each. For short-term CRISPR/Cas9 screens (Trp53 KO Eµ-Myc cell lines and BL2 cells), one flask was treated for 24 h with DMSO (vehicle control) and the other with a high dose of S63845 that kills 99% of parental lymphoma cells (IC99; 1 µM for Trp53 KO Eµ-Myc cell lines, 100 nM for BL2 cells), followed by flow sorting of live, drug-resistant cells. For longer-term screens (wildtype Eµ-Myc lymphoma cell lines), cells were treated with DMSO (vehicle control) or with suboptimal (100 nM = ≤ IC50) doses of S63845 every 2–3 days over a 2-week period. Cell pellets were collected and genomic DNA extracted using the DNeasy Blood and Tissue Kit (QIAGEN # 69506). DNA was amplified using library-specific primers with indexing overhangs allowing for Illumina sequencing. Samples were pooled and sequenced on the Illumina NextSeq 550. MAGeCK v0.5.9.4 [55] was used to map reads to sgRNAs and quantify sgRNA enrichment. For all CRISPR/Cas9 screens, genes with FDR < 0.05 were considered to be significant hits.
Targeted CRISPR/Cas9 gene editing
CRISPR/Cas9 gene editing was employed to generate Trp53, Bak or Bax single KO, and Bak/Bax double KO mouse Eµ-Myc and/or human BL2 lymphoma cell lines. Cells harbouring a non-targeting sgRNA were also generated and used as ‘control’ cells in all experiments using CRISPR/Cas9-dervied cell lines. Lentiviruses containing sgRNAs in the doxycycline-inducible FgH1tUTG backbone (Addgene Plasmid #70183) were produced and cells were transduced with these vectors using established methods [15]. Successfully transduced GFP-positive cells were sorted as described above. sgRNA sequences used are listed in Supplementary Table 1. To induce sgRNA expression, culture medium was supplemented with 1 µg/mL doxycycline hyclate (Sigma #D9891) each day for 5 days. Efficient knockout of target proteins was confirmed by western blotting.
Western blotting
Cell pellets were resuspended in radioimmunoprecipitation assay (RIPA) buffer containing protease inhibitors (Roche #11836145001) and incubated on ice for 25 min. Centrifugation was performed at 13,300 rpm for 10 min at 4 °C and supernatant containing proteins was collected. Protein levels were quantified using the Pierce BCA Protein Assay Kit (Thermo Fisher, #23225) and 10 µg was run on a 10% NuPAGE Bis-Tris 1.5 mm gel in 2-(N-morpholino)-ethanesulfonic acid (MES) running buffer. Proteins were transferred onto nitrocellulose membranes using the iBlot2 (Thermo Fisher). Membranes were blocked at room temperature (RT) for 1 h in phosphate-buffered saline containing 0.1% Tween-20 (PBST) and 5% skim milk. Membranes were rinsed in PBST and incubated overnight at 4 °C in primary antibody diluted in PBST containing 5% bovine serum albumin. The next day, membranes were washed in PBST and incubated for 1 h at RT in HRP-conjugated secondary antibodies diluted in PBST containing 5% skim milk. Membranes were washed again in PBST and imaged by the addition of Immobilon Forte (Merck Millipore #WBLUF0500) on a ChemiDoc system (Bio-Rad). Comparison to Precision Plus Protein Dual Color Standard (Bio-Rad #1610374) was used to estimate protein weight. Antibodies used for western blotting are listed in Supplementary Table 2. Uncropped blots are shown in the supplementary “original data” file 2.
Apoptosis assays
Cells were plated at 3 × 104 cells/well of a flat-bottom 96-well plate in triplicate and drugs were added to give the indicated final concentrations. The following drugs were used: S63845 (MCL-1 inhibitor; Active Biochem #A-6044), A-1331852 (BCL-XL inhibitor; AbbVie, provided by Dr G. Lessene, WEHI), ABT-199 (BCL-2 inhibitor; Active Biochem #A-1231), ABT-737 (BCL-2, BCL-XL and BCL-W inhibitor; Active Biochem #A-1002), etoposide (Ebewe Interpharma), ionomycin (Sigma-Aldrich #I9657), cisplatin (Hospira Australia), doxorubicin (Ebewe Interpharma), vincristine (Sigma-Aldrich #V8879), Nutlin-3a (Cayman Chemicals #18585) and 5’azacytidine (Sigma-Aldrich #A2385). Cells were incubated with the drugs at 37 °C for 24 h. Plates were centrifuged and cells stained in Annexin V binding buffer (10 mM HEPES pH 7.4, 140 mM NaCl, 2.5 mM CaCl2) containing Annexin V-Alexa Fluor 647 (1:1000, made in house) and 1 µg/mL propidium iodide (PI; Sigma-Aldrich #P4170). Viable cells (Annexin V and PI double negative) were quantified on an LSR II W flow cytometer (BD Biosciences) and data were analysed in FlowJo v10.8 (BD Life Sciences). For each sample, the proportion of live cells was normalised to the DMSO-treated (vehicle, control) sample for each cell line. Non-linear regression (curve fit) analysis was used to generate dose–response curves and calculate IC50 values in GraphPad Prism v9. All data are presented as mean ± standard deviation of 2–3 independent experiments. One-way ANOVA with Dunnett’s post hoc test was used to compare control and treated or parental and drug-resistant groups.
Derivation of Eµ-Myc/Bax KO and Eµ-Myc/Bak KO lymphoma cell lines
All experiments involving animals were performed in accordance with the guidelines set out by the WEHI Animal Ethics Committee. All mice were on a C57BL/6 background and bred at WEHI. Mice deficient for BAX [56] or BAK [44] were crossed with Eµ-Myc mice [17]. Offspring were monitored for lymphoma and euthanised when they reached the ethical endpoint as determined by trained animal technicians according to the WEHI Animal Ethics Committee guidelines. Enlarged organs (spleen, lymph nodes, thymus) were harvested, homogenised and filtered through a 100 µm strainer to generate a single cell suspension. Eµ-Myc/Bax−/− (Bax KO) and Eµ-Myc/Bak−/− (Bak KO) lymphoma cell lines were cultured as described above.
Generation of MCL-1-targeting BH3-mimetic drug-resistant lymphoma cell lines
To generate drug-resistant Eµ-Myc lymphoma cell lines, cells were cultured in increasing doses of S63845 over time. Cells were allowed to recover to >70% viability before higher doses of the drug were added. Cells were cultured until they could withstand doses of S63845 10–50× the IC50 concentrations of the parental cell lines. Two independent culturing campaigns were undertaken. For each campaign, cell lines were generated that could resist 1 µM S63845 (resistant cell lines 1.1 and 2.1). These cell lines were then cultured further in increasing doses of S63845 until they could withstand 5 µM S63845 (resistant cell lines 1.2 and 2.2). Resistance to S63845 was confirmed by short-term apoptosis assays as described above.
Next-generation sequencing of mouse Bax gene coding regions
Genomic DNA was isolated from parental and S63845-resistant cells using the DNeasy Blood and Tissue Kit (QIAGEN). Primers tiling the murine Bax promoter and exon sequences were designed using Primer Blast and modified with overhangs to allow for the indexing of products for sequencing on the Illumina platform. Primer sequences are listed in Supplementary Table 3. For each primer pair, 200 ng of genomic DNA was PCR amplified using GoTaq Green Mix (Promega #M712) along with 0.25 µM of each primer. The following cycling conditions were used: denaturation at 95 °C for 3 min, followed by 35 cycles of 95 °C for 30 s, 60 °C for 30 s and 72 °C for 30 s. The final extension was performed at 72 °C for 5 min. Products were pooled and cleaned up using AmPure XP beads (Beckman Coulter #A63880) and sequenced on an Illumina MiSeq. Sequencing reads from S63845-resistant cells were aligned to reads from parental cells using the R package DECIPHER [57] in order to identify mutations in Bax coding regions.
Quantitative reverse-transcriptase PCR (qRT-PCR)
To obtain intact cells for qRT-PCR, cells were incubated with 25 μM of the broad-spectrum caspase inhibitor Q-VD-OPh (MedChem Express # HY-12305) for 15 min prior to treatment with 5’azacytidine or Nutlin-3a. After 24 h, cell pellets were collected and resuspended in 0.5 mL TRIzol reagent (Thermo Fisher #15596026). RNA was isolated according to the manufacturer’s instructions. cDNA was prepared using the Superscript III First Strand Synthesis System (Thermo Fisher #18080051) according to the manufacturer’s instructions. qRT-PCR reactions were performed using TaqMan Fast Advanced Master Mix (Thermo Fisher, #4444963) according to the manufacturer’s instructions. TaqMan probes to detect mouse Puma/Bbc3 (Mm00519268_m1), Noxa/Pmaip1 (Mm00451763_m1), Bax (Mm00432050_m1) and Hmbs (Mm01143545_m1) were used (Thermo Fisher). Reactions were run on a Quantstudio 12 K Flex Real-Time PCR System (Thermo Fisher). Cycle threshold (Ct) values for each gene were normalised to the housekeeping control gene (Hmbs) Ct value for that sample and all data are presented relative to the DMSO-treated parental cell line. All qRT-PCR data are presented as mean ± standard deviation of two independent experiments.
Data availability
Data are available upon request to Gemma L. Kelly (gkelly@wehi.edu.au).
References
Singh R, Letai A, Sarosiek K. Regulation of apoptosis in health and disease: the balancing act of BCL-2 family proteins. Nat Rev Mol Cell Biol. 2019;20:175–93.
Cory S, Adams JM. The Bcl2 family: regulators of the cellular life-or-death switch. Nat Rev Cancer. 2002;2:647–56.
Riedl SJ, Salvesen GS. The apoptosome: signalling platform of cell death. Nat Rev Mol Cell Biol. 2007;8:405–13.
Roberts AW, Wei AH, Huang DCS. BCL2 and MCL1 inhibitors for hematologic malignancies. Blood. 2021;138:1120–36.
Diepstraten ST, Anderson MA, Czabotar PE, Lessene G, Strasser A, Kelly GL. The manipulation of apoptosis for cancer therapy using BH3-mimetic drugs. Nat Rev Cancer. 2022;22:45–64.
Merino D, Kelly GL, Lessene G, Wei AH, Roberts AW, Strasser A. BH3-mimetic drugs: blazing the trail for new cancer medicines. Cancer Cell. 2018;34:879–91.
Souers AJ, Leverson JD, Boghaert ER, Ackler SL, Catron ND, Chen J, et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat Med. 2013;19:202–8.
Roberts AW, Davids MS, Pagel JM, Kahl BS, Puvvada SD, Gerecitano JF, et al. Targeting BCL2 with venetoclax in relapsed chronic lymphocytic leukemia. N Engl J Med. 2016;374:311–22.
Konopleva M, Pollyea DA, Potluri J, Chyla B, Hogdal L, Busman T, et al. Efficacy and biological correlates of response in a phase II study of venetoclax monotherapy in patients with acute myelogenous leukemia. Cancer Discov. 2016;6:1106–17.
Klanova M, Kazantsev D, Pokorna E, Zikmund T, Karolova J, Behounek M, et al. Anti-apoptotic MCL1 protein represents critical survival molecule for most Burkitt lymphomas and BCL2-negative diffuse large B-cell lymphomas. Mol Cancer Ther. 2022;21:89–99.
Daly T, Ippolito T, Gu JJ, Mavis C, Torka P, Hernandez-Ilizaliturri FJ, et al. MCL-1 inhibition by the selective MCL-1 inhibitor AMG-176 induces in vitro activity against Burkitt lymphoma cell lines and synergistically enhances the cytotoxic effect of chemotherapy and BH3 mimetics. Blood. 2019;134:5303.
Diepstraten ST, Chang C, Tai L, Gong JN, Lan P, Dowell AC, et al. BCL-W is dispensable for the sustained survival of select Burkitt lymphoma and diffuse large B-cell lymphoma cell lines. Blood Adv. 2020;4:356–66.
Wang MY, Li T, Ren Y, Shah BD, Lwin T, Gao J, et al. MCL-1 dependency as a novel vulnerability for aggressive B cell lymphomas. Blood Cancer J. 2021;11:14.
Kelly GL, Grabow S, Glaser SP, Fitzsimmons L, Aubrey BJ, Okamoto T, et al. Targeting of MCL-1 kills MYC-driven mouse and human lymphomas even when they bear mutations in p53. Genes Dev. 2014;28:58–70.
Aubrey BJ, Kelly GL, Kueh AJ, Brennan MS, O’Connor L, Milla L, et al. An inducible lentiviral guide RNA platform enables the identification of tumor-essential genes and tumor-promoting mutations in vivo. Cell Rep. 2015;10:1422–32.
Aubrey BJ, Brennan MS, Diepstraten ST, Wang Z, Chang C, Herold MJ, et al. Loss of TRP53 reduces but does not overcome dependency of lymphoma cells on MCL-1. Cell Death Differ. 2022;29:1074–6.
Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, et al. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–8.
Merino D, Whittle JR, Vaillant F, Serrano A, Gong J-N, Giner G, et al. Synergistic action of the MCL-1 inhibitor S63845 with current therapies in preclinical models of triple-negative and HER2-amplified breast cancer. Sci Transl Med. 2017;9:eaam7049.
Nangia V, Siddiqui FM, Caenepeel S, Timonina D, Bilton SJ, Phan N, et al. Exploiting MCL1 dependency with combination MEK + MCL1 inhibitors leads to induction of apoptosis and tumor regression in KRAS-mutant non-small cell lung cancer. Cancer Discov. 2018;8:1598–613.
Kotschy A, Szlavik Z, Murray J, Davidson J, Maragno AL, Le Toumelin-Braizat G, et al. The MCL1 inhibitor S63845 is tolerable and effective in diverse cancer models. Nature. 2016;538:477–82.
Lasica M, Anderson MA. Review of venetoclax in CLL, AML and multiple myeloma. J Pers Med. 2021;11:463.
Roberts AW, Ma S, Kipps TJ, Coutre SE, Davids MS, Eichhorst B, et al. Efficacy of venetoclax in relapsed chronic lymphocytic leukemia is influenced by disease and response variables. Blood. 2019;134:111–22.
DiNardo CD, Tiong IS, Quaglieri A, MacRaild S, Loghavi S, Brown FC, et al. Molecular patterns of response and treatment failure after frontline venetoclax combinations in older patients with AML. Blood. 2020;135:791–803.
Thijssen R, Diepstraten ST, Moujalled D, Chew E, Flensburg C, Shi MX, et al. Intact TP-53 function is essential for sustaining durable responses to BH3-mimetic drugs in leukemias. Blood. 2021;137:2721–35.
Chen X, Glytsou C, Zhou H, Narang S, Reyna DE, Lopez A, et al. Targeting mitochondrial structure sensitizes acute myeloid leukemia to venetoclax treatment. Cancer Discov. 2019;9:890–909.
Nechiporuk T, Kurtz SE, Nikolova O, Liu T, Jones CL, D’Alessandro A, et al. The TP53 apoptotic network is a primary mediator of resistance to BCL2 inhibition in AML cells. Cancer Discov. 2019;9:910–25.
Savona MR, Rathmell JC. Mitochondrial homeostasis in AML and gasping for response in resistance to BCL2 blockade. Cancer Discov. 2019;9:831–3.
Wei AH, Strickland SA Jr, Hou JZ, Fiedler W, Lin TL, Walter RB, et al. Venetoclax combined with low-dose cytarabine for previously untreated patients with acute myeloid leukemia: results from a Phase Ib/II study. J Clin Oncol. 2019;37:1277–84.
Chua CC, Roberts AW, Reynolds J, Fong CY, Ting SB, Salmon JM, et al. Chemotherapy and Venetoclax in Elderly Acute Myeloid Leukemia Trial (CAVEAT): a Phase Ib dose-escalation study of venetoclax combined with modified intensive chemotherapy. J Clin Oncol. 2020;38:3506–17.
Koike-Yusa H, Li Y, Tan EP, Velasco-Herrera Mdel C, Yusa K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol. 2014;32:267–73.
Ngu H, Takiar R, Phillips T, Okosun J, Sehn LH. Revising the treatment pathways in lymphoma: new standards of care—how do we choose? Am Soc Clin Oncol Educ Book. 2022;42:629–42.
Seshadri T, Kuruvilla J, Crump M, Keating A. Salvage therapy for relapsed/refractory diffuse large B cell lymphoma. Biol Blood Marrow Transplant. 2008;14:259–67.
Igata E, Inoue T, Ohtani-Fujita N, Sowa Y, Tsujimoto Y, Sakai T. Molecular cloning and functional analysis of the murine BAX gene promoter. Gene. 1999;238:407–15.
Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–7.
Thijssen R, Tian L, Anderson MA, Flensburg C, Jarratt A, Garnham AL, et al. Single-cell multiomics reveal the scale of multi-layered adaptations enabling CLL relapse during venetoclax therapy. Blood. 2022;140:2127–41.
Blombery P, Lew TE, Dengler MA, Thompson ER, Lin VS, Chen X, et al. Clonal hematopoiesis, myeloid disorders and BAX-mutated myelopoiesis in patients receiving venetoclax for CLL. Blood. 2022;139:1198–207.
Deng Y, Diepstraten ST, Potts MA, Giner G, Trezise S, Ng AP, et al. Generation of a CRISPR activation mouse that enables modelling of aggressive lymphoma and interrogation of venetoclax resistance. Nat Commun. 2022;13:4739.
Blombery P, Anderson MA, Gong JN, Thijssen R, Birkinshaw RW, Thompson ER, et al. Acquisition of the recurrent Gly101Val mutation in BCL2 confers resistance to venetoclax in patients with progressive chronic lymphocytic leukemia. Cancer Discov. 2019;9:342–53.
Guieze R, Liu VM, Rosebrock D, Jourdain AA, Hernandez-Sanchez M, Martinez Zurita A, et al. Mitochondrial reprogramming underlies resistance to BCL-2 inhibition in lymphoid malignancies. Cancer Cell. 2019;36:369–84.e13.
Moujalled DM, Brown FC, Pomilio G, Anstee NS, Litalien V, Thompson ER, et al. Acquired mutations in BAX confer resistance to BH3 mimetics in acute myeloid leukemia (Conference Abstract). 62nd ASH Annual Meeting and Exposition. 2020;Session 604:263.
Rahmani M, Nkwocha J, Hawkins E, Pei X, Parker RE, Kmieciak M, et al. Cotargeting BCL-2 and PI3K induces BAX-dependent mitochondrial apoptosis in AML cells. Cancer Res. 2018;78:3075–86.
Moujalled DM, Brown FC, Chua CC, Dengler MA, Pomilio G, Anstee NS, et al. Acquired mutations in BAX confer resistance to BH3-mimetic therapy in acute myeloid leukemia. Blood. 2022:blood.2022016090.
Thomalla D, Beckmann L, Grimm C, Oliverio M, Meder L, Herling CD, et al. Deregulation and epigenetic modification of BCL2-family genes cause resistance to venetoclax in hematologic malignancies. Blood. 2022;140:2113–26.
Lindsten T, Ross AJ, King A, Zong W, Rathmell JC, Shiels HA, et al. The combined functions of proapoptotic Bcl-2 family members Bak and Bax are essential for normal development of multiple tissues. Mol Cell. 2000;6:1389–99.
Ke FFS, Vanyai HK, Cowan AD, Delbridge ARD, Whitehead L, Grabow S, et al. Embryogenesis and adult life in the absence of intrinsic apoptosis effectors BAX, BAK, and BOK. Cell. 2018;173:1217–30.e17.
Hsu Y-T, Wolter KG, Youle RJ. Cytosol-to-membrane redistribution of Bax and Bcl-XL during apoptosis. Proc Natl Acad Sci USA. 1997;94:3668–72.
Wolter KG, Hsu YT, Smith CL, Nechushtan A, Xi XG, Youle RJ. Movement of Bax from the cytosol to mitochondria during apoptosis. J Cell Biol. 1997;139:1281–92.
Robin AY, Iyer S, Birkinshaw RW, Sandow J, Wardak A, Luo CS, et al. Ensemble properties of BAX determine its function. Structure. 2018;26:1346–59.e5.
Llambi F, Wang YM, Victor B, Yang M, Schneider DM, Gingras S, et al. BOK is a non-canonical BCL-2 family effector of apoptosis regulated by ER-associated degradation. Cell. 2016;165:421–33.
Echeverry N, Bachmann D, Ke F, Strasser A, Simon HU, Kaufmann T. Intracellular localization of the BCL-2 family member BOK and functional implications. Cell Death Differ. 2013;20:785–99.
Zhai D, Jin C, Huang Z, Satterthwait AC, Reed JC. Differential regulation of BAX and BAK by anti-apoptotic BCL-2 family proteins BCL-B and MCL-1. J Biol Chem. 2008;283:9580–6.
Soderquist RS, Crawford L, Liu E, Lu M, Agarwal A, Anderson GR, et al. Systematic mapping of BCL-2 gene dependencies in cancer reveals molecular determinants of BH3 mimetic sensitivity. Nat Commun. 2018;9:3513.
Tahir SK, Smith ML, Hessler P, Rapp LR, Idler KB, Park CH, et al. Potential mechanisms of resistance to venetoclax and strategies to circumvent it. BMC Cancer. 2017;17:399.
Brinkmann K, Ng AP, de Graaf CA, Strasser A. What can we learn from mice lacking pro-survival BCL-2 proteins to advance BH3 mimetic drugs for cancer therapy? Cell Death Differ. 2022;29:1079–93.
Li W, Xu H, Xiao T, Cong L, Love MI, Zhang F, et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 2014;15:554.
Knudson CM, Tung KSK, Tourtellotte WG, Brown GAJ, Korsmeyer SJ. Bax-deficient mice with lymphoid hyperplasia and male germ cell death. Science. 1995;270:96–9.
Wright ES. RNAconTest: comparing tools for noncoding RNA multiple sequence alignment based on structural consistency. RNA. 2020;26:531–40.
Acknowledgements
The authors thank all members of the Blood Cells and Blood Cancer Division at The Walter and Eliza Hall Institute (WEHI) for their support and advice; Marco Herold and Lin Tai for CRISPR/Cas9 constructs; Lina Happo, Clare Scott and Suzanne Cory for Eµ-Myc/Bax KO and Eµ-Myc/Bak KO lymphoma samples; Simon Monard and his team at the WEHI Flow Cytometry Unit; Stephen Wilcox and his team at the WEHI Genomics Facility; and Robin Anderson for antibodies.
Funding
This work was supported by fellowships and grants from the Australian National Health and Medical Research Council (NHMRC) (Program Grant GNT1113133 to RMK and AS, Research Fellowship GNT1116937 to AS, Project Grant GNT1143105 to AS, Ideas Grants GNT 2002618 and GNT2001201 to GLK, Synergy Grants GNT2011139 to GLK and GNT 2010275 to AS), the Leukemia & Lymphoma Society of America (Specialized Center of Research [SCOR] grant no. 7015-18 to RMK, AS and GLK), Victorian Cancer Agency (MCRF Fellowship 17028 to GLK and ECRF Fellowship 21006 to STD), CASS Foundation Grants (to STD and JELM), the estate of Anthony (Toni) Redstone OAM (AS and GLK), the Craig Perkins Cancer Research Foundation (GLK), the Dyson Bequest (GLK) and the Harry Secomb Foundation (GLK), and operational infrastructure grants through the Victorian State Government Operational Infrastructure Support (OIS) and Australian Government NHMRC Independent Research Institute Infrastructure Support (IRIIS) Schemes. Open Access funding enabled and organized by CAUL and its Member Institutions.
Author information
Authors and Affiliations
Contributions
STD, AS and GLK designed the study. STD, SY and JELM performed the experiments. STD analysed and interpreted the data. ZW helped with generating cell lines. AS and GLK supervised the study. STD drafted the manuscript. RMK provided reagents and critical review of the manuscript. AS and GLK edited and revised the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
All authors are employees of WEHI which receives milestone and royalty payments related to venetoclax. AS and GLK have received research funding from Servier.
Ethics approval
All experiments with animals were approved by the Walter and Eliza Hall Institute Animal Ethics Committee and conducted in accordance with said committee guidelines.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Diepstraten, S.T., Young, S., La Marca, J.E. et al. Lymphoma cells lacking pro-apoptotic BAX are highly resistant to BH3-mimetics targeting pro-survival MCL-1 but retain sensitivity to conventional DNA-damaging drugs. Cell Death Differ 30, 1005–1017 (2023). https://doi.org/10.1038/s41418-023-01117-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-023-01117-0
- Springer Nature Limited
This article is cited by
-
Venetoclax resistance in acute lymphoblastic leukemia is characterized by increased mitochondrial activity and can be overcome by co-targeting oxidative phosphorylation
Cell Death & Disease (2024)
-
The birth of death, 30 years ago
Cell Death & Differentiation (2024)
-
BCL-W makes only minor contributions to MYC-driven lymphoma development
Oncogene (2023)