Abstract
In this report, we evaluated the effect of the pasteurization (P) process of mother’s own milk (MOM) on the miRNA content of extracellular vesicles (EVs) and its impact on innate immune responses. Differences in size or particle number were not observed upon pasteurization of MOM (PMOM). However, significant differences were observed in the EV membrane marker CD63 and miRNA profiles. miRNA sequencing identified 33 differentially enriched miRNAs between MOMEV and PMOMEV. These changes correlated with significant decreases in the ability of PMOMEV to modulate IL-8 secretion in intestinal Caco2 cells where only MOMEV were able to decrease IL-8 secretion in presence of TNFα. While EVs from MOMEV and PMOMEV were both able to induce a tolerogenic M2-like phenotype in THP-1 macrophages, a significant decrease in the transcript levels of IL-10 and RNA sensing genes was observed with PMOMEV. Together, our data indicates that pasteurization of MOM impacts the integrity and functionality of MOMEV, decreasing its EVs-mediated immunomodulatory activity. This data provides biomarkers that may be utilized during the optimization of milk processing to preserve its bioactivity.
Similar content being viewed by others
Introduction
Human milk (herein referred to as Mother’s own milk, MOM) is a complex source of nutrition and metabolic-endocrine regulators that program the signaling system of infants in a very personalized manner. The bioactive components responsible for these benefits include a wide variety of cytokines, hormones, immunoglobulins, miRNAs, and bacterial cells that are transmitted from mother to child1, 2. Our group and others have reported that around 200 different bacterial species have been identified in MOM, suggesting that these may not be simply contaminants but integral components that play a beneficial role in development3, 4. MOM provides important benefits to preterm infants, including immune benefits, nutrition, protection against infections (especially necrotizing enterocolitis), and overall, a decreased risk of mortality5.
In the absence of MOM, the American Academy of Pediatrics (AAP) recommends using pasteurized donor human milk (DHM) over formula in very preterm infants6. Current practice in the Neonatal Intensive Care Unit (NICU) at Shands Children’s Hospital, (University of Florida), is to provide DHM to premature infants of gestational age less than 30 weeks. DHM is obtained from pooled donors and pasteurized, rendering the milk nearly devoid of personalized live commensal bacteria7. Furthermore, pasteurization of DHM has also been reported to significantly decrease the levels of sIgA, alkaline phosphatase, bile salt stimulated lipase, lactoferrin and leptin8, 9. Nonetheless, the role of the pasteurization process on the diversity of microRNA (miRNAs) has not been evaluated until now. miRNAs are small, non-coding RNAs (around 22-nucleotides in length) that have been found in bacteria, animals, and some viruses. More than 3000 recognized or new miRNAs have been identified in human milk10, 11, most of which have been implicated in immune functions.miRNAs are secreted within milk exosomes or extracellular vesicles (EV). EVs have an endocytic origin and are around 30–150 nm (diameter) in size. They are surrounded by a lipid bilayer membrane and contain a rich cargo composed of proteins, DNA, RNA, peptides, and lipid-derivatives12, 13. These nanostructures have been isolated from many biological fluids including MOM. Among the rich cargo, miRNAs are considered an outstanding feature due to their role as modulators of a wide variety of processes14, 15. It has been reported that miRNAs can regulate more than 30% of human genes16. Thus, the transfer of exosomes and their miRNA cargo from mother to child contributes to the benefits provided by breastfeeding. miRNAs contained in MOM EVs have high stability at low pH, suggesting that these regulators could survive the gastric transit, reaching the intestine where they could modulate the host’s immune system17. Recently, it was shown that pasteurized or milk subjected to high pressure milk processing introduced shifts in the abundancy of several miRNA18. This article overcomes a limitation identified in previous reports where the effect of pasteurization on the functionality of miRNAs contained in the exosomes was not evaluated. The most abundant miRNAs found in MOM target several processes important for infant development (reviewed in19). Some of those are: miR-182-5P, miR-148a-3p, let-7f-5p and miR-22-3p, targeting genes involved in lipid metabolism; miR26a and miR181b involved in glucose metabolism; miR21 and miR-200 family participating in tight junction stability in the gut; miR-29b and the let-7 family affecting neurogenesis; and the miR29 and miR-148a-3p implicated in epigenetic regulation. Furthermore, miRNAs miR-181a, miR223, miR146b-5p, and miR155 play important roles in the development of both innate and acquired immune systems.
The preservation of these bioactive components in MOM is highly important in preterm infants, where MOM feedings reduce the incidence and severity of infections, especially necrotizing enterocolitis (NEC) and late onset sepsis (LOS)20, 21. As indicated above MOM feedings have shown superior epidemiological performance when compared to DHM and formula. In this report, we have evaluated the effect of the pasteurization process of MOM on the miRNA content of MOM EVs and their impact on innate immune response in human cell lines. We found that pasteurization of MOM impacts the integrity and the miRNA diversity in the EV cargo. Furthermore, biomarkers of host responses that may be followed during the milk processing to evaluate its bioactivity were identified.
Results
Pasteurization does not affect the EVs yield of MOM
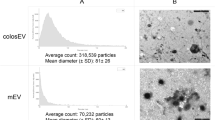
To evaluate the role of pasteurization on the stability of miRNAs in human milk, we first developed PMOM (as a proxy for DHM) in which the components are identical to MOM, except that it has been subjected to pasteurization (see “Material and methods” for details). Next, the EVs enriched fractions of MOM and PMOM were assessed for yield and size distribution. A comparable yield of EVs was observed between MOMEV and PMOMEV with an average concentration of 1.6 × 1010 ± 9.6 × 108 and 1.7 × 1010 ± 1 × 109 EVs per mL of milk, respectively (Suppl. Fig. 1). MOMEV and PMOMEV also showed a similar size distribution (200 nm in average diameter), and negative zeta potential (MOMEV = − 13.4 mV, PMOMEV = − 16.64 mV, respectively). However, immunoblots using the exosome common markers CD63 and CD9 showed that only CD9 was detected in MOMEV and PMOMEV. On the contrary, CD63 was observed in MOMEV samples but was absent in PMOMEV, indicating damage or degradation during the pasteurization process. The ER marker Calnexin was not detected in either EVs samples, however the apolipoprotein A1 was detectable in both MOMEV and PMOMEV, indicating that lipoproteins were co-purified with human milk EVs (Fig. 1).
MOMEV and PMOMEV are enriched with different miRNA cargo
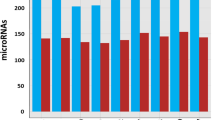
Global miRNA RNAseq was performed on MOMEV and PMOMEV enriched samples. A total of 166 miRNA were identified with 33 miRNAs of those being differentially enriched. It was found that 9 miRNAs were enriched in PMOMEV while 24 were enriched in MOMEV (Fig. 2, Suppl. Table 1). To further understand the potential impact of the differences observed between MOMEV and PMOMEV, we utilized the microRNA target filter tool using the Ingenuity Pathway Analysis software (IPA) (Table 1). Of the differentially enriched mRNAs, 25 were predicted to target 201 messenger RNAs. About fifty percent of those genes (92 mRNAs), are involved in five different pathways: AMPK signaling, NF-κB signaling, STAT3 pathway, T Cell Receptor signaling, and IL-15 production (Fig. 3). The analyses of those five pathways showed that they share 28 genes that may be affected by the miRNAs differentially enriched in MOMEV and PMOMEV.
Summary of the miRNA target filter Analysis using IPA Software. The mRNA predicted targets of the miRNA enriched in MOMEV and PMOMEV were filtered using a high confidence level. The pathways involved were filtered by cell proliferation and immune modulators. The network was built with the different effectors targeted by the 33-miRNA enriched in MOMEV and PMOMEV. Highlighted in black ovals are the 5 different pathways affected, in bold font and highlighted in purple are the 28 effectors shared by these different pathways. The pathways and targets were generated using QIAGEN Ingenuity Target Explorer (QIAGEN, Inc., https://targetexplorer.ingenuity.com/).
Specifically, the miRNAs enriched in MOMEV target 41 mRNA while the miRNAs enriched in PMOMEV target 51 genes (Suppl. Table 2). To integrate and visualize the IPA results obtained, we used Circos plot to combine in a single plot the potential pathways affected by miRNA from both MOMEV and PMOMEV (Fig. 4). As shown in the Circos plot, only has-let-7b-5p, has-miR-16-5p, has-miR-193a-5p, and has-miR-423-5p identified in PMOMEV are predicted to have an impact on these pathways while 14 miRNAs from MOMEV are predicted to impact the mRNA involved in the same pathways (Figs. 3 and 4, Table 1 and Suppl. Table 2). Based on these predictions, miRNAs enriched in PMOMEV may have a greater influence in the regulation of the transcription factor NF-κB signaling pathway miRNAs enriched in MOMEV may have a greater influence in the regulation of IL-15 signaling pathway (Fig. 4). However, both pathways are essential in the development and modulation of multiple aspects of the innate and adaptive immune system22, 23. These results suggest that miRNA differentially enriched in MOMEV and PMOMEV may affect the immune system through different but overlapping mechanisms.
EVs from MOM and PMOM stimulate IL-8 secretion in Caco2 cells
Based on the previous predictions, the impact of enriched MOMEV and PMOMEV fractions was evaluated on the nuclear factor kappa B (NF-κB) pathway by following the induction of IL-8 expression24. As controls, whole MOM and PMOM as well as exosome free supernatants were tested. A total of 24 fresh MOM samples were pooled into 3 groups and their ability to stimulate IL-8 secretion in Caco2 cells was evaluated. Differential fractionation was performed to evaluate whole milk, EV free supernatant and EV enriched fractions. It was found that while all the treatments induced secretion of IL-8 in Caco2 cells compared to the control, significantly higher secretion levels of IL-8 were observed in Caco2 cells treated with MOM 2%, supernatant and MOMEV when compared to their corresponding PMOM fractions (Fig. 5A). The results obtained with the supernatants were expected as the milk contains a variety of cytokines that may nonspecifically stimulate IL8 secretion.
Effect of human milk EVs on stimulation of the innate immune response in Caco-2 epithelial cell line. (A–B) ELISA quantification of IL-8 in Caco2 epithelial cells after 6 h of treatment with A) different milk fractions or B) with human milk enriched EV fractions in presence and absence of TNFα. Untreated Caco2 cells were used as control. (C–E) Relate expressions of AKT (C), IGF1R (D), and STAT3 (E) are shown as fold change relative to the vehicle control. The experiments were performed with biological and technical triplicates. One way ANOVA were performed, different letters indicate statistical significance at p < 0.05.
The effect of the human milk EVs was further investigated in a proinflammatory environment by adding TNFα to Caco2 cells in presence or absence of EVs. It was found that MOMEV had similar secretion levels of IL-8 as the TNFα control, while the addition of TNFα to Caco2 in presence of MOMEV was able to decrease IL-8 secretion. In contrast, PMOMEV showed trends (albeit not significant p value = 0.055) to increase IL-8 in presence of TNFα in Caco2 cells, (Fig. 5B). TNFα is a positive regulator of IL-8 gene expression through NF-κB, which is essential for IL-8 gene transcription24. These results are in agreement with predicted regulatory effect of PMOMEV miRNAs on the NF-κB signaling pathway (Fig. 4).
Another predicted signaling pathway affected by the miRNAs enriched in both MOMEV and PMOMEV is STAT3 Pathway (Figs. 3 and 4). The Signal Transducer and Activator of Transcription (STAT) 3 pathway is involved in survival, cell growth, and immune response25. This pathway is rigorously regulated by Janus Kinase (JAK) and Epidermal Growth Factor Receptor (EGFR). We identified EGFR as one of the targets regulated by hsa-miR-16-5p enriched in PMOMEV. In order to elucidate and further understand if the EVs from human milk were able to differentially modulate these pathways, we determined the mRNA expression of some key effectors such as AKT, STAT3, and IGF-1 receptor, in Caco2 cells treated with MOMEV or PMOMEV. A significant decrease in the expression of IGF-1 receptor was observed with both EVs treatments while no significant differences were observed in the expression of AKT compared to the control (Fig. 5C–D). Additionally, a 3.5-fold increase was observed in the expression of STAT3 after treating Caco2 cells with MOMEV. While the expression of STAT3 was higher in the cells treated with PMOMEV, no significant differences were observed when compared to the control (Fig. 5E). However, when STAT3 was evaluated by Western blot, no significant differences were observed between the treatments and the control (Supplementary Fig. 2). These results suggest that regulation of STAT3 transcript could be modulated in a hsa-miR-16-5p through EGFR repression in Caco2 cells treated with PMOMEV.
Differential M2 tolerogenic differentiation of THP-1 human macrophages by MOMEV and PMOMEV
The potential impact of the enrichment of has-let-7b-5p in PMOMEV was evaluated on the mRNA expression levels of IL1026 using the THP-1 PMA-activated macrophage cell line. Macrophages are broadly located in the human body playing a crucial role in regulating immune responses27. Once activated, macrophages can differentiate into two sub-types M1-like macrophages capable of proinflammatory responses and M2-like macrophages capable of tolerogenic responses28. To this end, THP-1 monocytes induced with PMA were treated with MOMEV and PMOMEV at 1010 particles/well for 6 h. The mRNA levels of IL-10, TNFα and IL-1β were quantified as signature markers of tolerogenic THP-1 (M2) macrophages. It was found that both MOMEV and PMOMEV induced a M2 phenotype when compared to the vehicle control. Nonetheless, THP-1 macrophages treated with MOMEV had significantly higher levels of IL-10 expression when compared to PMOMEV (Fig. 6A–C). These findings are in agreement with the higher abundance of has-let-7b-5p in PMOMEV.
Human milk EVs stimulate a M2-tolerogenic phenotype in THP-1 macrophages. The relative expression of IL-10 (A), TNFα (B) and IL1β (C) are shown relative to the vehicle control. The experiments were performed with biological and technical triplicates. ANOVA and mean comparisons were performed, different letters indicate statistical significance for p < 0.05.
As lipoproteins were observed with MOMEV and PMOMEV, their contribution to the M2b phenotype observed was investigated. LDL (Low-density lipoprotein) and HDL (High-density lipoprotein) lipoproteins from MOMEV and PMOMEV enriched fractions were extracted and tested at equivalent concentrations to those found in the EVs. The addition of MOMHDL or PMOMHDL significantly induced expression of TNF-α, IL1β and IL-10 when compared to the vehicle control (Fig. 6A–C). However, the induction levels were significantly lower than those compared to the MOMEV or PMOMEV. It was found that the addition of MOMLDL or PMOMLDL resulted in a similar response as the vehicle control for all the genes tested. These results indicate that lipoproteins found in the EV enriched fractions partially contribute to the tolerogenic M2b phenotype.
Next, we investigated the impact of milk EVs on the IL-15 production pathway (Figs. 3 and 4). IL-15 is a master regulator playing a crucial role in inflammatory and protective immune response29, 30. IL-15 has pivotal role as a signal molecule shared by several pathways like JAK/STAT pathway, Ras/MEK/MAPK pathway, PI3K/Akt/mTOR pathway and NF-κB pathway31. Interestingly, significant expression levels of IL-15 mRNA were observed in THP-1 macrophages treated with MOMEV but not with PMOMEV (Fig. 7 A) or with purified lipoproteins (Suppl. Fig. 4). These results are in agreement with the predicted regulatory role of has-let-7b-5p and has-miR-16-5p, enriched in PMOMEV, on the NF-κB signaling pathway.
Human milk EVs stimulate gene expression of RNA sensing signaling in THP-1 macrophages. The relative expressions of IL-15 (A), IL29 (B), INFα (C), RIG1 (D) and OAS2 (E) are shown as fold change relative to the vehicle control. The experiments were performed with biological and technical triplicates. Single way ANOVA and mean comparisons were performed, different letters indicate statistical significance at p < 0.05.
IL-15 in humans is further regulated by IFN-γ (also known as IL-29) which is part of the IL-10-family of cytokines. This type III interferon exhibits similar type I interferon-like RNA sensing properties resulting in activation of the JAK/STAT pathway32, 33. Additional inducers of interferon are the sensing of RNA by retinoic acid-inducible gene I (RIG-I)34 and 2’-5’-oligoadenylate synthetase 2 (OAS2). Therefore, to further evaluate the impact of differential miRNA cargo in human milk EVs, in RNA sensing pathways and interferons, the mRNA levels of IL29, OAS2, RIG-I and INF-α were determined. Although both treatments, MOMEV and PMOMEV, induced higher expression of these genes when compared to the control, significant higher levels of OAS2, INF-α and IL29 were observed in THP-1 macrophages treated with MOMEV compared to PMOMEV for p < 0.05 (Fig. 7). Altogether, these results indicate that the miRNA cargo of human milk EVs could be compromised after the pasteurization process decreasing or changing the regulatory effect exerted by human milk miRNA.
MOMEV differentially affects AMP, IKBα and STAT3 signaling pathways
The impact of MOMEV and PMOMEV on AMPK signaling, NF-κB signaling (by following the protein basal level and activation of the IkB kinase), and STAT3 pathways, was confirmed by evaluating the protein levels in THP-1 macrophages. It was found that the basal level abundancy of ERK and IkBα were significantly higher in MOMEV when compared to the control (Fig. 8). STAT3 showed a similar trend albeit not statistically significant. PMOMEV showed significantly lower concentrations of STAT3 and ERK when compared to MOMEV while IkBα showed a similar trend albeit not statistically significant. The quantification of the activating phosphorylation’s in ERK (P-Thr202/Tyr204 ERK/ERK) showed a significant decrease in PMOMEV when compared to MOMEV. No significant differences were observed for P-S727 STAT3/STAT3 between MOMEV, PMOMEV and the vehicle control (Fig. 8). These results are in agreement with the stronger stimulation of innate immune responses observed with MOMEV.
Effect of MOMEV and PMOMEV on AMP, IKBα and STAT3 signaling pathways. (A) Summary panel of the Western blots performed after incubation of THP-1 macrophages in presence or absence of MOMEV and PMOMEV. The results of biological triplicates are shown. (B–F) Quantification of the results obtained with (B) total IkBα, (C) total STAT3, (D) STAT3 phosphorylation S-727, (E) total ERK, and (F) ERK phosphorylation Thr202/Tyr204. β-actin was used as a loading control. Quantification of band intensity was performed with ImageJ. One way ANOVA were performed, different letters indicate statistical significance at p < 0.05.
Discussion
Many mothers that deliver preterm do not produce sufficient volumes of MOM35, and as a result, DHM is provided for these infants due to the increased benefits over formula6. However, the pasteurization process of DHM significantly reduces the availability of bioactive molecules as well as personalized live commensal bacteria9. In this report, we evaluated the impact of pasteurization on human milk on EVs integrity and miRNA cargo. We found that while similar yields and charge of EVs were obtained for MOM and PMOM EVs, a significant decrease in the protein marker CD63 was observed in PMOMEV, while CD9 was detectable in both EVs samples. Other ER markers such as calnexin were not detected on either sample while the Apolipoprotein A1 was present in both MOMEV and PMOMEV. Lipoproteins and EVs are both in the submicron scale and overlap in size distributions which translates frequently in co-purification of EVs and lipoproteins36, 37. The presence of lipoproteins could explain the comparable yield and size between MOMEV and PMOMEV samples. Recent reports in bovine milk-derived EVs evaluated the impact of several industrial processes on EV integrity. Similar to our findings, pasteurization and homogenization significantly affected the levels of CD63 while CD9 was heat stable38. Other studies have shown that the stability of exosomes is highly affected by low temperatures as well39.
Our RNAseq approach also identified significant changes to the miRNA diversity in EVs of MOM upon pasteurization. These results suggest that the decrease in protein stability of the EVs was translated into changes in the miRNA cargo in human milk. The stability and shifts in the miRNA cargo upon heat treatments procedures such as pasteurization and ultra-heat treated (UHT) milk has been studied in human and bovine milk with conflicting results38. Smyczynska et al.18 observed more severe effects of pasteurization of DHM, using Holder Pasteurization (HoP) than with High-Pressure Processing (HPP). A 302-fold decrease in the yield of exosomes was observed with (HoP), with a complete loss of RNA fragments of length typical for miRNA and piRNA. High pressure processing DHM showed less detrimental effect than pasteurization in human milk18. However, the impact of these results on host responses were not evaluated. Some studies have reported a significant effect in the abundance and stability of miRNA in bovine milk38, 40, while others have shown that pasteurization does not have a significant effect41, 42. These conflicting results may be explained by the differences in methods used for their analyses.
We determined the differential enrichment of 33 miRNA between MOMEV and PMOMEV. Moreover, the main five pathways predicted to be impacted by the differentially enriched miRNA share multiple genes resulting in overlapping effects on immune stimulation and cell proliferation. To evaluate the impact of the potential loss in integrity or decreased abundancy of specific miRNA, we used intestinal epithelial and macrophage cell lines to measure key gene markers. The miRNA enriched in MOMEV and PMOMEV target 25 genes involved in NF-κB signaling pathway. Of those, only six genes are shared among MOMEV and PMOMEV (CD40, CXCR5, PIK3R3, TGFBR1, TGFBR2 and TLR4). We hypothesized that the differential enrichment of miRNA after pasteurization would affect signaling through the NF-κB pathway. To this end, we followed the expression of IL-8 in human epithelial cells24. Stimulation of IL-8 secretion was observed with all the fractions of human milk tested (human milk, supernatant free of EVs and EVs), however, significant difference in the stimulation of IL-8 secretion was observed when fractions from PMOM were compared with their correspondent MOM fractions. Similar findings have been observed in the stimulation of IL-8 secretion after heat treatment of bovine and goat milk EVs43, 44. Likewise, a reduction in the expression of IL-6 was observed in a mice model of necrotizing enterocolitis (NEC) when comparing raw and pasteurized human milk EVs45.
Another predicted pathway showing overlapping effector genes was the MAPKs and PI3K/AKT. To further investigate the effects of MOMEV and PMOMEV in Caco2 cells, we evaluated the expression levels of 3 key genes involved in these pathways. Lower expression levels of IGF1R were observed with both MOMEV and PMOMEV treatment in Caco2 cells. However, only MOMEV showed statistically significant differences. IGF1R activation mediates signaling cascades through MAPKs and PI3K/AKT46, 47 impacting many cellular responses including cell proliferation, differentiation, and survival46, 47. We found that the posttranscriptional regulators of IGF1R, hsa-miR-16-5p and hsa-let-7b-5p, were enriched in PMOMEV, consistently with the lower levels in IGF1R mRNA observed. However, the decrease in expression did not reach statistical significance. The indirect compensatory effect of increased expression levels of STAT3 observed could explain those observations.
The beneficial effects of human milk on immune stimulation have been largely described1, 2, however little is known about the immune stimulatory properties of human milk EVs. In bovine milk, the lactation-related differential expression of miRNAs suggests that the miRNA produced in the mammary glands may have a specific function48. Since many of the enriched miRNA in human milk EVs identified in this work are predicted to impact immune modulatory pathways, we evaluated the impact of MOMEV and PMOMEV on the stimulation of a M2-like tolerogenic phenotype in macrophages. Interestingly, a stronger stimulation of IL-10 (as gene expression levels) was observed in macrophages treated with MOMEV compared to PMOMEV. These results concur with the predicted activation of the canonical NF-κB signaling pathway through has-let-7b-5p and has-miR-16-5p enrichment quantified in PMOMEV. While the tolerogenic M2-like polarization of macrophages observed in this work has been described earlier for other miRNAs17, 49, macrophage differentiation and responses could be tissue- and species-dependent. In example, inflammatory M1-like macrophage differentiation, characterized by high levels of IL-6, TNFα, IL-12/23 and decreased IL-10, was observed with bovine milk-derived EVs (BEVs) using a mice model for agricultural dust exposure50. In contrast, BEVs administration in two mouse models for arthritis reduced serum levels of MCP-1 and IL-6 correlated with delays in the onset of arthritis and diminished cartilage pathology and bone marrow inflammation51.
A remarkable finding was the induction of IL15 in macrophages treated with MOMEV. This cytokine has protective roles towards viral and bacterial infections52, 53. IL15 has been also successfully used as adjuvant in many antiviral vaccines23. The stimulation of IL15 positively correlated with higher expression levels of the interferons IL29, INFα, and the RNA sensing genes RIG1 and OSA2 in MOMEV when compared to PMOMEV. The potential role of milk miRNA in viral interference was explored through in silico methods in the context of the SARS-CoV-2 pandemic. It was found that some of the abundant miRNA in milk (miR29a, miR21 and miR181), can interfere with replication of a wide variety of viruses such as HIV, enterovirus 71 and influenza. However, scarce information is available on the mechanisms that mediate these cross-kingdom interactions11, 19. Together, these results suggest that human milk EVs may play a significant role in stimulation of RNA sensing pathways to trigger an antiviral response that may be diminished by the pasteurization process.
There is substantial evidence that milk exosomes can be absorbed and delivered to peripheral tissues where they can exert their regulatory effect (For review see Zempleni et al.)12. Therefore, the differences observed in the bioactivity of MOM after pasteurization raise concerns regarding the potential impacts of these processes on health outcomes. While the benefits of MOM have been widely reported, the benefits provided by specific bioactive components such as EVs and their cargo in human milk is unknown. It has been shown that physiological concentrations of bovine milk miRNA can affect gene expression in vivo and in cell cultures (Peripheral Blood Mononuclear Cells, HEK-293 Kidney)54. These reports suggest that a decrease in MOM EVs concentration or in their integrity affecting their cargo bioactivity can potentially have an impact on the health of infants. The most significant difference observed in this work was a decrease in the stimulation of IL15 as well as RNA sensing genes by PMOMEV. The reduced stimulation observed in innate immune responses maybe translated into a decreased response to viral infections. These findings, combined with the reported decreases in numerous proteins (lactoferrin, lysozyme)55, immunoglobulin, and cytokines (like IL-7)56 reported by others9, 57, may significantly impact the overall immune bust usually provided by MOM.
The results presented here highlight the need to optimize processes in human milk banks in order to preserve the potential bioactivity of all components in human milk while maximizing its biosafety. There is no direct substitute for the nutritional and immune benefits that MOM provides, but its proven benefits highlight the need for attempting to replicate these factors in alternative feedings as closely as possible. In this work, we identified mechanistic effectors that may be utilized as biomarkers for process optimization. The decreased immunomodulatory activity of pasteurized milk and its derived EVs observed needs to be addressed in future studies, in order to establish better processing strategies and to provide our VLBW infants with the best and personalized nutrition possible.
Material and methods
Milk collection and EV enrichment
For the RNAseq extractions, samples were collected using a sterile Symphony® double breast pump kit at a single expression session with an electric hospital Symphony® breast pump (Medela, McHenry, IL). The protocols used in this study for sample collection were reviewed and approved through the University of Florida Institutional Review Board (IRB RB201400527), a written informed consent was obtained from each donor mother. All methods here were performed in accordance with the relevant guidelines and regulations of the University of Florida Institutional Review Board. Beyond standard hand washing and pumping per NICU protocol, no breast hygiene preparation was performed. The sample was assigned a de-identified subject number, then immediately transported on ice to the microbiology lab for further processing. For the functional analyses using human cell lines, 24 milk samples we obtained from the NICU. Sets of 8 samples (total of 3 sets) were pooled. For both experiments each pool of milk was divided into two fractions, one fraction was immediately processed (MOM) for EV enrichment (MOMEV), while the other half was pasteurized to mimic DHM (P-MOM) to obtain PMOMEV, following the HMBANA protocol (HMBANA 2020). Briefly, milk was heated at constant temperature 65 °C for 30 min and cooled down immediately after in an ice bath. After pasteurization milk was further processed for EV enrichment as follows. MOM and PMOM fractions were centrifuged at 2246 g for 15 min at 4 °C to remove the fat, following a 45 min centrifugation at 15900 g to eliminate cell debris. Milk supernatants were further filtered sequentially using nitrocellulose filters of 11 um, 6 um, 2.5 um, 0.45 um and 0.2 um. After filtration, EVs were enriched by ultracentrifugation at 207,888 g for 2 h. The EVs were washed twice with PBS, quantified, and stored in aliquots at − 80 °C.
EV physical characterization and quantification
Nanoparticle tracking analysis (NTA) using a NanoSight NS300 (Malvern Instruments Ltd, Malvern, UK) was utilized to quantify the EVs as well as to determine its size distribution. Videos were recorded for 60 s (five times), with the camera level at 15, and analyzed with NTA software 4.3 (Malvern instruments Ltd, Malvern, UK). An average yield of 1012 exosomes per uL was obtained. Dynamic light scattering was performed to measure the zeta potential of the EV suspensions using a Zetasizer ultra particle analyzer (Malvern Instruments Ltd, Malvern, UK). The samples were diluted 1:1000 with distilled water. The measurements were conducted in biological and technical triplicates at 25 °C.
RNA extraction, sequencing, and data analysis
RNA extractions were performed from three pools of MOMEV and PMOMEV samples using mirVana™ miRNA Isolation Kit (Invitrogen), with small RNA enrichment from total RNA according to manufacturer’s instructions. Library construction with fragments around 20–75 bp in length and sequencing using Ion Torrent sequencing platform was performed by PrimBio (PrimBio Research Institute LLC, Exton, PA). FastQC was used to filter high quality reads58. For the data analysis Kallisto v0.46.159 was used to create an index and map the high-quality raw reads using a reference transcriptome for no coding RNA through EnsDb.Hsapiens.v86 2.99.0 package in RStudio60, 61. Differential abundance analysis of miRNA was performed using limma 3.52.4, edgeR 3.38.4 and SVA 3.44.0 packages in RStudio62,63,64. All the graphics were generated using ggplot2 3.3.6 R package65.
Protein extractions and Western Blot
Aliquots normalized to the same particle concentration of MOMEV and PMOMEV were used for protein extraction and quantification using the Pierce™ BCA Protein Assay Kit (Thermo Scientific, Rockford, IL). Briefly, total proteins were extracted from EVs using Radio Immunoprecipitation Assay Buffer (RIPA) containing 150 mM NaCl, 50 mM Tris (pH 8), 1% Triton X-100, and 0.1% sodium dodecyl sulfate (SDS), with Halt™ protease inhibitor cocktail (Thermo Fisher, Waltham, MA, USA). The EVs homogenates were centrifuged at 12,000 g for 10 min, at 4 °C and the protein concentration was measured following the manufacturer instruction.
Lipoproteins HDL (High-Density lipoprotein) and LDL (Low-Density lipoprotein) were purified from human milk EVs suspensions using LDL/VLDL and HDL purification kit (STA-608 ultracentrifugation free) following manufacturer protocol (Cell Biolabs, INC). Lipoproteins were solubilized in equal volume of PBS as EVs suspensions.
Human milk EVs were analyzed for exosome surface proteins by Western blot. Anti-CD63 (ab134045) and anti-CD9 (ab263019) were used as positive control. To investigate the presence of endoplasmic reticulum proteins (ER) or lipoproteins co-enriched in the EV preparations, anti-Calnexin (ab133615) and anti-Apolipoprotein A I (ab227455) was used, respectively. Appropriate secondary antibodies were used, and detection conducted using enhanced chemiluminescence reagent (Genesee Scientific, San Diego, CA).
Cell lines propagation, treatment and western Blot
Human Intestinal Caco-2 and monocyte THP-1 cell lines were obtained from ATCC (Gaithersburg, MD, USA). Caco-2 cells were cultured at 37 °C in Eagle’s minimum essential medium (EMEM), supplemented with 15% heat inactivated fetal bovine serum (FBS) (Invitrogen), 2% of penicillin and streptomycin solution containing 10,000 units of penicillin and 10 mg of streptomycin/ml (Sigma-Aldrich, Saint Louis, MO) in a humidified atmosphere (5% CO2 and 95% air). For the experiments, 1 × 106 Caco-2 cells per well were seeded in 6-well plates. The cells were treated with vehicle control (10 uL of EMEM media), 2% of defatted MOM (v/v) in EMEM, 2% defatted PMOM, 2% MOM supernatant (EVs free), 2% PMOM supernatant (exosome free), 1010 MOMEV in EMEM, or 1010 PMOMEV. After 24 h of incubation, culture supernatants were collected for IL-8 (BD OptEIA™) by enzyme-linked immunosorbent assay (ELISA) and the cells collected for RNA extractions and/or protein extractions. THP-1 Cells were cultured in RPMI 1640 medium supplemented with 10,000 units of penicillin, 10 mg of streptomycin, and 10% heat inactivated FBS. For the experiments, THP-1 cells were seeded in 6-well plates at 1 × 106 cells/well and activated by adding 100 nM phorbol 12-myristate-13-acetate (PMA), for 48 h at 37 °C. The cells were treated with 1010/mL of MOMEV or PMOMEV for 6 h. Supernatants and cells were processed as indicated above. For protein extractions, RIPA buffer containing 150 mM NaCl, 50 mM Tris (pH 8), 1% Triton X-100, and 0.1% sodium dodecyl sulfate (SDS), with Halt™ protease inhibitor cocktail (Thermo Fisher, Waltham, MA, USA) was used. The cell homogenates were centrifuged at 12,000 g for 10 min, at 4 °C and the protein concentration was measured using Pierce™ BCA Protein Assay Kit (Thermo Fisher Scientific, Waltham, USA). For western blots, primary antibodies against AKT (#9272), pAKT-T308 (#13038), pAKT-S473 (#9271), STAT3 (#4904), pSTAT3-S727 (#94994), p44/42 MAPK (Erk ½ #4695), pp44/42 MAPK (Erk1/2)-Thr202/Tyr204 (#9101), IGF-1R (#9750S) from Cell Signaling Technology, and IkB-alpha (MAB4299 R&D SYTEMS) were used. Anti-GAPDH (ab9485 abcam) and anti-βactin (#8457 Cell Signaling Technology) were used as loading controls. All the experiments were performed with biological and technical triplicates. Data was analyzed using ANOVA, and Tukey’s ‘Honest Significant Difference’ method was used to assign statistical significance for a p < 0.05. All the graphics and statistical analysis were generated using RStudio61.
qRT-PCR and mRNA expression
RNA was isolated from cell lines using RNeasy® Mini Kit following the manufacturer’s protocol (QIAGEN, Germantown, MD). DNA was removed by treatment with DNase (QIAGEN, Germantown, MD) according to the manufacturer’s protocol. RNA quality was monitored on 1% agarose gels, and RNA quantification was performed using Thermo Scientific Nanodrop One Microvolume UV–vis spectrophotometer (Thermo Fisher Scientific, Grand Island, NY). qRT-PCR was performed as described66. Primer sequences used to determine relative transcript abundance are listed in Suppl. Table 3.
Data availability
miRNA raw data generated in this study was deposited in the NCBI Sequence Read Archive (SRA) under BioProject ID PRJNA930463. GEO accession GSE225840 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE225840. Token yxwhykosdbgjxyp.
References
Mueller, N. T., Bakacs, E., Combellick, J., Grigoryan, Z. & Dominguez-Bello, M. G. The infant microbiome development: Mom matters. Trends Mol. Med. 21, 109–117. https://doi.org/10.1016/j.molmed.2014.12.002 (2015).
Hanson, L. Å., Korotkova, M. & Telemo, E. Breast-feeding, infant formulas, and the immune system. In Annals of Allergy, Asthma and Immunology Vol. 90 59–63 (American College of Allergy, Asthma and Immunology, 2003).
Hunt, K. M. et al. Characterization of the diversity and temporal stability of bacterial communities in human milk. PLoS ONE 6, e21313 (2011).
Cacho, N. T. et al. Personalization of the microbiota of donor human milk with mother’s own milk. Front. Microbiol. 8, 1470 (2017).
Mosca, F. & Giannì, M. L. Human milk: Composition and health benefits. Pediatr. Med. Chir. 39, 47–52 (2017).
Abrams, S. A., Landers, S., Noble, L. M. & Poindexter, B. B. Donor human milk for the high- risk infant: Preparation, safety, and usage options in the United States. Pediatrics 139, e20163440 (2017).
Radmacher, P. G. & Adamkin, D. H. Fortification of human milk for preterm infants. In Seminars in Fetal and Neonatal Medicine Vol. 22 30–35 (Elsevier, 2017). https://doi.org/10.1016/j.siny.2016.08.004.
Chang, J. C. et al. Influence of prolonged storage process, pasteurization, and heat treatment on biologically-active human milk proteins. Pediatr. Neonatol. 54, 360–366 (2013).
Torrez Lamberti, M. F. et al. Metabolomic profile of personalized donor human milk. Molecules 25, 5783 (2020).
Alsaweed, M., Lai, C. T., Hartmann, P. E., Geddes, D. T. & Kakulas, F. Human milk miRNAs primarily originate from the mammary gland resulting in unique miRNA profiles of fractionated milk. Sci. Rep. 6, 1–3 (2016).
Hatmal, M. M. et al. Immunomodulatory properties of human breast milk: MicroRNA contents and potential epigenetic effects. Biomedicines 10, 1219. https://doi.org/10.3390/biomedicines (2022).
Zempleni, J. et al. Biological activities of extracellular vesicles and their cargos from bovine and human milk in humans and implications for infants. J. Nutr. 147, 3–10 (2017).
van Bergenhenegouwen, J. et al. Extracellular vesicles modulate host-microbe responses by altering TLR2 activity and phagocytosis. PLoS ONE 9, e89121 (2014).
Hashimoto, Y., Akiyama, Y. & Yuasa, Y. Multiple-to-multiple relationships between microRNAs and target genes in gastric cancer. PLoS ONE 8, e62589 (2013).
Kim, V. N., Han, J. & Siomi, M. C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 10, 126–139. https://doi.org/10.1038/nrm2632 (2009).
Melnik, B. C. The pathogenic role of persistent milk signaling in mTORC1-and milk-microRNA-driven type 2 diabetes mellitus. Curr. Diabetes Rev. 11, 46–62 (2015).
Kosaka, N., Izumi, H., Sekine, K. & Ochiya, T. MicroRNA as a new immune-regulatory agent in breast milk. Silence 1, 1–8 (2010).
Smyczynska, U. et al. Impact of processing method on donated human breast milk microRNA content. PLoS ONE 15, e0236126 (2020).
Leroux, C., Chervet, M. L. & German, J. B. Perspective: Milk microRNAs as important players in infant physiology and development. Adv. Nutr. 12, 1625–1635. https://doi.org/10.1093/advances/nmab059 (2021).
Cortez, J. et al. Maternal milk feedings reduce sepsis, necrotizing enterocolitis and improve outcomes of premature infants. J. Perinatol. 38, 71–74 (2018).
Goulet, O. Potential role of the intestinal microbiota in programming health and disease. Nutr. Rev. 73, 32–40 (2015).
Liu, T., Zhang, L., Joo, D. & Sun, S. C. NF-κB signaling in inflammation. Signal Transduct. Target. Ther. 2, 1–9. https://doi.org/10.1038/sigtrans.2017.23 (2017).
Perera, P. Y., Lichy, J. H., Waldmann, T. A. & Perera, L. P. The role of interleukin-15 in inflammation and immune responses to infection: Implications for its therapeutic use. Microbes Infect. 14, 247–261. https://doi.org/10.1016/j.micinf.2011.10.006 (2012).
Matsushima, K., Yang, D. & Oppenheim, J. J. Interleukin-8: An evolving chemokine. Cytokine 153, 155828. https://doi.org/10.1016/j.cyto.2022.155828 (2022).
Hirano, T., Ishihara, K. & Hibi, M. Roles of STAT3 in mediating the cell growth, differentiation and survival signals relayed through the IL-6 family of cytokine receptors. www.nature.com/onc.
Aday, S. et al. Bioinspired artificial exosomes based on lipid nanoparticles carrying let-7b-5p promote angiogenesis in vitro and in vivo. Mol. Ther. 29, 2239–2252 (2021).
Davies, L. C., Jenkins, S. J., Allen, J. E. & Taylor, P. R. Tissue-resident macrophages. Nat. Immunol. 14, 986–995. https://doi.org/10.1038/ni.2705 (2013).
Yunna, C., Mengru, H., Lei, W. & Weidong, C. Macrophage M1/M2 polarization. Eur. J. Pharmacol. 877, 173090. https://doi.org/10.1016/j.ejphar.2020.173090 (2020).
Waldmann, T. A. & Tagaya, Y. The multifaceted regulation of interleukin-15 expression and the role of this cytokine in NK cell differentiation and host response to intracellular pathogens. Annu. Rev. Immunol 17, 19–49 (1999).
Budagian, V., Bulanova, E., Paus, R. & Bulfone-Paus, S. IL-15/IL-15 receptor biology: A guided tour through an expanding universe. Cytokine Growth Factor Rev. 17, 259–280. https://doi.org/10.1016/j.cytogfr.2006.05.001 (2006).
Patidar, M., Yadav, N. & Dalai, S. K. Interleukin 15: A key cytokine for immunotherapy. Cytokine Growth Factor Rev. 31, 49–59. https://doi.org/10.1016/j.cytogfr.2016.06.001 (2016).
Chesler, D. A. & Reiss, C. S. The role of IFN-in immune responses to viral infections of the central nervous system. Cytokine Growth Factor Rev. 13, 441–454 (2002).
Witte, K., Witte, E., Sabat, R. & Wolk, K. IL-28A, IL-28B, and IL-29: Promising cytokines with type I interferon-like properties. Cytokine Growth Factor Rev. 21, 237–251. https://doi.org/10.1016/j.cytogfr.2010.04.002 (2010).
Tatematsu, M., Funami, K., Seya, T. & Matsumoto, M. Extracellular RNA sensing by pattern recognition receptors. J. Innate Immun. 10, 398–406. https://doi.org/10.1159/000494034 (2018).
Asztalos, E. V. Supporting mothers of very preterm infants and breast milk production: A review of the role of galactogogues. Nutrients 10, 600. https://doi.org/10.3390/nu10050600 (2018).
Taylor, D. D. & Shah, S. Methods of isolating extracellular vesicles impact down-stream analyses of their cargoes. Methods 87, 3–10. https://doi.org/10.1016/j.ymeth.2015.02.019 (2015).
Wu, M. et al. Separating extracellular vesicles and lipoproteins via acoustofluidics. Lab Chip 19, 1174–1182 (2019).
Kleinjan, M. et al. Regular industrial processing of bovine milk impacts the integrity and molecular composition of extracellular vesicles. J. Nutr. 151, 1416–1425 (2021).
Zhang, Y. et al. Exosome: A review of its classification, isolation techniques, storage, diagnostic and targeted therapy applications. Int. J. Nanomed. 15, 6917–6934. https://doi.org/10.2147/IJN.S264498 (2020).
Golan-Gerstl, R. et al. Characterization and biological function of milk-derived miRNAs. Mol. Nutr. Food Res. 61, 1700009 (2017).
Melnik, B. C. & Schmitz, G. Exosomes of pasteurized milk: Potential pathogens of Western diseases 06 Biological Sciences 0601 Biochemistry and Cell Biology. J. Transl. Med. 17, 1–33 (2019).
Kirchner, B., Pfaffl, M. W., Dumpler, J., von Mutius, E. & Ege, M. J. MicroRNA in native and processed cow’s milk and its implication for the farm milk effect on asthma. J. Allergy Clin. Immunol. 137, 1893-1895.e13 (2016).
Benmoussa, A. et al. Concentrates of two subsets of extracellular vesicles from cow’s milk modulate symptoms and inflammation in experimental colitis. Sci. Rep. 9, 1–16 (2019).
Mecocci, S. et al. In vitro evaluation of immunomodulatory activities of goat milk Extracellular Vesicles (mEVs) in a model of gut inflammation. Res. Vet. Sci. 152, 546–556 (2022).
Miyake, H. et al. Human breast milk exosomes attenuate intestinal damage. Pediatr. Surg. Int. 36, 155–163 (2020).
Siddle, K. Signaling by insulin and IGF receptors: Supporting acts and new players. J. Mol. Endocrinol. 47, R1. https://doi.org/10.1530/JME-11-0022 (2011).
Riedemann, J. & Macaulay, V. M. IGF1R signaling and its inhibition. Endocr. Relat. Cancer 13, S33 (2006).
Li, Z., Liu, H., Jin, X., Lo, L. & Liu, J. Expression profiles of microRNAs from lactating and non-lactating bovine mammary glands and identification of miRNA related to lactation. BMC Genomics 13, 1–15 (2012).
Alsaweed, M., Hartmann, P. E., Geddes, D. T. & Kakulas, F. Micrornas in breastmilk and the lactating breast: Potential immunoprotectors and developmental regulators for the infant and the mother. Int. J. Environ. Res. Public Health 12, 13981–14020 (2015).
Nordgren, T. M. et al. Bovine milk-derived extracellular vesicles enhance inflammation and promote M1 polarization following agricultural dust exposure in mice. J. Nutr. Biochem. 64, 110–120 (2019).
Arntz, O. J. et al. Oral administration of bovine milk derived extracellular vesicles attenuates arthritis in two mouse models. Mol. Nutr. Food Res. 59, 1701–1712 (2015).
Schluns, K. S. & Lefrançois, L. Cytokine control of memory T-cell development and survival. Nat. Rev. Immunol. 3, 269–279. https://doi.org/10.1038/nri1052 (2003).
Yajima, T. et al. Memory phenotype CD8+ T cells in IL-15 transgenic mice are involved in early protection against a primary infection with Listeria monocytogenes. Eur. J. Immunol. 31, 757–766 (2001).
Baier, S. R. et al. MicroRNAs are absorbed in biologically meaningful amounts from nutritionally relevant doses of cow milk and affect gene expression in peripheral blood mononuclear cells, HEK-293 kidney cell cultures, and mouse livers. J. Nutr. 144(10), 1495–1500. https://doi.org/10.3945/jn.114.196436 (2014).
Paulaviciene, I. J. et al. The effect of prolonged freezing and holder pasteurization on the macronutrient and bioactive protein compositions of human milk. Breastfeed. Med. 15(9), 583–588. https://doi.org/10.1089/bfm.2020.0219 (2020).
Espinosa-Martos, I. et al. Bacteriological, biochemical, and immunological modifications in human colostrum after Holder pasteurization. J. Pediatr. Gastroenterol. Nutr. 56(5), 560–568. https://doi.org/10.1097/MPG.0b013e31828393ed (2013).
O’Connor, D. L. et al. Human milk pasteurization: Benefits and risks. Curr. Opin. Clin. Nutr. Metab. Care 18(3), 269–275. https://doi.org/10.1097/MCO.0000000000000160 (2015).
Andrews, S. FASTQC. A quality control tool for high throughput sequence data (2010).
Bray, N. L., Pimentel, H., Melsted, P. & Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 (2016).
Rainer, J. EnsDb.Hsapiens.v86: Ensembl based annotation package. R package version 2.99.0 (2017).
R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/ (2018).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2009).
Smyth, G. K. et al. limma: Linear Models for Microarray and RNA-Seq Data User’s Guide.
Leek, J. T. et al. Package ‘sva’ Title Surrogate Variable Analysis. https://git.bioconductor.org/packages/sva (2022).
Wickham, H. ggplot2 (Springer, New York, 2009).
Teixeira, L. D. et al. Lactobacillus johnsonii N6.2 and blueberry phytophenols affect lipidome and gut microbiota composition of rats under high-fat diet. Front. Nutr. 8, 757256 (2021).
Acknowledgements
We are grateful for all the mothers who generously provided milk samples for the study and our dedicated neonatal nurses for taking excellent care of our patients. We acknowledge Sharon Perry for her technical assistance. We would also like to acknowledge Alexandra Cuaycal and Reagan Beliakoff for the critical reading of the manuscript.
Funding
Research reported in this publication was supported by the National Institutes of Health (NIH 1R01NR016964). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author information
Authors and Affiliations
Contributions
G.L.L., L.A.P. and M.F.T.L. conceptualized the study. M.F.T.L. and C.F.G. contributed to the sample processing, performed the experiments and data analyses. M.F.T.L. wrote the original draft. G.L.L., C.F.G. and L.A.P. reviewed and edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Torrez Lamberti, M.F., Parker, L.A., Gonzalez, C.F. et al. Pasteurization of human milk affects the miRNA cargo of EVs decreasing its immunomodulatory activity. Sci Rep 13, 10057 (2023). https://doi.org/10.1038/s41598-023-37310-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-37310-x
- Springer Nature Limited