Abstract
Timely and effective diagnosis of fungal keratitis (FK) is necessary for suitable treatment and avoiding irreversible vision loss for patients. In vivo confocal microscopy (IVCM) has been widely adopted to guide the FK diagnosis. We present a deep learning framework for diagnosing fungal keratitis using IVCM images to assist ophthalmologists. Inspired by the real diagnostic process, our method employs a two-stage deep architecture for diagnostic predictions based on both image-level and sequence-level information. To the best of our knowledge, we collected the largest dataset with 96,632 IVCM images in total with expert labeling to train and evaluate our method. The specificity and sensitivity of our method in diagnosing FK on the unseen test set achieved 96.65% and 97.57%, comparable or better than experienced ophthalmologists. The network can provide image-level, sequence-level and patient-level diagnostic suggestions to physicians. The results show great promise for assisting ophthalmologists in FK diagnosis.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Introduction
Fungal keratitis (FK) is a serious ocular infection occurring in the cornea, which has been known as one of the leading causes of visual impairment1,2. It is gaining increasing attention around the world, especially in developing countries due to a higher incidence rate3,4,5,6,7,8. FK often occurs due to corneal trauma and wearing contact lens9,10,11, and can cause serious complications, such as corneal perforation and endophthalmitis. However, their clinical features are not distinctive enough, and as a result fungal keratitis can be misdiagnosed as bacterial or parasitic keratitis1. Therefore, early diagnosis is critical for instituting timely and proper treatment to improve the curative effect and the prognosis of patients, reducing the risk of irreversible vision loss.
The traditional keratitis diagnostic methods include corneal scraping and fungal culture. Corneal scraping brings pain to patients and increases the risk of secondary injury in the cornea. Moreover, fungal culture takes a long time and has a relatively low sensitivity, especially for infections in the deep corneal stroma3,12. Shotgun metagenomics is a new DNA sequencing method to identify complete taxonomical and functional profile of an organism from little volume of the sample. However, it’s sensitivity, reference standards for downstream analysis, costs, turnover time are limitations for routine clinical practice13,14,15. In contrast, the use of in vivo confocal microscopy (IVCM) enables non-invasive and prompt eye examinations16. Ophthalmologists can check for corneal conditions almost at any depth by IVCM and make a diagnosis for fungal keratitis based on observed fungal hyphae in the IVCM images. However, the nerve fibers, vessels, and dendritic cells could confuse ophthalmologists since some filiform texture regions have similar appearance features to fungal hyphae. Ophthalmologists need to gather extensive clinical experience to effectively distinguish fungi from confounding objects. Considering the high prevalence rate of fungal keratitis in many countries, the number of qualified ophthalmologists is not sufficient to provide care for the large population, leading to delayed treatment and management for some patients. That may cause irreversible damage to their cornea and bring a high risk to public health. In this work, we aim to provide an automated FK detection system working on IVCM images to strengthen the capability of ophthalmologists in the diagnosis of fungal keratitis.
As one of the top breakthroughs in recent years, deep neural networks have greatly benefited the field of medical image analysis and have been applied in a variety of medical imaging modalities, including X-ray images, retina images, Magnetic Resonance Imaging (MRI), and Computerized Tomography (CT)17,18,19,20. They have shown dominant performance in automatic disease detection and lesion region segmentation21,22 due to their inherent capability of learning complex features directly from raw image data. In the last decade, convolutional neural networks including ResNet-like frameworks18,23 have shown great power in extracting spatial features of medical images and yielded impressive results. With the advances of attention mechanisms including become another popular deep networks for image analysis tasks, such as classification, segmentation, and object detection24. Prior works on diagnosis of fungal keratitis using IVCM images have employed traditional image recognition methods25 and deep convolutional neural networks26,27 to detect fungi using visual features. However, those methods are often hampered by the lack of large-scale FK IVCM image datasets and the limited capability of their learning models. Although these methods have shown promising performance in their FK diagnosis experiments, their generalizability is yet to be further validated.
Our research is motivated by the real clinical process. The main aim of the IVCM image-based FK diagnosis is to identify fungal hyphae structures and to distinguish them from other structures in the cornea, such as nerve fibers and vessels. We observe that the diagnosis of FK in clinical practice does not only rely on a single IVCM image. During the real clinical diagnosis, experienced ophthalmologists look carefully at a set of IVCM images of the same patient and give the final decision based on the observed spatial structure of hyphae. It is a decision based on the combination of all the visual feature observations of a group of images of the patient. In this work, we propose to explore the relationship among multiple IVCM images of the same patient captured in sequence for automated FK diagnosis. Such images tend to be spatially neighboring and cover related regions, and we develop a new deep architecture based on transformer modules with a higher capability of extracting spatially correlative features.
In this study, we present and validate our deep learning framework for automated fungal keratitis diagnosis, which contains two stages. In stage 1, we train a deep neural network with a single IVCM image as its input, which is able to detect fungal keratitis at the image level. We utilize recent transformer-based modules28 to effectively extract the filiform texture features and identify the images with hyphae structures. In stage 2, we train a multi-instance deep network that takes a set of neighboring IVCM images belonging to the same patient as input and predicts a diagnostic conclusion for the image set. Since datasets used in previous work are either unavailable or too small, we built a new large-scale dataset suitable for our two-stage training. And we also collected images from separate patients for validation and testing, to allow evaluation at image, sequence and patient levels.
Results
Performance of the first stage network
We evaluate the image-level diagnostic performance of the first stage network using the stage 1 test dataset from FK-IMG (Fungal Keratitis Image Dataset, detailed description in Page. 7) that contains 8,568 images, including 3,815 positive images and 5,383 negative images. To find the best backbone for effectively extracting image features, we compared several image classification networks, including ResNet18, ResNet34, PoolFormer, and SwinTransformer. The classification performances of these backbone networks are reported in Table 1, where we compare them using specificity, sensitivity, accuracy, and AUC (Area Under the Curve) scores, based on 95% confidence intervals. SwinTransformer achieves the overall best performance, with the highest sensitivity, accuracy and AUC score, and the second best specificity, just after PoolFormer, so we chose SwinTransformer as our backbone model.
Performance of the second stage network
To evaluate the performance of the second stage network on image sequences, we first compare its performance against a naive method based on the prediction results of single images by the stage 1 network, where a sequence will be labeled as positive if at least one of its images is identified as positive. The stage 2 test dataset for evaluation contains images of 20 positive patients and 17 negative patients from FK-SEQ (Fungal Keratitis Image-Sequence Dataset, detailed description in Page. 7)). We use the aforementioned index-based strategy to select the neighboring images to build the sequence dataset. Here, we use Seq.k to denote the dataset with an image sequence length of k in the following evaluation. We compared performance under different lengths of image sequences, where the Seq.5 test dataset contains 2,411 negative groups and 4,508 positive groups, the Seq.7 test dataset contains 2,330 negative groups and 4,981 positive groups and the Seq.9 test dataset contains 2,257 negative groups and 5,361 positive groups, more test datasets with different sequence lengths are shown in Table 2.
Take image sequences of length 7 (Seq.7) as an example. As shown in Table 3, the baseline using SwinTransformer as the first stage backbone achieves overall highest performance, better than baselines with alternative backbones in stage 1 network for the sensitivity of 95.34% (94.72–95.91%), accuracy of 94.42% (93.87–94.93%) and AUC score of 0.9864 (0.9845–0.9883), and the baseline with PoolFormer backbone achieves the highest specificity of 92.45% (91.30–93.35%) among all the baselines. When reporting performance, we show the mean and confidence intervals. Our stage 2 network better utilizes sequence-information through multi-instance learning29, and achieves clearly better performance than baselines: specificity of 96.65% (95.84–97.35%), sensitivity of 97.57% (97.10–97.98%), accuracy of 97.28% (96.88–97.64%), and AUC score of 0.9950 (0.9938–0.9962). More results in different sequence lengths are shown in Table 3.
Performance of patient level diagnosis
As previously explained, we further extend the prediction from the sequence level to the patient level based on the stage 2 results. And we evaluate the diagnostic performance of our method at the patient level. Since some of the positive patients in FK-SEQ take IVCM images more than once, we group the images by patient and date, as patients’ circumstances may change over time. Therefore, the patient level test dataset contains 36 entities from 20 positive patients and 17 entities from 17 negative patients. Each entity includes the IVCM images taken from a single patient in one examination. The results of patient-level diagnosis are shown in Table 4. We list the patient diagnostic results of our naive solution using the stage 1 network and the stage 2 network. For the stage 2 network, we only label the patients as positive if the number of their predicted positive images is larger than a threshold \(\sigma\). We show the results under different values of \(\sigma\). As \(\sigma\) increases, the specificity increases and the sensitivity decreases slightly. Our system can also list all the suspicious images to ophthalmologists for further examination to avoid missing positive patients as much as possible.
Comparison with human experts
We further conducted an experiment to validate the effectiveness of our method by comparing its diagnostic performance with experienced ophthalmologists. We randomly selected a subset of the Seq.7 test dataset and invited four ophthalmologists with different levels of experience to diagnose FK given the image sequences. For each patient in our Seq.7 test dataset, we randomly selected five image sequences at most and built a subset with 249 image sequences, including 179 positive and 70 negative sequences. The performances of two junior ophthalmologists, two senior ophthalmologists and our deep network are shown in Table 5.
The binary classification task usually takes the probability of 50% as threshold to separate negative cases and positive cases, which tends to achieve a balance for specificity and sensitivity. Under this setting, our network achieves a higher sensitivity and a slightly lower specificity than ophthalmologists. The precision-recall curve in Fig. 1 shows that when we increase the probability threshold until the specificity rising to 100%, the sensitivity of our network remains at 96.65%. The results show that ophthalmologists usually do not diagnose normal or other cornea infections as fungal keratitis, but even the senior ophthalmologists miss some non-typical fungal keratitis cases. Our network achieves a higher sensitivity than human experts, with the ability to bring in higher specificity while preserving sensitivity by tuning a higher threshold, showing great promise in assisting ophthalmologists for FK diagnosis.
Discussion
The proposed two-stage deep learning framework achieved high sensitivity and specificity in FK diagnosis. Although the first stage network has already shown great performance in identifying FK-related visual features to label single IVCM images, the relatively high false positive rate on single images leads to more misdiagnosis at the patient level. Instead of just formulating the diagnosis process as a single-image-based binary classification task, we employ the stage 2 network to combine the information of a group of images from the same patient for prediction, further improving the sensitivity and specificity. Our experiments show that the proposed deep learning framework generates promising results in assisting ophthalmologists for timely and effective fungal keratitis diagnosis.
All the related prior works only considered the fungal keratitis diagnosis problem in IVCM images as a binary classification task on single images. However, our experiments show that false positive instances predicted by a single image classification network are very common for negative patients without fungal keratitis. It demonstrates that there could still be some filiform textures like nerve fibers or vessels that cannot be distinguished from fungal hyphae. The relatively high false positive rate leads to a low specificity if we directly apply the single image results to the diagnosis for a patient. We have tried several state-of-the-art binary classification network architectures, and empirically no existing deep network architecture appears to be able to solve the low specificity issue. Therefore, considering the real clinical diagnosis process, making decisions based on the relationship among a group of images is needed for better diagnosis performance.
In a clinical diagnosis process, an ophthalmologist usually screens a patient’s cornea by IVCM in different locations and makes the diagnostic conclusion based on their observations of all the IVCM images. An experienced ophthalmologist usually roughly checks the cornea, locates the suspected region, and takes more images to find the lesions caused by fungal keratitis and fungal hyphae. Besides the fungal hyphae features observed in single images, they also take into account spatial information by checking nearby images to better distinguish hyphae from other filiform textures. Once the acquired information is adequate to conclude whether fungi infect this region, the ophthalmologist will move to the next suspected region for further inspection to measure the level of infection for this patient. Therefore, we consider that our stage 2 network based on multi-instance learning can better simulate a real clinical diagnosis process. Our network takes an image sequence of neighboring images as input and explores the relationships between them by an attention mechanism, which can combine the complementary information provided by different images for the same patient when learning how to make the final prediction. To the best of our knowledge, it is the first two-stage deep architecture to use image sequence information in automated fungal keratitis diagnosis. The results have shown that our second stage network increases the specificity and the sensitivity compared to the naive method based on the image-level results. It has shown great potential to assist ophthalmologists in real-world clinical practice.
In our two-stage framework, the second stage network can correct the false positive instances predicted by the first stage network to get higher specificity. We show two examples of false positive images corrected by the second stage network in Fig. 2. Figure 2a shows incorrectly predicted positive images containing filiform textures and messy regions. Figure 2b shows the generated image sequences containing the false positive images and their neighboring images, which are then fed into the second stage network. Although the false positive images may have some suspicious filiform textures, the neighboring images are normal and have no fungal hyphae features. Then the second stage network can collect the information of all the images in the sequence and label the whole sequence as negative, correcting the prediction of the first stage. In Table 3, we show the performance with different image sequence lengths. With an increasing sequence length, the sensitivity of our stage 2 network increases slightly, but the specificity declines. Our experiments show that longer sequences could not provide a significant improvement in diagnostic performance, which is similar to the real clinical process where the ophthalmologist usually takes a few images in one region and then moves the microscopy to another region.
We inspect our prediction results in comparison with human experts at the image sequence level. When using the more balanced threshold (\(P=0.5\)), in all the 249 image sequences, our deep network only misdiagnoses five positive and four negative cases. We note that the five positive cases misdiagnosed by our method are also misdiagnosed by the four ophthalmologists, which may be caused by their confusing visual features that are hard to be distinguished by both human experts and our deep network. Overall, the human experts tend to be more conservative and missed more positive cases, leading to a lower sensitivity than our method. One of the four negative cases incorrectly predicted by our deep network is also misdiagnosed by a junior ophthalmologist. Our precision-recall curve in Fig. 1 shows that the predicted probability of these misdiagnosed negative cases is still lower than that of most positive cases, so that we can get a specificity of 100% with a sensitivity of 96.65% with a high probability threshold (\(P=0.63\)). Notably, our setting of the experiment only provides image sequences to human experts, while in clinical settings, experienced ophthalmologists will gather more information (e.g. corneal images taken by a slit lamp, patients’ symptom and patients’ experiences) to make final diagnosis. Limited information from image sequences may be the reason why human experts got relative lower performance in our experiments, but the results still demonstrate that our network can assist ophthalmologists to avoid missing suspicious positive cases.
During the real clinical process, it is important to ensure that the negative patients are not misdiagnosed, as the anti-fungal medicine is expensive and toxic, which would put extra burden on the patients’ finance and health. Compared with human experts, our network is shown to be able to achieve a higher specificity while maintaining a higher sensitivity when setting a higher probability threshold. Therefore, our network shows great promise in assisting ophthalmologists.
In the diagnostic process of fungal keratitis, an ophthalmologist normally makes an overall decision after inspecting all the captured IVCM images. In the inference phase, our deep learning framework can take all the patient’s IVCM images by separating them into sequences, and provides an overall probability of fungal keratitis, with the most suspected images of the patients listed. Therefore, besides automatically producing a diagnostic decision, our method can also play an assistive role for ophthalmologists by validating the ophthalmologists’ diagnostic conclusion and generating a confidence value for a suspected case. The experiments have shown that the ophthalmologists usually get higher specificity and our network gets higher sensitivity. The ophthalmologists assisted by our network could pay more attention to those listed suspected cases and avoid missing atypical fungal keratitis as much as possible.
There are also several limitations in our work. Firstly, the second stage network takes the predicted positive images to build the image sequence in the inference phase so that the false negative images predicted in the first stage cannot be further addressed in the second stage. Future study needs to focus more on correcting false negative instances from the first stage. Secondly, due to the relatively small patient number in our dataset, it is hard to fully validate the robustness of the deep learning framework in patient level diagnosis. More external clinical data are needed for further study. Thiredly, Our method is only trained and evaluated on the image captured by “HRT III/RCM Heidelberg Engineering, Germany”. The quality and form of images captured by other devices may influence the performance of current methods. Finally, ophthalmologists know the depth of each image when examining the cornea, but that information is lost in our dataset. Since our system is not trained using that prior knowledge, we may have some misdiagnosis cases that could be potentially fixed by incorporating the depth information of IVCM images.
In conclusion, we proposed a deep learning framework for diagnosing fungal keratitis using IVCM images, which not only analyzes the features of single images, but also explores how to effectively combine visual features of a group of images to make better diagnostic decisions. Our method of leveraging a sequence of images for automatic fungal keratitis diagnosis is a more reasonable solution, which is similar to the real clinical process of making diagnostic conclusions for a patient. Our experiments also show a promising potential of our method in assisting ophthalmologists to diagnose fungal keratitis and evaluating the confidence of a diagnostic conclusion.
Methods
In this study, we aim to provide a deep learning framework to conduct fungal keratitis diagnostic tasks like human experts. Therefore, our framework is not only designed for detecting FK infections in a single image, but is also capable of making diagnostic decisions by combining the features of multiple images for a patient.
Datasets preparation
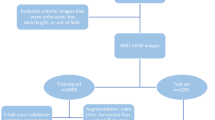
The IVCM image dataset that we used for training and validating our two-stage deep networks was collected from 2013 to 2021, which contains 96,632 IVCM images from 377 patients. Examples of positive and negative IVCM images are shown in Fig. 3. All of the IVCM images in FK-IMG and FK-SEQ datasets were captured by IVCM (HRT III/RCM Heidelberg Engineering, Germany) in Wuhan Aier Hankou Eye Hospital, Beijing Aier Intech Eye Hospital, and Chengdu Aier East Eye Hospital. Images were stored in JPEG or BMP with a resolution of \(384 \times 384\) pixels. The positive patients were diagnosed with fungal keratitis on the basis of their positive corneal scraping microscopy examination results, or positive fungal cultures. The images were each identified and labeled by two experienced ophthalmologists. The two ophthalmologists were asked to review all the images independently. If the diagnosis of the two ophthalmologists was inconsistent, the image was submitted to another experienced ophthalmologist for a final decision. Because our networks in two stages require image data and continuous image sequence data respectively, we separated our collected images to form two different datasets, named FK-IMG and FK-SEQ, to support training and evaluation at both image and sequence levels, and meet the requirements in different stages. FK-IMG is built for stage 1 network, which contains 12,228 images with positive labels from the samples of 163 patients, and each positive image has fungal hyphae that can be seen as the main features and diagnostic criteria of fungal keratitis. As the stage 1 task is performed at the image level, we require individual images to have the correct labels. Since some of the IVCM images of positive patients can still be negative, such images are excluded from the dataset to ensure image-level correctness. FK-IMG also includes 16,417 IVCM images with the negative label from 88 patients with no signs of fungal infection. FK-SEQ contains continuous image sequences taken by IVCM. There are 57,020 original IVCM images from 68 positive patients and 10,967 IVCM images from 58 negative patients. All the original images captured for each patient are included in FK-SEQ without dropping negative images. The images came from diagnosed fungal keratitis patients and were taken during clinical processes on different dates. We group the images by the date they were taken, so that each patient in FK-SEQ dataset may have more than one group of images. In FK-SEQ, there are a great number of negative images from positive patients, as the fungal hyphae usually exist only in some areas of the cornea. All the images were collected from the real clinical diagnostic process.
To properly train and evaluate deep models, we split the IVCM images of FK-IMG and FK-SEQ into training set, validation set and test set at the patient level. We use the FK-IMG dataset for the training and evaluation of stage 1. In stage 1, we randomly selected 151 patients (60%) to build the training set, including 7,946 positive images from 98 patients and 9,573 negative images from 53 negative patients. A set of images from 26 patients (10%) is randomly selected as the validation set, including 2,558 images. Another group of 74 patients (30%) is selected for the evaluation of stage 1, including 8,568 images. In stage 2, we utilize the FK-SEQ dataset for training, validation, and testing. We randomly select 35 negative patients and 41 positive patients from FK-SEQ as our training data in stage 2. For validation, we use seven positive patients and six negative patients. The images of the remaining 20 positive patients and 17 negative patients are used to build the test set. More details of our datasets and the distribution of the positive/negative samples are reported in Table 6.
Network architecture
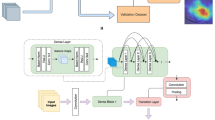
Our framework contains two stages, which learn to extract features and predict diagnostic decisions. In the first stage, we train an image-level deep neural network to extract features from a single IVCM image and detect whether fungal keratitis can be observed in that image. The second stage aims to give a comprehensive consideration by combining all the learned features from a set of IVCM images from the same patient. We train a multi-instance network to learn the relationships between IVCM images in this stage, which takes a sequence of neighboring images as input. The patient-level diagnosis pipeline is constructed by aggregating the results from the two-stage networks, which combines the image-sequence level results to obtain the final patient-level diagnostic result. We show the architecture of the 2-stage deep networks and illustrate the diagnostic process at image level, sequence level and patient level in Fig. 4.
Stage 1: image level diagnosis network
We leverage the recently developed SwinTransformer28 as the backbone of our image-level deep neural network and train it for the binary classification task. We use transfer learning in our stage 1 training, where the pretrained SwinTransformer weights in ImageNet22k30 are transferred to our backbone network as an initialization of the trainable parameters. The training dataset is denoted by \(\{{\mathscr {X}}_i, y_i\}(i \in \{1,2,\ldots ,N\})\), where \({\mathscr {X}}_i \in {\mathbb {R}}^{H \times W}\) represents the grayscale image captured by the confocal microscope and \(y_i \in \{0, 1\}\) represents the annotation indicating whether the i-th image belongs to the positive or negative group of fungal keratitis. The pipeline of our image-level diagnosis network is shown at the top of Fig. 4. The input of the network is the image \({\mathscr {X}}_i\), which is then processed by the pretrained SwinTransformer network to extract the image feature \(v_i\). The extracted feature \(v_i\) is subsequently fed into the linear classifier, which outputs the diagnostic result.
Stage 2: image sequence level diagnosis network
Considering that ophthalmologists often take a few images around the suspicious regions in the cornea during the real examination, the neighboring images captured in a sequence often contain additional fungal hyphae features. For this purpose, we take the images captured at similar times and regions by the ophthalmologists during the cornea examination. When captured images are recorded sequentially, such images can be easily located by taking the nearest images in the captured sequence, e.g. based on image indices. In the training stage, we build such input sequences by taking nearest images for each image of a patient. For negative training samples, the image sequences are all selected from negative patients. For positive samples, the images are all selected from the patients with fungal keratitis and each image sequence has at least one positive image.
As shown at the middle of Fig. 4, the second stage network uses the trained backbone network of stage 1 to extract the features of the IVCM image, followed by a transformer-based network29,31,32 to learn the relationships among the image features. The aggregated sequence feature vector is then processed by a linear classifier predicting the positive/negative labels. The implementation of the stage 2 Transformer-based network, designed to process image sequences, is shown in Fig. 5. We denote the image sequence dataset as \(\{({\mathscr {X}}_i^1, {\mathscr {X}}_i^2, \ldots , {\mathscr {X}}_i^S; y_i)\}\), where the sequence length is S and \(y_i \in \{0, 1\}\) represents the label of the i-th sequence indicating whether the sequence contains fungal hyphae. The feature matrix \({\mathscr {V}}_i = (v_i^1, v_i^2, \ldots , v_i^S)\) extracted by the stage 1 feature backbone, is then processed by the Transformer-based network. We remove the position embedding module of the original transformer architecture in the stage 2 network since we cannot treat the sequence as an ordered set of elements. The relationship features between neighboring images are extracted using the four-layer Transformer block, which is described by the following equations:
where \({\mathscr {V}}_i^{(l)}\) represents the output feature matrix of the l-th layer, \(MSA(\cdot )\) represents the multi-head self-attention module, \(FF(\cdot )\) represents the feed-forward module, and \(LN(\cdot )\) represents the layer normalization module. The output feature matrix \({\mathscr {V}}^{out}\) is a sequence of feature vectors with a length of S. In order to obtain the final sequence feature that represents the relationships between neighboring images, we apply a max-pooling layer to aggregate \({\mathscr {V}}^{out}\).
The training of the two-stage feature extraction and diagnostic networks is regarded as a binary classification problem, and the networks are optimized using the cross-entropy loss function. Specifically, the loss function is defined as:
where \(y_i\) represents the label of the image or image sequence, and \({\hat{y}}_i\) represents the predicted probability of the network classifying it as fungal-positive.
Patient level diagnosis pipeline
Our networks are trained both at the image level (Stage 1) and image sequence level (Stage 2). In practice, we can further use our model to perform patient level diagnosis. As shown in the bottom of Fig. 4, the images of each patient are first processed by the first stage network to get image-level visual feature identification results. The predicted positive images are then selected with their neighboring images (defined by image indices) to generate a set of image sequences. The stage 2 network processes the image sequences to get sequence-level diagnostic predictions. We set a threshold \(\sigma\) for automatic diagnosis: The patient will be diagnosed as having fungal keratitis if there are at least \(\sigma\) image sequences predicted as positive by the second stage network. Using this scheme, our network can get higher specificity while increasing the threshold or get higher sensitivity while decreasing the threshold.
Preparation for training the networks
The original input IVCM images are grayscale images at a resolution of \(384 \times 384\). We first normalize the input images by mean and standard deviation calculated from the training data. Because the first stage backbone is initialized by a pre-trained SwinTransformer model on ImageNet-22k from pytorch-image-models33, whose inputs are RGB images with a resolution of \(224 \times 224\), we resize the IVCM images to \(224 \times 224\) and average the weights of the first convolutional layer into one input channel. We also use data augmentation by randomly flipping the images and changing the brightness, contrast and saturation.
During the training process of the two stages, our training datasets have imbalance data between two categories. To balance the data in two categories, we resample the images by a predefined weight, which is equal to the reciprocal of the total image number of the corresponding category in the training set. To alleviate the possible overfitting to the training data, we choose the model trained at the epoch that achieves the best performance on the validation set in our training process.
Statistical analysis
The fungal keratitis diagnosis is a binary classification task. Therefore, we evaluate the performance of the proposed deep learning framework by sensitivity, specificity, and AUC score. We calculate the 95% confidence intervals of sensitivity and specificity by Clopper-Pearson intervals34. We calculate the AUC score, the area under the receiver operating characteristic curve, and the 95% confidence intervals of the AUC score by bootstrapping35. The deep learning framework and statistical analysis are built on Python (version 3.6.9). The network architecture, training and test process are built on PyTorch (version 1.9.0), PyTorch-lightning (version 1.5.10) and Jittor36 (version 1.3.4.1). The accuracy, sensitivity, specificity, and AUC score are calculated by sklearn (version 0.24.2) and torchmetrics (0.7.2).
Ethics declarations
This study was conducted in compliance with the Declaration of Helsinki and approved by the ethics committee of Wuhan Aier Hankou Eye Hospital, Beijing Aier Intech Eye Hospital and Chengdu Aier East Eye Hospital. Informed consent was waived by the ethics committee of Wuhan Aier Hankou Eye Hospital, Beijing Aier Intech Eye Hospital and Chengdu Aier East Eye Hospital because of the retrospective nature of the study and anonymized usage of images.
Data availability
The IVCM images used for the study are not publicly available because of privacy protection. All data supporting the findings of this study are available from the corresponding authors for non-commercial and academic purposes.
Code availibility
The code for training and evaluating the two-stage neural network is available on Github: https://github.com/IGLICT/Fungal_Keratitis_Classification.
References
Garg, P., Roy, A. & Roy, S. Update on fungal keratitis. Curr. Opin. Ophthalmol. 27, 333–339 (2016).
Suman, S., Kumar, A., Saxena, I. & Kumar, M. Fungal keratitis: Recent advances in diagnosis and treatment. Infect. Eye Dis. Recent Adv. Diagn. Treatment 55, 5772 (2021).
Niu, L. et al. Fungal keratitis: Pathogenesis, diagnosis and prevention. Microb. Pathog. 138, 103802 (2020).
Wahyuningsih, R. et al. Serious fungal disease incidence and prevalence in Indonesia. Mycoses 64, 1203–1212 (2021).
Brown, L., Leck, A. K., Gichangi, M., Burton, M. J. & Denning, D. W. The global incidence and diagnosis of fungal keratitis. Lancet. Infect. Dis 21, e49–e57 (2021).
Bezerra, F. M., Höfling-Lima, A. L. & Oliveira, L. A. Fungal keratitis management in a referral cornea center in Brazil. Rev. Bras. Oftalmol. 79, 315–319 (2020).
Ting, D. S. J., Ho, C. S., Deshmukh, R., Said, D. G. & Dua, H. S. Infectious keratitis: An update on epidemiology, causative microorganisms, risk factors, and antimicrobial resistance. Eye 35, 1084–1101 (2021).
Pei, Y. et al. Microbiological profiles of ocular fungal infection at an ophthalmic referral hospital in southern china: A ten-year retrospective study. Infect. Drug Resist. 15, 3267 (2022).
Yildiz, E. H. et al. Alternaria and paecilomyces keratitis associated with soft contact lens wear. Cornea 29, 564–568 (2010).
Garg, P. Fungal, mycobacterial, and nocardia infections and the eye: An update. Eye 26, 245–251 (2012).
Stapleton, F. The epidemiology of infectious keratitis. Ocular Surf. 28, 351–63 (2021).
Shukla, P., Kumar, M. & Keshava, G. Mycotic keratitis: An overview of diagnosis and therapy. Mycoses 51, 183–199 (2008).
Borroni, D. et al. Shotgun metagenomic sequencing in culture negative microbial keratitis. Eur. J. Ophthalmol. 33, 1589–1595 (2023).
Borroni, D. Granulicatella adiacens as an unusual cause of microbial keratitis: A metagenomic approach. Ocul. Immunol. Inflamm. 30, 1550–1551 (2022).
Parekh, M. et al. Shotgun sequencing to determine corneal infection. Am. J. Ophthalmol. Case Rep. 19, 100737 (2020).
Bakken, I. M. et al. The use of in vivo confocal microscopy in fungal keratitis: Progress and challenges. Ocul. Surf. 24, 103–118. https://doi.org/10.1016/j.jtos.2022.03.002 (2022).
Çallı, E., Sogancioglu, E., van Ginneken, B., van Leeuwen, K. G. & Murphy, K. Deep learning for chest x-ray analysis: A survey. Med. Image Anal. 72, 102125 (2021).
Lin, D. et al. Application of comprehensive artificial intelligence retinal expert (care) system: A national real-world evidence study. Lancet Digit. Health 3, e486–e495 (2021).
Lundervold, A. S. & Lundervold, A. An overview of deep learning in medical imaging focusing on mri. Z. Med. Phys. 29, 102–127 (2019).
Domingues, I. et al. Using deep learning techniques in medical imaging: A systematic review of applications on ct and pet. Artif. Intell. Rev. 53, 4093–4160 (2020).
Wang, R. et al. Medical image segmentation using deep learning: A survey. IET Image Proc. 16, 1243–1267 (2022).
Suganyadevi, S., Seethalakshmi, V. & Balasamy, K. A review on deep learning in medical image analysis. Int. J. Multimedia Inf. Retrieval 11, 19–38 (2022).
He, K., Zhang, X., Ren, S. & Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 770–778 (2016).
Guo, M.-H. et al. Attention mechanisms in computer vision: A survey. Comput. Visual Media 8, 1–38 (2022).
Wu, X., Tao, Y., Qiu, Q. & Wu, X. Application of image recognition-based automatic hyphae detection in fungal keratitis. Aust. Phys. Eng. Sci. Med. 41, 95–103 (2018).
Liu, Z. et al. Automatic diagnosis of fungal keratitis using data augmentation and image fusion with deep convolutional neural network. Comput. Methods Programs Biomed. 187, 105019 (2020).
Lv, J. et al. Deep learning-based automated diagnosis of fungal keratitis with in vivo confocal microscopy images. Ann. Transl. Med.8 (2020).
Liu, Z. et al. Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision, 10012–10022 (2021).
Shao, Z. et al. Transmil: Transformer based correlated multiple instance learning for whole slide image classification. Adv. Neural Inf. Process. Syst.34 (2021).
Krizhevsky, A., Sutskever, I. & Hinton, G. E. Imagenet classification with deep convolutional neural networks. Commun. ACM 60, 84–90 (2017).
Vaswani, A. et al. Attention is all you need. Adv. Neural Inf. Process. Syst.30 (2017).
Dosovitskiy, A. et al. An image is worth 16x16 words: Transformers for image recognition at scale (2020). arXiv: 2010.11929.
Wightman, R. Pytorch image modelshttps://doi.org/10.5281/zenodo.4414861. https://github.com/rwightman/pytorch-image-models (2019).
Newcombe, R. G. Two-sided confidence intervals for the single proportion: Comparison of seven methods. Stat. Med. 17, 857–872 (1998).
Carpenter, J. & Bithell, J. Bootstrap confidence intervals: When, which, what? a practical guide for medical statisticians. Stat. Med. 19, 1141–1164 (2000).
Hu, S.-M., Liang, D., Yang, G.-Y., Yang, G.-W. & Zhou, W.-Y. Jittor: A novel deep learning framework with meta-operators and unified graph execution. Sci. China Inf. Sci. 63, 1–21 (2020).
Acknowledgements
This work was supported in part by the Aier-ICT Joint Laboratory for Digital Ophthalmology (No. SZYK202204).
Author information
Authors and Affiliations
Contributions
Y.C. and Q.Z. initiated the project and the collaboration. C.-P.L., W.D., Y.-P.X., L.-X.Z. and L.G. developed the network architectures, training, and testing setup. C.-P.L., W.D. and Q.Z. designed the clinical setup. C.L., J.L., F.C., D.C., S.S. and S.L. collected and labeled the datasets. C.-P.L., W.D. and Y.-P.X. analyzed the data. C.-P.L., W.D., Y.-P.X., M.Q., L.G., F.-L.Z and Y.-K.L. wrote the paper. All authors provided critical feedback to the manuscript. Y.-P.X. and L.-X.Z. deployed open source code. C.-P.L. and W.D. contributed equally.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, CP., Dai, W., Xiao, YP. et al. Two-stage deep neural network for diagnosing fungal keratitis via in vivo confocal microscopy images. Sci Rep 14, 18432 (2024). https://doi.org/10.1038/s41598-024-68768-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-68768-y
- Springer Nature Limited