Abstract
The expression of coinhibitory receptors, such as CTLA-4, on effector T cells is a key mechanism for the negative regulation of T-cell activation. However, the transcriptional regulation of CTLA-4 is not well understood. Zfp281, a C2H2 zinc finger protein, is a negative regulator of pluripotency maintenance of embryonic stem cells. Nevertheless, the function of Zfp281 in differentiated cells has not been studied. We generated Zfp281 conditional knockout mice in which the function of the Zfp281 gene was conditionally disrupted by the Cd4Cre transgene to study its impact on T cell function. Zfp281 had no effect on T-cell development, but CD4+ T cell activation and cytokine production were impaired due to diminished T-cell receptor signaling. Furthermore, Zfp281 deficiency inhibited in vivo T cell responses to Listeria monocytogenes infection. Using genome-wide expression profiling assays, we determined that Zfp281 repressed Ctla-4 expression by directly binding to GC-rich sites in its promoter, which inhibited the negative feedback of T cell activation. In line with this result, CTLA-4 blockade and shRNA knockdown partly rescued the reduced cytokine production caused by Zfp281 deficiency. These findings indicate that Zfp281 sustains CD4+ T lymphocyte activation by directly repressing Ctla-4 transcription.
Similar content being viewed by others
Introduction
T cells play an important role in adaptive immune responses to foreign antigens and pathogens. T-cell activation is tightly regulated by T-cell receptor (TCR) signaling. An effective T-cell response requires at least two signals.1,2 The first signal comes from TCR engagement with antigenic peptides bound to the major histocompatibility complex (MHC) on antigen presenting cells (APC). The second signal is provided by costimulatory molecules binding to ligands on APC. There are multiple coreceptors on T cells that provide either costimulatory or coinhibitory signals. Costimulatory signals promote and sustain T-cell responses, such as CD28.3 Coinhibitory signals downregulate immune responses, such as cytotoxic T-cell antigen 4 (CTLA-4).4 The balance of costimulatory and coinhibitory signals is crucial to maximize protective immune responses. After TCR engagement and the formation of the immunological synapse, intracellular signaling is rapidly initiated.5 The phosphorylation of immune receptor tyrosine based activation motif (ITAM) in the CD3 cytoplasmic domains leads to the recruitment of the kinase ZAP-70.6 Activated ZAP-70 phosphorylates the adaptor LAT, inducing downstream signaling cascades, including activation of ERK or transport of key transcription factors, such as NFAT, to the nucleus.2,7 These signaling events are necessary for cytokine production and the proliferation of T cells. Identifying the factors that impact effector functions is a central issue in T-cell immunology. Nevertheless, the critical factors involved in the activation and expansion of T cells remain incompletely characterized.
The C2H2 zinc finger family is the largest class of transcription factors in mammals. Many of these family members are key transcriptional regulators involved in immune cell development and function.8,9 Expression and regulation of these transcription factors are complicated and dynamic throughout T-cell development. Han et al. showed that Zfp335 regulates the expression of multiple genes that are involved in the late stage of naive T-cell maturation.10 Growth factor independence 1 (Gfi-1) has been identified as a C2H2 zinc finger transcriptional repressor and is critical for the maturation, activation and effector function of T lymphocytes.11
Another C2H2 zinc finger protein, Zfp281, was first cloned and identified as a transcriptional repressor that binds to a GC-rich promoter in human cells.12,13 It was later determined that Zfp281 plays an important role in regulating stem cell development and pluripotency. Zfp281 expression is relatively high in undifferentiated embryonic stem cells (ESCs) and decreases in differentiated ESCs.14,15,16 Zfp281-null ESCs have an enhanced self-renewal capacity and dysregulated pluripotency.17 Deletion of Zfp281 in neural stem cells promotes the reprogramming of pre-induced pluripotent stem cells (pre-iPSCs) to iPSCs.18 Moreover, when Zfp281 is silenced in umbilical cord blood-derived mesenchymal stem cells, they rapidly convert into osteoblasts.19 These data suggest a pivotal role of Zfp281 in pluripotency and development. Zfp281 regulates pluripotency by interacting with the core transcriptional regulatory network factors that control stemness.20 It has been shown that Zfp281 can bind to the promoters of targeted genes, such as Nanog, Oct4, and Sox2.16,18,21,22 Furthermore, using chromatin immunoprecipitation sequencing (ChIP-seq), Fidalgo et al.17 identified genome-wide Zfp281 binding sites in mouse ESCs and found that Zfp281 mainly functions as a repressor to restrict the expression of many stem cell pluripotency genes. They also showed that Zfp281 directly regulates Nanog expression by recruiting the NuRD repressor complex to the Nanog locus, which contributes to Nanog autoregulation in mouse ESCs. Although the functions of Zfp281 have mostly been studied in ESCs, Zfp281 mRNA is also ubiquitously expressed at low levels, with elevated expression levels in the placenta, kidney, liver and peripheral lymphocytes.12,16 However, the functions and roles of Zfp281 in differentiated cells have not been described.
Germ-line knockout of Zfp281 in mice results in peri-implantation lethality,17,23 which limits the study of its effects in differentiated cells. To understand the role of Zfp281 in T lymphocytes, we generated Zfp281 conditional knockout mouse strains using the Cre-loxP system. We investigated whether T-cell development and peripheral activation were impaired after Zfp281 deletion. To further validate the role of Zfp281 in the T-cell-mediated immune response in vivo, wild-type and Zfp281-deficient mice were infected with Listeria monocytogenes. We also evaluated downstream genes of Zfp281 in CD4+ T cells using RNA-Seq and ChIP to identify its direct targets during T-cell activation.
Materials and methods
Mice
Zfp281-targeted (Zfp281T) mice were generated by homologous recombination-mediated gene targeting in ESCs of strain 129 (Supplementary Fig. S1a).24 Zfp281T mice were crossed with FLPeR mice to delete the Neo cassette and generate Zfp281fl/fl mice. Zfp281fl/fl mice were then backcrossed into the C57BL/6 background for eight generations. Exon 1 and exon 2 in Zfp281 were deleted by Cre recombinase. To obtain Zfp281+/− mice, Zfp281T mice were crossed with PGKCre mice, which resulted in the widespread deletion of Zfp281.25 Zfp281+/− is normal and fertile; however, no Zfp281−/− mice were born from crossing with Zfp281+/− mice (Supplementary Fig. S1c).
FLPeR (JAX: 003946), PGKCre (JAX: 020811), and OT-II (JAX: 004194) TCR transgenic mice were purchased from Jackson Laboratory. Cd4Cre transgenic mice were generously provided by Dr Rémy Bosselut (National Institutes of Health, Bethesda, MD). Mice with the various genotypes studied were generated by the appropriate intercrosses or backcrosses and were analyzed between 6 and 8 weeks of age. All mice were housed in Zhejiang University Laboratory Animal Center under specific pathogen-free conditions. The animal experimental procedures were approved by the Animal Review Committee of Zhejiang University School of Medicine.
Flow cytometry and antibodies
For immunoblot analysis, we used the following antibodies. The anti-Zfp281 (ab101318) and anti-NFAT1 (25A10.D6. D2) antibodies were purchased from Abcam. Anti-ERK1-ERK2 (9102), which is an antibody against ERK1-ERK2 phosphorylated at Thr201 and Tyr204 (9101); anti-JNK1-JNK2 (56G8), which is an antibody against JNK1-JNK2 phosphorylated at Thr183 and Tyr185 (9251); anti-P38 (9212), which is an antibody against P38 phosphorylated at Thr180 and Tyr182 (9211); an antibody against ZAP-70 phosphorylated at Tyr319; and anti-histone H3 (9717S) were purchased from Cell Signaling Technology. Anti-PLC-γ1 (EP1898-7Y) and an antibody against PLC-γ1 phosphorylated at Tyr783 (EP1898Y) were purchased from Epitomics. For flow cytometry, we used the following antibodies from eBioscience: anti-mouse CD4 (RM4-5), CD8α (53-6.7), CD25 (PC61), CD44 (IM7), TCRβ (H57-597), CD69 (H1.2F3), CD62L (MEL-14) and CTLA-4 (UC10-4B9). Flow cytometry was performed with FACSCalibur and FACSAria II machines (BD Biosciences). Data were analyzed by FlowJo software (Tree Star, Inc.). Intracellular staining was performed using IC Fixation Buffer (eBiosciences). For T-cell stimulation, we used monoclonal anti-mouse CD3e (145-2c11) and anti-mouse CD28 (37.51, both from eBioscience) antibodies. For CTLA-4 blockade, we used an anti-CTLA-4 mAb antibody (9H10) from eBioscience.
Ca2+ flux
Sorted naive CD4+ T cells were first labeled for 1 h at 37 °C with 4 μg/mL Fluo-4 (Invitrogen) and then washed with ice-cold PBS and resuspended in PBS. Cells (2 × 106) were incubated with biotinylated anti-CD3 (5 μg/mL; 145-2C11; eBioscience) and anti-CD28 (5 μg/mL; 37.51; BioLegend) antibodies for 30 min on ice. Labeled cells were warmed for 20 min at room temperature and then crosslinked with streptavidin (25 μg/mL) immediately before flow cytometry. The mean fluorescence ratios were plotted after analysis with FlowJo software (Tree Star).
Upregulation assay for CD69 and CD25
Sorted naive CD4+ T cells from wild-type and Zfp281-deficient mice were unstimulated or stimulated with serial dilutions of a plate-bound anti-CD3 mAb overnight to assess the upregulation of early activation markers by flow cytometry. For OT-II-Zfp281fl/fl and OT-II-Zfp281fl/flCd4Cre mice, splenocytes were incubated for 5 h at 37 °C with various concentrations of ovalbumin (OVA) peptide in 96-well round-bottom plates. Cells were stained and analyzed after gating CD4+Vβ5+Vα2+ by flow cytometry as previously described.26
Listeria monocytogenes infection
L. monocytogenes (strain 10403S) was a gift from Dr Qibin Leng (Institute Pasteur of Shanghai, Chinese Academy of Sciences). Age- and sex-matched wild-type and Zfp281-deficient mice (6–8 weeks old) were infected i.v. with L. monocytogenes (3 × 105 CFU per mouse) and sacrificed after 7 days of infection to analyze the primary host response. Livers and spleens were homogenized in 10 mL 0.2% (vol/vol) Triton X-100 in PBS, and the organ homogenates were serially diluted and plated on streptomycin agar plates to determine the CFU of L. monocytogenes. In addition, splenocytes were harvested for flow cytometric analysis of listeriolysin O (LLO)-specific CD4+ T lymphocytes. In brief, splenocytes were stimulated with 5 μg/mL LLO190–201 peptide (NEKYAQAYPNVS) in the presence of a Golgi inhibitor, followed by intracellular IFN-γ staining and flow cytometry analysis.
Ctla-4 shRNA knockdown
Two shRNA-mir sequences targeting Ctla-4 were designed using the shERWOOD Algorithm, (shCtla-4-240, 5′-TGCTGTTGACAGTGAGCGACCAGTCTTCTCTGAAGCCATATAGTGAAGCCACAGATGTATATGGCTTCAGAGAAGACTGGGTGCCTACTGCCTCGGA-3′, and shCtla-4-734,5′- TGCTGTTGACAGTGAGCGCTACAACAGGGGTCTATGTGAATAGTGAAGCCACAGATGTATTCACATAGACCCCTGTTGTAATGCCTACTGCCTCGGA-3′), and then cloned into the pMLP vector. These two plasmids were transfected into the Plat-E packaging cell line as previously described,27 and then, the supernatant containing the retrovirus was collected after 3 days. Sorted naive CD4+ T cells, which were activated by 1 μg/mL coated anti-CD3 antibody for 24 h, were infected with the shCtla-4 retrovirus. After 48 h, cells were restimulated with the anti-CD3 mAb for another 24 h; then, the supernatant was collected and IL-2 production was analyzed by ELISA.
CTLA-4 blockade
Naive CD4+ T cells from OT-II-Zfp281fl/fl or OT-II-Zfp281fl/flCd4Cre mice were sorted and stimulated with 1 μg/mL OVA peptide in T-cell medium supplemented with or without 50 μg/mL anti-CTLA-4 mAb. APCs receiving 30 Gy of ionic irradiation were added at a ratio of 1:1 with naive CD4+ T cells. The supernatant was collected after 24 h and analyzed for IL-2 production by ELISA.
Luciferase reporter assay
The promoter of mouse Ctla-4 and its truncations were produced by PCR-based amplification and subcloned into the pGL3-Enhancer vector to form luciferase reporter plasmids. Human embryonic kidney (HEK293) cells were cotransfected with 100 ng of the luciferase reporter plasmid, 10 ng of the thymidine kinase promoter-Renilla luciferase reporter plasmid plus pCMV tag2b-Zfp281, or the control vector. After 48 h, luciferase activity was determined by the Dual-Luciferase Reporter Assay System (Promega, Cat. No. E10910) according to the manufacturer’s instructions.
ChIP and data analysis
ChIP assays were performed as previously described.28 Briefly, unstimulated and anti-CD3-stimulated naive CD4+ cells were crosslinked with 1% (wt/vol) formaldehyde for 10 min at room temperature, and glycine at a final concentration of 125 mM was added to inactivate formaldehyde. Crosslinked chromatin was sonicated in a 4 °C water bath using a Bioruptor UCD-200 sonicator to obtain DNA fragments between 200 and 500 bp. Chromatin from 10 × 106 cells was used for each ChIP experiment. Antibodies against Zfp281, H3K4me3 (Active Motif), and H3K27me3 (Abcam) were used. Immunoprecipitated DNA was purified using the QIAquick PCR Purification Kit (QIAGEN, Germany) according to the manufacturer’s protocol. Immunoprecipitated DNA was analyzed by q-PCR, and the sequences of the primers used for q-PCR are listed in the Supplemental Materials and Methods. Measurements were performed in triplicate, and error bars denote SDs.
Statistical analysis
The results are presented as the means ± SDs. Significant differences were calculated using the two-tailed unpaired Student’s t tests.
Results
Zfp281 is expressed in T lymphocytes but is not required for T-cell development
Zfp281 is a regulator of pluripotency of ESCs and is essential for embryogenesis.16,17,23 Furthermore, Zfp281 expression is more abundant in the kidney, liver, and peripheral blood lymphocytes than in other adult organs,12,16 suggesting that it might have potential roles besides regulating early embryo development. The q-PCR results showed that Zfp281 mRNA was ubiquitously expressed in adult tissues and was highly expressed in the heart, liver, kidney, thymus, and lymph nodes (Supplementary Fig. S2a), in agreement with previous reports.12,16 We observed that Zfp281 expression was abundant in the thymus and lymph nodes, which is where T cells are abundant. Therefore, we measured Zfp281 expression in different T-cell subsets. Zfp281 was most highly expressed in CD4+ T cells, especially in CD4+ single positive thymocytes, and, to a lesser extent, in peripheral CD4+ T cells (Fig. 1a). Immunoblot analysis further confirmed that the Zfp281 protein was more highly expressed in CD4+ T cells than in CD8+ T cells (Fig. 1b).
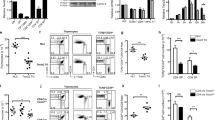
Zfp281 expression is induced by TCR stimulation in CD4+ T cells and does not affect T-cell development. a q-PCR detection of the Zfp281 expression levels in different T-cell subsets, including DN (Lin−CD4−CD8−), DP (CD4+CD8+), CD4 SP (TCRβ+CD4+CD8−), CD8 SP (TCRβ+CD4−CD8+) thymocytes and splenic CD4+, CD8+ T cells (n = 3). b Immunoblot detection of the Zfp281 protein in sorted CD4+, CD8+ splenic T cells. q-PCR (c) and immunoblot analysis (d) of Zfp281 in purified CD4+ splenic T cells stimulated with anti-CD3 and anti-CD28 mAbs for the indicated times. The results are presented relative to Actb expression (n = 3). N.S, non-specific band. e Flow cytometric analysis and quantification of the DN, DP, CD4 SP, and CD8 SP thymocyte subpopulations from Zfp281fl/fl (WT) and Zfp281fl/flCd4Cre (cKO) mice (n = 5). f Flow cytometric analysis of CD4 and CD8 expression in splenocytes from the indicated mice, and quantitation of the absolute cell numbers (n = 5). Data are presented as the mean ± s.d. A two-tailed, unpaired t-test was used. NS not significant. *P < 0.05, **P < 0.01, ***P < 0.001
Next, we evaluated whether Zfp281 was involved in T-cell activation. Primary CD4+ T and CD8+ T lymphocytes were isolated from the spleens of C57BL/6 (wild-type) mice and then stimulated with TCR stimuli (anti-CD3 mAb plus anti-CD28 mAb). The results showed that Zfp281 expression was induced in TCR-engaged CD4+ T cells (Fig. 1c, d), indicating that Zfp281 may be required for this process. We also observed a similar upregulation of Zfp281 in CD8+ T cells, which were stimulated like CD4+ T cells (Supplementary Fig. S2b). Collectively, these findings suggested that Zfp281 might be required for T-cell development and activation.
Nevertheless, Zfp281-null mice are embryonic lethal.17,23 To clarify the function of Zfp281 in T lymphocytes, we generated the conditional Zfp281 allele (Zfp281fl), in which exons 1 and 2 were flanked by loxP sites (Supplementary Fig. S1a). We verified that the first loxP insertion did not disrupt promoter activity (Supplementary Fig. S1b). Intercrossing Zfp281+/− mice (generated by crossing Zfp281T with PGKCre) did not generate Zfp281 deleted offspring, which verified the lethality of Zfp281-null mice (Supplementary Fig. S1c). To study the physiological role of Zfp281 in T lymphocytes, Zfp281fl/fl mice were crossed with Cd4Cre mice, leading to the deletion of functional Zfp281 protein at the DP stage of developing thymocytes,29,30 and Zfp281 was effectively deleted (Supplementary Fig. S1d, e). Zfp281 conditional knockout mice (Zfp281fl/flCd4Cre, cKO) did not show obvious abnormalities in thymocyte development or peripheral T-cell homeostasis (Fig. 1e, f) (Supplementary Fig. S3a, b). Furthermore, Zfp281-deficient mice did not display an obvious alteration of the number of regulatory T cells compared with wild type (Supplementary Fig. S3c). Together, these experiments identified Zfp281 as a gene that is highly expressed in T lymphocytes but is not required for T-cell development.
TCR-mediated responses are diminished in Zfp281-deficient mice
As mentioned above, TCR engagement induces Zfp281 expression in CD4+ T cells. To investigate the function of Zfp281 in CD4+ T-cell activation, we assessed the ability of Zfp281-deficient splenic CD4+ T cells to respond to TCR stimuli. The upregulation of the early activation marker CD25 was lower in Zfp281-deficient CD4+ T cells than in wild type (Fig. 2a). Similarly, CD69 expression was also lower in CD4+ T cells from Zfp281 conditional knockout mice (Fig. 2b). Further investigations showed that Zfp281-deficient CD4+ T cells produced less interferon-γ (IFN-γ) (Fig. 2c). In addition, IL-2 production in Zfp281-deficient CD4+ T cells was also decreased after stimulation (Fig. 2d). Furthermore, CD8+ T cells showed less upregulation of CD25 and CD69 in the absence of Zfp281 (Supplementary Fig. S4a, b). However, intracellular IFN-γ and IL-2 only showed mild changes in Zfp281-knockout CD8+ T cells (Supplementary Fig. S4c and d). We also determined whether CD4+ T-cell proliferation was impaired in cKO mice. Zfp281-deficient CD4+ T cells had a comparable proliferative capacity in response to anti-CD3 stimulation as wild type did (Supplementary Fig. S5a). In addition, Zfp281 deficiency had little effect on apoptosis of CD4+ T cells (Supplementary Fig. S5b).
TCR-induced activation and cytokine production are decreased in Zfp281-deficient CD4+ T cells. CD25 (a) and CD69 (b) staining on sorted naive CD4+ (CD44loCD62LhiCD4+CD25−) splenocytes from the indicated mice stimulated with the indicated concentration of the anti-CD3 mAb overnight. The numbers above the lines (left) indicate the percentage of CD25+ (a) or CD69+ (b) cells. The statistics of the percentage of CD25+ (a) and CD69+ cells are shown on the right; b the statistics of the percentage of T cells are shown on the left. ELISA detection of IFN-γ (c) and IL-2 (d) secreted by wild-type and Zfp281-deficient naive CD4+ T cells stimulated with the indicated concentration of the anti-CD3 mAb for 3 days. Data are representative of three independent experiments (mean ± s.d.). A two-tailed, unpaired t-test was used. NS not significant. *P < 0.05, **P < 0.01
The impaired TCR-mediated responses in Zfp281-deficient CD4+ T cells raised the question of whether the defect caused by Zfp281 loss was antigen-specific. To address this question, we crossed MHC class II-restricted TCR transgenic mice (OT-II) with Zfp281-deficient mice. T-cell development in Zfp281-deficient mice was normal in the OT-II background (Supplementary Fig. S6). Similarly, the upregulation of CD25 and CD69 expression were reduced in OT-II-Zfp281fl/flCd4Cre T cells in response to OVA peptide (Supplementary Fig. S7a, b). In addition, we also found that Zfp281-deficient OT-II CD4+ T cells stimulated with OVA had decreased production of IFN-γ and IL-2 compared with control (Zfp281fl/fl) OT-II cells (Supplementary Fig. S7c, d). Collectively, these results suggest that Zfp281 contributes to the TCR-mediated CD4+ T-cell response.
Zfp281 deficiency interrupts TCR signaling in naive CD4+ T cells
T-cell activation involves cascades of signaling events triggered by TCR stimuli, which is critical for survival, proliferation, and cytokine production.2 To further determine whether Zfp281 functions downstream of TCR, we analyzed intracellular signaling events in naive CD4+ T cells from wild-type and Zfp281-deficient mice after crosslinking TCR to the anti-CD3 plus anti-CD28 mAb. We observed less phosphorylation of the TCR-proximal signaling protein ZAP-70 in Zfp281-deleted CD4+ T cells (Fig. 3a). Activated ZAP-70 phosphorylates the adaptor LAT, resulting in the formation of LAT signalosomes. PLC-γ1 is one of the downstream components in this signaling cascade. Western blotting showed a slight decrease in pPLC-γ1 levels in Zfp281-deficient CD4+ T cells (Fig. 3a). We further detected other molecules involved in T-cell activation. CD4+ T cells from Zfp281-deficient mice displayed less phosphorylation of the kinases ERK1/2, JNK, and P38 (Fig. 3b). Moreover, naive CD4+ T cells from Zfp281-deficient mice had a much lower TCR-driven calcium flux than wild-type cells (Fig. 3c). Consistent with the decreased TCR-induced Ca2+ influx, Zfp281-deficient CD4+ T cells led to less NFAT1 localization in the nucleus upon TCR stimulation (Fig. 3d). Thus, these data indicate that Zfp281 plays an important role in TCR signaling pathways.
TCR signaling is interrupted in Zfp281-deficient CD4+ T cells. a Immunoblot analysis of total PLC-γ1 and phosphorylated (p-) PLC-γ1 and ZAP-70 in extracts of sorted naive CD4+ T cells from WT and cKO splenocytes that were stimulated for 0, 5, 10, and 20 min with the anti-CD3 mAb and anti-CD28 mAb. b Immunoblot analysis of total and phosphorylated (p-) ERK1/2, JNK and P38 in extracts of cells stimulated as in a. c Calcium flux in naive CD4+ T cells from Zfp281fl/fl and Zfp281fl/flCd4Cre mice after stimulation with anti-CD3 and anti-CD28 mAbs. d Immunoblot analysis of NFAT1 in nuclear extracts of naive CD4+ T cells stimulated as in a. The numbers below the lanes (a, b, d) indicate densitometry analysis. Data are representative of at least three independent experiments
Zfp281 promotes T-cell immune responses to Listeria infection
Zfp281-deficient T cells have impaired T-cell activation and reduced IFN-γ production, indicating that T-cell immune responses are defective after Zfp281 deletion. To further analyze the role of Zfp281 in CD4+ T-cell immune responses in vivo, we used the L. monocytogenes infection model. L. monocytogenes is known to induce strong T-cell responses, in particular, triggering IFN-γ production of CD4+ T cells.31 Age- and sex-matched wild-type and Zfp281-deficient mice were infected with L. monocytogenes. Seven days later, splenocytes were stimulated ex vivo with bacteria-specific peptide LLO for flow cytometric analysis. We found that in response to L. monocytogenes infection, both wild-type and Zfp281-deficient mice generated IFN-γ-producing CD4+ T cells specific to the LLO antigen (Fig. 4a). However, Zfp281-deficient mice had a lower frequency of IFN-γ-producing CD4+ T cells than wild-type mice (Fig. 4a, b). Moreover, the serum IFN-γ concentration significantly decreased in L. monocytogenes-infected Zfp281-deficient mice compared with wild-type mice (Fig. 4c). Consistent with the diminished T-cell responses, Zfp281-deficient mice had a significantly higher bacterial load in their spleens and livers (Fig. 4d), suggesting a diminished ability to clear bacteria. These results indicate the positive role of Zfp281 in regulating the activation and inflammatory responses of CD4+ T cells.
Zfp281 deficiency impairs the T-cell response to L. monocytogenes infection. a Flow cytometric analysis of antigen (LLO)-specific IFN-γ–producing CD4+ T cells in the spleens of WT and cKO mice sacrificed on day 7 after L. monocytogenes infection. The numbers adjacent to the outlined areas indicate the percentage of IFN-γ–producing CD4+ T cells (n = 5 for each group). b The frequency of CD4+ IFN-γ+ T cells as indicated in a. c Serum IFN-γ levels detected by ELISA. d L. monocytogenes titer in spleens and livers. CFU, colony-forming units. Data are representative of three independent experiments (mean ± s.d.). A two-tailed, unpaired t-test was used. NS not significant. *P < 0.05, ***P < 0.001
Zfp281 controls the negative feedback of T-cell activation
To further understand the biology of Zfp281 in CD4+ T cells, we characterized the transcriptional profile of TCR-triggered WT and Zfp281-knockout CD4+ T cells using RNA sequencing (RNA-Seq). Using a fold change of 1.5 and P < 0.05 as cut-off parameters, we identified 246 upregulated genes in Zfp281-deficient cells and 157 downregulated genes (Fig. 5a). Using gene ontology (GO) analysis, we found a significant enrichment of genes associated with synapse organization and T-cell-related differentiation and activation (Fig. 5b). The transcripts that negatively regulated T-cell activation (e.g., Ctla-4, Tigit, Foxj1) were significantly upregulated in cKO CD4+ T cells, while the transcripts that were positively related to T-cell activation were downregulated, such as Thy-1 and Tnfsf14 (Fig. 5c).
Zfp281-deficient CD4+ T cells show decreased expression of T-cell activation-related genes. a Scatter plot of the expression of all genes in WT and Zfp281-deficient naive CD4+ T cells stimulated with the anti-CD3 mAb for 4 h in the presence of APC detected by RNA-Seq analysis. Downregulated genes are indicated in blue, upregulated genes are indicated in red. b Gene ontology (GO) analysis of the differentially expressed genes as shown in a. Blue, downregulated genes; red, upregulated genes. c Heat map comparing the indicated mRNA transcripts. d q-PCR analysis of the Ctla-4 transcript levels in naive CD4+ T cells from WT and cKO mice unstimulated or stimulated with coated anti-CD3 (5 μg/mL) and soluble anti-CD28 (3 μg/mL) for 4 h. e CTLA-4 expression in Tconv cells with or without plate-bound anti-CD3 stimulation for 8 h. f Detection of CTLA-4 in Tconv cells from mice with the OT-II background treated with or without the OVA peptide for 8 h. Data are representative of three independent experiments (mean ± s.d.). A two-tailed, unpaired t-test was used. NS not significant. *P < 0.05, **P < 0.01
To determine the direct targets of Zfp281 that are related to the defective activation of Zfp281-deficient CD4+ T cells, we focused on a set of genes that negatively regulated T-cell activation and had upregulated mRNA expression in Zfp281-deficient T cells. One of the most prominent genes was Ctla-4, a negative costimulator of T-cell activation.4 The RNA-Seq data showed that Zfp281 deletion led to increased Ctla-4 expression in activated CD4+ T cells (Fig. 5c), which was validated by q-PCR (Fig. 5d). Using flow cytometry, Zfp281-knockout mice were found to have a higher proportion of CTLA-4+CD4+ T cells after anti-CD3 stimulation, while no significant difference in resting T cells was observed (Fig. 5e). Zfp281-knockout CD4+ T cells in the OT-II background were also found to have a higher proportion of CTLA-4+ after OVA-specific stimulation (Fig. 5f). Unlike conventional CD4+ T cells, in which CTLA-4 is induced after T-cell activation, Tregs continuously express CTLA-4, which is a key receptor for controlling the suppressive function of Treg.32 CTLA-4 expression in Tregs showed no obvious difference between WT and cKO mice (Supplementary Fig. S8a–c). In addition, Zfp281-deficient CD8+ T cells also had increased expression of CTLA-4-positive cells after anti-CD3 stimulation (Supplementary Fig. S8d).
Zfp281 binds to the CTLA-4 promoter to inhibit its transcription activity
Zfp281 is a transcription factor that plays both transcriptionally active and repressive roles in ESCs. It has been reported that Zfp281 binds to GC-rich sequences of target genes to regulate their transcriptional activities and gene expression.16 Therefore, we compared the sequences of the Ctla-4 promoter among various species, including mice, rats, rabbits, and humans. Two GC-rich sites similar to known Zfp281 binding sites were found at the −291 and −397 loci (Supplementary Fig. S9a). The sequences of the two sites were relatively highly conserved in mammals (Supplementary Fig. S9a), suggesting that they are potential targets of Zfp281. Subsequently, a series of luciferase reporter plasmids encoding Ctla-4 promoters with different lengths were constructed to investigate whether Zfp281 could repress the transcription of Ctla-4 and to determine the regions of the Ctla-4 promoter targeted by Zfp281. We found that Zfp281 could significantly inhibit the transcriptional activity of the Ctla-4 promoter (−500) and (−1309) constructs but was unable to repress the luciferase activity of the −100 construct. These results indicate that Zfp281 might inhibit CTLA-4 expression by directly binding to GC-rich sites located in the −500 region of the Ctla-4 promoter (Supplementary Fig. S9b). To better identify the binding sites of Zfp281 on the Ctla-4 promoter and clarify whether the two conserved GC-rich sites were Zfp281 binding sites, we constructed single- or double-deleted luciferase reporter plasmids, as shown in Fig. 6a. The results showed that Zfp281 suppressed the luciferase activity of the Ctla-4 (−500 bp) construct and either of the one-site deleted constructs but had no effect on the luciferase activity of the two-site deleted construct (Fig. 6a). These results suggest that Zfp281 may inhibit CTLA-4 by directly binding to the two sites. Direct binding of Zfp281 to these two presumed gene loci was confirmed by ChIP-qPCR in wild-type CD4+ T cells under unstimulated and stimulated conditions (Fig. 6b). We also analyzed the chromatin modifications of the Ctla-4 promoter using ChIP-qPCR. The results showed that there was a significant decrease in H3K27me3 marks on the two sites of the Ctla-4 promoter in Zfp281-deficient CD4+ T cells, while the amount of H3K4me3 modification increased in the absence of Zfp281 (Fig. 6c, d). Therefore, Zfp281 suppresses Ctla-4 transcription by directly binding the Ctla-4 promoter.
Ctla-4 is a functional target gene of Zfp281, and Zfp281 inhibits the transcriptional activity of Ctla-4 by binding its promoter. a Zfp281 inhibits the luciferase activity of the Ctla-4 promoter. (Left) Schematic of the Ctla-4 promoter with the indicated constructs. (Right) HEK293 cells were infected for 48 h with luciferase reporter plasmids as shown on the left as well as a construct that encodes Zfp281 or the empty vector (Mock), and then, cells were harvested to detect luciferase activity. ChIP analysis of Zfp281 (b), H3K4me3 (c), and H3K27me3 (d) in wild-type and Zfp281-deficient CD4+ T cells unstimulated (Top) and stimulated with the anti-CD3 mAb (Bottom), followed by q-PCR detection of the specific enrichment of each modification at the Ctla-4 promoter. e Naive CD4+ T cells sorted from WT and cKO mice were stimulated with 1 μg/mL coated anti-CD3 mAb for 24 h and then infected with the shCtla-4 or shNC retrovirus. After 48 h, the cells were restimulated with the anti-CD3 mAb for another 24 h, and the supernatant was collected for IL-2 detection by ELISA. The IL-2 level in the shCtla-4 supernatant was normalized to that in the shNC supernatant. f CTLA-4 blockade assay. Data are representative of three independent experiments (mean ± s.d.). A two-tailed, unpaired t-test was used. NS not significant. *P < 0.05, **P < 0.01
Next, we determined whether impaired T-cell activation could be rescued after knocking down CTLA-4 using shRNA. We designed shRNA that targeted Ctla-4 and determined the knockdown efficiency at the mRNA and protein levels (Supplementary Fig. S9c and d). As shown in Fig. 6e, IL-2 expression could be restored in Zfp281-knockout CD4+ T cells transfected with Ctla-4 shRNA. Zfp281-knockout CD4+ T cells in the OT-II background also showed an increase in IL-2 production when treated with the OVA peptide in the presence of the anti-CTLA-4 mAb (Fig. 6f).
Discussion
In this study, we determined that Zfp281 is a regulator of CD4+ T-cell activation in addition to its well-recognized role in maintaining ESC pluripotency. The loss of Zfp281 did not affect T-cell development, as reflected by the normal frequencies of T-cell subsets. However, Zfp281 was required for proper activation and immune responses of CD4+ T cells. Zfp281 deficiency impaired T-cell activation in vitro and inhibited T-cell responses in vivo to L. monocytogenes infection. Using RNA-Seq, we found that Zfp281 deficiency affected genes related to T-cell activation and differentiation, and Ctla-4 was one of its targets. Further studies using luciferase reporter assays and ChIP-qPCR showed that Zfp281 bound to GC-rich sites of the Ctla-4 promoter and inhibited the transcription of Ctla-4. Thus, Zfp281 is required for the appropriate induction of T-cell activation.
Zfp281 is expressed in the pluripotent state and repressed upon differentiation to embryoid bodies.14,21 In addition, Zfp281 has been found to be more abundant in adult liver, kidney, and peripheral blood lymphocytes,12,16 indicating that it has other functions in addition to regulating early embryo development. However, the role of Zfp281 in the immune system remains poorly understood. Our results showed a similar tissue expression profile of Zfp281 as previous studies (Supplementary Fig. S2a). Zfp281 is highly expressed in the thymus, which attracted our attention. To explore whether Zfp281 was involved in T-cell development, we constructed Zfp281 conditional knockout mice using the Cre-loxP system. Zfp281 conditional knockout mice showed normal T-cell development (Fig. 1e, f), although Zfp281 was highly expressed in thymocytes, especially in CD4 SP cells (Fig. 1a). Other factors might compensate for the impairment caused by Zfp281 deficiency. Zinc finger protein 148 (Zfp148) is a Krüpple type transcription factor that shares 91% amino acid sequence similarity and 79% DNA sequence identity with Zfp281.12 Furthermore, Zfp148 and Zfp281 both bind to GC-rich regulatory regions of their target genes, indicating that they may complement the activities of each other. Reizis et al.33 found that the anti-Zfp148 antibody specifically blocked the formation of the Pre-TCR alpha complex and LckB complex, suggesting a possible role of Zfp148 in T-cell development by targeting specific genes. However, Zfp148-deficient mice did not show visible defects in T-cell development.34 In our cases, the Zfp148 mRNA levels were not significantly changed in Zfp281-deficient thymocytes and splenic CD4+ T cells (Supplementary Fig. S1f). In addition, Zfp148 expression could not be induced by TCR stimulation (Supplementary Fig. S2c). Whether Zfp281 and Zfp148 can compensate for each other in regard to T-cell development and their functions needs to be explored further.
The gene expression profile results showed that Zfp281 was also highly expressed in peripheral T cells (Fig. 1a). Splenic CD4+ T cells expressed more Zfp281 than CD8+ T cells. Due to this observation, we paid more attention to peripheral CD4+ T cells. TCR stimulation induced Zfp281 expression at the mRNA and protein levels in CD4+ T cells. CD4+ T cells from Zfp281 knockout mice showed impaired activation (Fig. 2). It is worth noting that Zfp281 was similarly induced in CD8+ T cells upon TCR stimulation (Supplementary Fig. S2b). These observations suggest a potential role for Zfp281 in CD8+ T-cell activation. Indeed, we found that CD8+ T-cell activation was also impaired after Zfp281 deletion, but the phenotype was mild (Supplementary Fig. S4).
Previous studies have identified that Zfp281 acts as a repressor of the maintenance of stem cell pluripotency by directly binding to key regulators, such as Nanog and Oct4, and the predicted targets of Zfp281 in ESCs have been shown to be considerably enriched for the repressive H3K27me3 modification.17,18,35 It has been reported that targeted genes are regulated by Zfp281 at both the transcriptional and posttranscriptional levels. Zfp281 cooperates with epigenetic control mechanisms, such as histone acetylation and methylation complexes, to exert transcriptional activity in ESCs or during epiblast maturation.36,37,38 According to the RNA-Seq analysis results, we found that more genes with altered expression in Zfp281-knockout CD4+ T cells tended to be upregulated (Fig. 5a), suggesting that Zfp281 might function as a repressor in CD4+ T cells. Therefore, we mainly focused on genes that negatively regulated T-cell activation and had upregulated expression in Zfp281-deficient T cells (Fig. 5c). Then, we performed sequence alignments and analyses of the −1000 bp promoter of these genes to identify the Zfp281 binding motif. Four of genes (Sox4, Foxj1, Ctla-4, and Ccr9) contain relatively conserved potential binding sites for Zfp281 (data not shown). SOX4 has been reported to be an upstream factor of Zfp28139 Furthermore, Malhotra et al.40 found that SOX4 positively regulates TCR signaling during iNKT differentiation. Although Foxj1 has been identified as a repressor of T-cell activation and autoreactivity,41 we determined that its expression in CD4+ T cells was less abundant.
CTLA-4 downregulates immune responses and has been identified to be an immune checkpoint for cancer immunotherapy.42 In this study, CTLA-4 expression was upregulated in the absence of Zfp281. Moreover, we found a significant decrease of the H3K27me3 modification on the Ctla-4 promoter in Zfp281-deficient CD4+ T cells, while the H3K4me3 modification was increased in the absence of Zfp281 (Fig. 6c, d). To date, the transcriptional regulation of Ctla-4 gene expression is only partially understood. The transcriptional regulation of Ctla-4 gene expression may depend on NFAT because modulation of the NFAT level is directly correlated with CTLA-4 expression. Furthermore, it has been shown that NFAT binds the Ctla-4 promoter region.43 Wu et al.44 demonstrated that Foxp3 is a direct activator of the Ctla-4 gene. In addition, GATA3 binds to the Ctla-4 proximal promoter in bortezomib-treated CD4+ T cells, and GATA3 expression can enhance Ctla-4 promoter activity in a dose-dependent manner.45 In this study, Ctla-4 expression increased after Zfp281 deletion, which may have been a direct effect of the absence of Zfp281. Wang et al.16 identified a 6-bp GC-rich domain as the common binding motif for Zfp281. We found two relatively conserved GC-rich regions in the Ctla-4 promoter (Supplementary Fig. S9a). Further studies using luciferase reporter assays and ChIP-qPCR showed that Zfp281 could bind the two sites and inhibit the transcription of Ctla-4 (Fig. 6a, b). However, we do not exclude the possibility that other differentially expressed genes may be targets of Zfp281.
In summary, we identified previously unrecognized functions of Zfp281: it maintains CD4+ T-cell activation and function by directly binding the Ctla-4 promoter. The immunomodulatory property of Zfp281 in CD4+ T cells identified in our study suggests that Zfp281 expression might increase in cases of CD4+ T-cell-mediated immunopathology, such as a bacterial infection. In the future, it will be important to better understand the role and regulation of Zfp281 in diverse physiological and pathological processes in more detail. This knowledge will potentially serve as a basis for understanding CD4+ T-cell-mediated diseases and exploring therapeutic approaches.
Accession numbers
The RNA-Seq data sets were deposited in GEO with the accession number GSE135968.
References
Chen, L. & Flies, D. B. Molecular mechanisms of T cell co-stimulation and co-inhibition. Nat. Rev. Immunol. 13, 227–242 (2013).
Smith-Garvin, J. E., Koretzky, G. A. & Jordan, M. S. T cell activation. Annu. Rev. Immunol. 27, 591–619 (2009).
Linsley, P. S. & Ledbetter, J. A. The role of the CD28 receptor during T cell responses to antigen. Annu. Rev. Immunol. 11, 191–212 (1993).
Krummel, M. F. & Allison, J. P. CD28 and CTLA-4 have opposing effects on the response of T cells to stimulation. J. Exp. Med. 182, 459–465 (1995).
Brownlie, R. J. & Zamoyska, R. T cell receptor signalling networks: branched, diversified and bounded. Nat. Rev. Immunol. 13, 257–269 (2013).
Chan, A. C., Iwashima, M., Turck, C. W. & Weiss, A. ZAP-70: a 70 kd protein-tyrosine kinase that associates with the TCR zeta chain. Cell 71, 649–662 (1992).
Guy, C. S. & Vignali, D. A. Organization of proximal signal initiation at the TCR:CD3 complex. Immunol. Rev. 232, 7–21 (2009).
Brayer, K. J. & Segal, D. J. Keep your fingers off my DNA: protein-protein interactions mediated by C2H2 zinc finger domains. Cell Biochem. Biophys. 50, 111–131 (2008).
Razin, S. V., Borunova, V. V., Maksimenko, O. G. & Kantidze, O. L. Cys2His2 zinc finger protein family: classification, functions, and major members. Biochemistry 77, 217–226 (2012).
Han, B. Y. et al. Zinc finger protein Zfp335 is required for the formation of the naïve T cell compartment. Elife 3, e03549 (2014).
Moroy, T. & Khandanpour, C. Growth factor independence 1 (Gfi1) as a regulator of lymphocyte development and activation. Semin. Immunol. 23, 368–378 (2011).
Law, D. J., Du, M., Law, G. L. & Merchant, J. L. ZBP-99 defines a conserved family of transcription factors and regulates ornithine decarboxylase gene expression. Biochem. Biophys. Res. Commun. 262, 113–120 (1999).
Lisowsky, T. et al. Identification of human GC-box-binding zinc finger protein, a new Krüppel-like zinc finger protein, by the yeast one-hybrid screening with a GC-rich target sequence. FEBS Lett. 453, 369–374 (1999).
Brandenberger, R. et al. Transcriptome characterization elucidates signaling networks that control human ES cell growth and differentiation. Nat. Biotechnol. 22, 707–716 (2004).
Wei, C. L. et al. Transcriptome profiling of human and murine ESCs identifies divergent paths required to maintain the stem cell state. Stem Cells 23, 166–185 (2005).
Wang, Z. X. et al. The transcription factor Zfp281 controls embryonic stem cell pluripotency by direct activation and repression of target genes. Stem Cells 26, 2791–2799 (2008).
Fidalgo, M. et al. Zfp281 functions as a transcriptional repressor for pluripotency of mouse embryonic stem cells. Stem Cells 29, 1705–1716 (2011).
Fidalgo, M. et al. Zfp281 mediates Nanog autorepression through recruitment of the NuRD complex and inhibits somatic cell reprogramming. Proc. Natl Acad. Sci. USA 109, 16202–16207 (2012).
Seo, K. W. et al. ZNF281 knockdown induced osteogenic differentiation of human multipotent stem cells in vivo and in vitro. Cell Transpl. 22, 29–40 (2013).
Hahn, S. & Hermeking, H. ZNF281/ZBP-99: a new player in epithelial-mesenchymal transition, stemness, and cancer. J. Mol. Med. 92, 571–581 (2014).
Wang, D. et al. Identification of pluripotency genes in the fish medaka. Int. J. Biol. Sci. 7, 440–451 (2011).
Wang, J. et al. A protein interaction network for pluripotency of embryonic stem cells. Nature 444, 364–368 (2006).
Huang, X. et al. Zfp281 is essential for mouse epiblast maturation through transcriptional and epigenetic control of Nodal signaling. Elife 6, e3333 (2017).
P., L., N., A. & N., G. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 13, 476–484 (2003).
Lallemand, Y., Luria, V., Haffner-Krausz, R. & Lonai, P. Maternally expressed PGK-Cre transgene as a tool for early and uniform activation of the Cre site-specific recombinase. Transgenic Res. 7, 105–112 (1998).
Yachi, P. P., Ampudia, J., Gascoigne, N. R. & Zal, T. Nonstimulatory peptides contribute to antigen-induced CD8-T cell receptor interaction at the immunological synapse. Nat. Immunol. 6, 785–792 (2005).
Saitoh, T., Nakano, H., Yamamoto, N. & Yamaoka, S. Lymphotoxinry peptides contribute to antigen-induced CD8-T cell rec. FEBS Lett. 532, 45–51 (2002).
Lee, T. I., Johnstone, S. E. & Young, R. A. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat. Protoc. 1, 729–748 (2006).
Lee, P. P. et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity 15, 763–774 (2001).
Sawada, S., Scarborough, J. D., Killeen, N. & Littman, D. R. A lineage-specific transcriptional silencer regulates CD4 gene expression during T lymphocyte development. Cell 77, 917–929 (1994).
Wakil, A. E., Wang, Z. E., Ryan, J. C., Fowell, D. J. & Locksley, R. M. Interferon gamma derived from CD4(+) T cells is sufficient to mediate T helper cell type 1 development. J. Exp. Med 188, 1651–1656 (1998).
Takahashi, T. et al. Immunologic self-tolerance maintained by CD25(+)CD4(+) regulatory T cells constitutively expressing cytotoxic T lymphocyte-associated antigen 4. J. Exp. Med. 192, 303–310 (2000).
Reizis, B. & Leder, P. Expression of the mouse pre-T cell receptor α gene is controlled by an upstream region containing a transcriptional enhancer. J. Exp. Med. 189, 1669–1678 (1999).
Nilton, A. et al. Zinc finger protein 148 is dispensable for primitive and definitive hematopoiesis in mice. Plos ONE 8, e70022 (2013).
Kim, J., Chu, J., Shen, X., Wang, J. & Orkin, S. H. An extended transcriptional network for pluripotency of embryonic stem cells. Cell 132, 1049–1061 (2008).
Wang, Y. et al. A permissive chromatin state regulated by ZFP281-AFF3 in controlling the imprinted Meg3 polycistron. Nucleic Acids Res. 45, 1177–1185 (2017).
Dai, Q. et al. Striking a balance: regulation of transposable elements by Zfp281 and Mll2 in mouse embryonic stem cells. Nucleic Acids Res. 45, 12301–12310 (2017).
Fidalgo, M. et al. Zfp281 coordinates opposing functions of Tet1 and Tet2 in pluripotent states. Cell Stem Cell 19, 355–369 (2016).
Scharer, C. D. et al. Genome-wide promoter analysis of the SOX4 transcriptional network in prostate cancer cells. Cancer Res. 69, 709–717 (2009).
Malhotra, N. et al. SOX4 controls invariant NKT cell differentiation by tuning TCR signaling. J. Exp. Med. 215, 2887–2900 (2018).
Lin, L., Spoor, M. S., Gerth, A. J., Brody, S. L. & Peng, S. L. Modulation of Th1 activation and inflammation by the NF-κB repressor Foxj1. Science 303, 1017–1020 (2004).
Li, X., Song, W., Shao, C., Shi, Y. & Han, W. Emerging predictors of the response to the blockade of immune checkpoints in cancer therapy. Cell Mol. Immunol. 16, 28–39 (2019).
Gibson, H. M. et al. Induction of the CTLA-4 gene in human lymphocytes is dependent on NFAT binding the proximal promoter. J. Immunol. 179, 3831–3840 (2007).
Wu, Y. et al. FOXP3 controls regulatory T cell function through cooperation with NFAT. Cell 126, 375–387 (2006).
Gibson, H. M. et al. Impaired proteasome function activates GATA3 in T cells and upregulates CTLA-4: relevance for Sezary syndrome. J. Investig. Dermatol. 133, 249–257 (2013).
Acknowledgements
We thank Dr Xiaolong Liu (Shanghai Institutes for Biological Sciences, Chinese Academy of Science) for his generous gifts of the cell lines. We thank Yingying Huang, Yiwei Li, and Jiajia Wang (Zhejiang University) for helping with the cell sorting; Yu Zhang and Rui Ma (Zhejiang University) for feeding the mice; and Breanna Laine Breaux (Stanford University) for improving the grammar and writing of the paper. This work was supported in part by grants from the National Basic Research Program of China 973 Program (2015CB943301), the National Natural Science Foundation of China (81830006, 31670887, 31870874, and 31800734), the Zhejiang provincial Key Project of Research and Development (2019C0304), the Zhejiang Natural Science Foundation (LQ16H030003), and the Zhejiang Science and Technology Program (2017C37117 and 2017C37170).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made.
The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
To view a copy of this licence, visit https://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Guo, J., Xue, Z., Ma, R. et al. The transcription factor Zfp281 sustains CD4+ T lymphocyte activation through directly repressing Ctla-4 transcription. Cell Mol Immunol 17, 1222–1232 (2020). https://doi.org/10.1038/s41423-019-0289-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41423-019-0289-y
- Springer Nature Limited
Keywords
This article is cited by
-
A new marker constructed from immune-related lncRNA pairs can be used to predict clinical treatment effects and prognosis: in-depth exploration of underlying mechanisms in HNSCC
World Journal of Surgical Oncology (2023)
-
Transcriptional regulation of the immune checkpoints PD-1 and CTLA-4
Cellular & Molecular Immunology (2022)
-
Engineering micro oxygen factories to slow tumour progression via hyperoxic microenvironments
Nature Communications (2022)
-
Optineurin modulates the maturation of dendritic cells to regulate autoimmunity through JAK2-STAT3 signaling
Nature Communications (2021)