Abstract
Siderophores are specialized molecules produced by bacteria and fungi to scavenge iron, a crucial nutrient for growth and metabolism. Catecholate-type siderophores are mainly produced by bacteria, while hydroxamates are mostly from fungi. This study investigates the capacity of nine hydroxamate-type siderophores from fungi and Streptomyces to facilitate iron acquisition by the human pathogen Pseudomonas aeruginosa. Growth assays under iron limitation and 55Fe incorporation tests showed that all nine siderophores promoted bacterial growth and iron transport. The study also aimed to identify the TonB-dependent transporters (TBDTs) involved in iron import by these siderophores. Using mutant strains lacking specific TBDT genes, it was found that iron is imported into P. aeruginosa cells by FpvB for coprogen, triacetylfusarinine, fusigen, ferrirhodin, and ferrirubin. Iron complexed by desferioxamine G is transported by FpvB and FoxA, ferricrocin-Fe and ferrichrycin-Fe by FpvB and FiuA, and rhodotoluric acid-Fe by FpvB, FiuA, and another unidentified TBDT. These findings highlight the effectiveness of hydroxamate-type siderophores in iron transport into P. aeruginosa and provide insights into the complex molecular mechanisms involved, which are important for understanding microbial interactions and ecological balance.
Similar content being viewed by others
Introduction
Siderophores are specialized small molecules produced by bacteria and fungi, to facilitate the acquisition of iron, an essential nutrient1. Indeed, iron plays a key role in biology as a cofactor for numerous enzymes and proteins involved in critical cellular processes, including oxygen transport, DNA synthesis, and energy metabolism2. As iron is often present in limited concentrations because sequestered by host organisms in the context of infections or immobilized in insoluble forms in the environment at neutral pH and in aerobic conditions, the ability to produce and utilize siderophores is a key adaptation for microorganisms. This capability influences their competitiveness for iron resources and, consequently, their survival and growth in different environments3,4.
Import of iron into Gram negative bacteria by siderophores involves several molecular steps. The stable ferri-siderophores complexes formed in the bacterial environment are imported from the environment into the periplasm by specific outer membrane transporters called TonB-dependent transporters (TBDT)5. It is an active transport depending on the inner membrane TonB molecular machinery, composed of the proteins TonB, ExbB, and ExbD6,7,8. This TonB complex couples the proton motive force of the cytoplasmic membrane to the TBDTs located in the outer membrane, and that way energizes the transport process6,7,8. TonB interacts with TBDTs that bind iron-siderophore complexes, initiating a series of conformational changes in the TBDT allowing the release of the bound ferri-siderophore complex into the periplasm. Once inside the periplasm, two scenarios are possible. Either the ferric iron is released from the siderophore in the bacterial periplasm or the ferri-siderophore complex is transported further into the cytoplasm via specific inner membrane transporters9.
In iron chelation by siderophores, iron is typically surrounded by six coordinating oxygens belonging mostly to hydroxamate, catecholate, or carboxylate groups present in the siderophore structure1,10. Consequently, siderophores can be broadly classified into four families based on their functional groups chelating iron1,10. One major family is composed of siderophores having only hydroxamic acid functional groups for ferric iron chelation. Notable examples include ferrichrome and ferrichrysin11,12. Another prominent family consists of catecholate siderophores, featuring catechol or its derivatives as the iron-binding moiety. Enterobactin, produced by enteric bacteria, is the archetype siderophore of this family13. Carboxylate siderophores, such as citrate and staphyloferrin, represent another group that utilizes carboxylic acid groups for iron coordination14,15,16. In addition, there are mixed-function siderophores that incorporate multiple different iron-binding moieties within the same molecule, like pyoverdine produced by Pseudomonas aeruginosa17, which is composed of a catechol group and two hydroxamate groups for iron chelation, or aerobactin, schizokinen and arthrobactin, which are mixed α-carboxylate/hydroxamate type siderophores18,19,20. In their general chemical structures, siderophores exhibit remarkable diversity across microbial species, reflecting adaptation to specific ecological niches and iron availability in their respective environments.
Tris-hydroxamate siderophores are produced mostly by fungi21,22,23. Interestingly, bacteria have evolved the ability to exploit these fungal tris-hydroxamate siderophores for their own iron acquisition strategies. This interkingdom iron-scavenging system underscores the complexity and adaptability of microbial communities, showcasing how organisms collaborate to overcome nutritional challenges in their ecosystems.
In the present work, we have investigated the ability of the opportunist pathogen P. aeruginosa to use hydroxamate siderophores produced by Fungi and Streptomyces to access iron. It is well known that P. aeruginosa produces two siderophores pyoverdine and pyochelin and is able in addition to use many siderophores produced by other microorganisms (xenosiderophores)24. The possibility to use diverse xenosiderophores is due to the presence in the genome of P. aeruginosa of diverse genes encoding TBDTs25,26. Currently, the only hydroxamate-type xenosiderophores known to facilitate iron import into P. aeruginosa cells, with their corresponding TBDT identified, are ferrichrome, nocardamine and desferrioxamine B, bisucaberin and the α-carboxylate/hydroxamate mixed siderophores aerobactin, schizokinen and arthrobactin27,28,29,30,31. The TBDTs involved are FiuA and FpvB for ferrichrome, FoxA for nocardamine and bisucaberin, FoxA and FpvB for DFOB29, and ChtA for aerobactin, schizokinen and arthrobactin27,28,29,30,31. Additionally, P. aeruginosa can utilize coprogen, ferrirubin, ferricrocin, and ferrichrysin to access iron, though the specific TBDTs involved have not yet been identified32.
In the present study we demonstrated that nine hydroxamate-type siderophores effectively stimulated P. aeruginosa growth and transported iron into P. aeruginosa cells, with varying efficiencies and tried to identify the TBDTs involved. The results also indicated that four TBDTs, FpvB, FiuA, FoxA and an additional unidentified TBDTs, are involved in facilitating iron import by these different siderophores, but with different transport specificities. Proteomic and RT-qPCR approaches showed that some of these siderophores, when present in the environment of P. aeruginosa are able to induce the transcription and expression of fiuA and foxA.
Results
Description of the xenosiderophores used in the present study
Catecholate-type siderophores are exclusively produced by bacteria, while hydroxamates are predominantly produced by Fungi but are also found in several bacterial species, including actinomycetes 23,33,34. The family of hydroxamate-type siderophores can be classified into 5 subfamilies based on their chemical structure, including rhodotorulic acid, coprogen-, fusarinine-, ferrioxamine-, and ferrichrome-type siderophores35. Among the commercially available siderophores (Biophore Research Product), we selected several representatives from each of the 5 subfamilies for our study. Their chemical structures are presented in Fig. 1.
Chemical structures of the hydroxamate siderophores tested in the present study. For ferricrocin, ferrichrysin, ferrirhodin and ferrirubin, the differences in their chemical structures are highlighted in pink. Ferrirhodin and ferrirubin have the same chemical structures except that ferrirhodin has three cys-anhydromevalonyl groups and ferrirubin three trans-anhydromevalonyl groups.
Rhodotorulic acid (RHODO) possesses a unique chemical structure featuring a pyridine ring and a long acyl chain. Its iron-chelating ability is attributed to the two hydroxamate groups within the molecule. RHODO is produced by species of yeasts of the genus Rhodotorula36,37. Coprogens are a subfamily of siderophores produced by fungi, and different species or strains of fungi may produce coprogens with distinct structures. The general features comprise a pyridine ring in a trimeric linear tris-hydroxamate backbone, with specific modifications or substitutions leading to variations in the chemical structure among different coprogens. Here, we tested the coprogen produced by Neurospora crassa38. Triacetylfusarinine C (TAFC) and fusigen belong to the fusarin subfamily and share the same trimeric cyclic ester backbone with three endo-hydroxamate groups for iron coordination but differ in specific modifications or substitutions in the molecule39. TAFC is produced by species of the genus Aspergillus40, while fusigen is produced by Ascomycota, such as species of the genus Fusarium, and by Basidiomycota, such as species of the genus Laccaria41,42,43, Desferrioxamine G (DFOG) is a linear tris-hydroxamate siderophore with the same tris-hydroxamate backbone as the well-known desferrioxamine E, also called nocardamine, except that in the case of nocardamine, it is a cyclic tris-hydroxamate siderophore. DFOG is produced by Hafnia alvei44. We have also tested ferricrocin, ferrichrysin, ferrirhodin, and ferrirubin, having a chemical structure composed of a peptide macrocycle as the one found in the ferrichrome and with three exo-hydroxamate groups for iron chelation. Ferrichrome is a cyclic hexapeptide composed of three glycines and three acylated ∂-N-hydroxyornithyl residues, with the hydroxamate groups of the ornithines chelating iron. In contrast, ferricrocin is a cyclic hexapeptide composed of two glycines, one serine, and three modified ornithine residues with hydroxamate groups. Ferrichrysin, ferrirhodin, and ferrirubin all share a cyclic hexapeptide composed of one glycine, two serines, and three modified ornithine residues. The differences lie in the modification of the acetylated ornithines, as shown in Fig. 1. Ferricrocin, ferrichrysin, ferrirhodin, and ferrirubin are produced by species of the genus Aspergillus such as A. fumigatus or A. ochraceus45,46,47. Ferrirhodin is also produced by plant pathogens of the phylum Ascomycota such as Fusarium sacchari or Botrytis cinerea48,49. The affinities for iron of these nine different siderophores are summarized in Table S1.
All the xenosiderophores tested were able to supply P. aeruginosa with iron
We initially assessed the ability of the siderophores presented in Fig. 1 to function as xenosiderophores for iron acquisition by P. aeruginosa. Two distinct approaches were employed. The first is based on the capacity to promote the growth of P. aeruginosa in an iron-deficient medium, while the second involves the incorporation of 55Fe. Both assays have been comprehensively described in previously published studies and have proven effective31,50,51,52. For the growth assay, we employed a strain of P. aeruginosa (∆pvdF∆pchA) incapable of producing its own siderophores, pyoverdine, and pyochelin, due to the deletion of two genes encoding enzymes involved in the biosynthesis of these siderophores53. This mutant is cultivated in CAA medium, which is iron-deficient (approximately 20 nM iron concentration54), and in the presence of 10 µM of the tested siderophore. The excess siderophore chelates all the iron present in the CAA medium. If the bacteria are able to use the siderophore, growth is observed, as with pyochelin (PCH), one of the two siderophores produced by P. aeruginosa (Fig. 2A). If the bacteria cannot utilize the siderophore for iron acquisition, complete growth inhibition is observed. Bacillibactin, for instance, serves as an illustration as it completely inhibits the growth of P. aeruginosa under the conditions of the test culture52. For the nine xenosiderophores we tested, we observed stimulation of P. aeruginosa growth, indicating that these molecules can be used as siderophores by this pathogen (Fig. 2 data with strain ∆pvdF∆pchA).
Growth of ∆pvdF∆pchA mutants in the presence of RHODO, coprogen, DFOG, TAFC, fusigen, ferricrocin, ferrichrysin, ferrirhodin, or ferrirubin. ∆pvdF∆pchA and its corresponding fpvB, fiuA and foxA deletion mutants were grown in CAA medium in the absence (growth kinetics in blue) or presence (kinetics in orange) of 10 µM pyochelin (PCH), RHODO, coprogen, DFOG, TAFC, fusigen, Ferricrocin, ferrichrysin, ferrirhodin, and ferrirubin. Growth was followed by monitoring the optical density at 600 nm. Error bars were calculated from three independent biological replicates. Since a growth stimulation was observed for some siderophores tested, the OD scales are not the same in all the strains tested.
The results were further validated by the 55Fe incorporation assay. In this test, siderophore-55Fe complexes were formed with each of the different siderophores from Fig. 1 by mixing the siderophores with 55Fe as described in the Experimental Procedures. These complexes were incubated with ∆pvdF∆pchA cells, and radioactivity incorporated into the bacteria was measured over time. All nine tested molecules demonstrated the ability to transport 55Fe into P. aeruginosa cells, albeit with variations in efficiency (Fig. 3A). RHODO imported around 210 pmol.mL-1.OD600 nm-1 of 55Fe when incubated during 3 h with ∆pvdF∆pchA cells, ferricrocin 360 pmol.mL-1.OD600 nm-1, ferrichrysin, 250 pmol.mL-1.OD600 nm-1, and TAFC 50 pmol.mL-1.OD600 nm-1. All other xenosiderophores tested transported between 95 and 150 pmol.mL-1.OD600 nm-1. These uptakes of 55Fe are due to an active transport and not to diffusion via porins, as the presence of carbonyl cyanide m-chlorophenyl hydrazone (CCCP), an inhibitor of bacterial proton motive force, inhibits the transport.
55Fe uptake by P. aeruginosa strains mediated by RHODO, coprogen, DFOG, TAFC, fusigen, ferricrocin, ferrichrysin, ferrirhodin, or ferrirubin. P. aeruginosa ∆pvdF∆pchA cells were grown in iron-restricted CAA medium in the presence of 10 µM RHODO, coprogen, DFOG, TAFC, fusigen, ferricrocin, ferrichrysin, ferrirhodin, and ferrirubin to induce expression of the corresponding uptake pathway. Afterwards, bacteria were incubated with 500 nM RHODO-55Fe, coprogen-55Fe, TAFC-55Fe, fusigen-55Fe, DFOG-55Fe, ferricrocin-55Fe, ferrichrysin-55Fe, ferrirhodin-55Fe, and ferrirubin-55Fe. The amount of 55Fe taken up by the bacteria was measured as a function of time. As a control, the experiment was repeated in the presence of the protonophore CCCP (200 μM). The experiment was also repeated with the corresponding fpvB, fiuA and foxA deletion mutants of ∆pvdF∆pchA. Error bars were calculated from three independent biological replicates. In the case of fusigen, a high level of radioactivity in the cell pellet was already observed at time zero due to the precipitation of either uncomplexed 55Fe or fusigen-55Fe complexes.
In conclusion, both approaches demonstrated that the nine hydroxamate-type siderophores can effectively transport iron into P. aeruginosa cells.
FpvB, FiuA, and FoxB are the TonB-dependent transporters (TBDTs) involved in the import of iron by the 9 hydroxamate siderophores tested
The growth assay under iron-deficient conditions also serves to identify the TBDTs responsible for iron import by the tested siderophores. To achieve this, strains of P. aeruginosa that are incapable of producing pyoverdine and pyochelin and are mutated for one or more gene(s) encoding a (or several) TBDT(s) must be employed. If the deletion of a gene encoding a TBDT affects bacterial growth in the presence of a specific siderophore, it implies that the tested siderophore can no longer import iron into the bacteria because it utilizes the mutated TBDT. If growth inhibition is total, it indicates that this mutated TBDT is the sole participant in the iron import mediated by the tested siderophore. If partial inhibition is observed, one or more additional TBDTs are involved. In addition to this growth test, we also used the 55Fe incorporation test with strains mutated for one or more genes encoding one or several TBDTs.
For RHODO, the results obtained are intricate. Deletion of either fpvB or fiuA alone has no effect on bacterial growth (Fig. 2B). In contrast, the double mutation fpvB/fiuA almost completely inhibits growth, but a very low residual growth seems to persist. The growth inhibition observed with the double fpvB/fiuA mutation indicates that both TBDTs encoded by these genes are involved in iron import by RHODO. If one is absent, the other can compensate without affecting bacterial growth. FoxA deletion has no additional effect on growth inhibition. Concerning the import of 55Fe, double deletions of fiuA and fpvB do not completely inhibit 55Fe import (Fig. 3), suggesting that another unidentified TBDT is also involved. We have tested the deletion of some other genes encoding TBDTs (chtA and PA0151), but without success (Figure SM1). In conclusion, iron is transported into P. aeruginosa cells by FpvB, FiuA, and another unidentified TBDT. However, iron import by this third TBDT does not seem very effective in supporting bacterial growth, as the deletion of fiuA and fpvB nearly inhibits bacterial growth.
For coprogen, TAFC, fusigen, ferrirhodin and ferrirubin, complete growth inhibition is observed with the sole deletion of the fpvB gene, indicating that the ferric complexes of these five xenosiderophores enter P. aeruginosa cells through the TBDT FpvB (Fig. 2C,E–G,I). These results are confirmed by the 55Fe import test (Fig. 3). When fpvB is deleted, no incorporation of 55Fe is observed with any of these five xenosiderophores. FpvB is the only TBDT involved in iron import by coprogen, TAFC, fusigen, ferrirhodin and ferrirubin.
For DFOG, the single deletion of fpvB or foxA had no effect on the bacterial growth. A complete growth inhibition is observed only for the double mutant foxA/fpvB, indicating that both TBDTs are involved in the import of DFOG-Fe complexes, and if one is absent, the other can replace it (Fig. 2D). The 55Fe import tests is consistent with these results (Fig. 3). The deletion of both foxA and fpvB is required to completely inhibit bacterial growth.
For ferricrocin and ferrichrysin, complete growth inhibition is observed for the double mutant fiuA and fpvB, and not for the corresponding single mutants, indicating that both TBDTs are responsible for iron import by ferricrocin and ferrichrysin, and one can replace the other if absent (Fig. 2H,J). The 55Fe import tests are consistent with these results (Fig. 3), there is total inhibition with the double mutation fiuA/fpvB. But it is interesting to note that the simple deletion of fpvB does not affect the import of iron by these two siderophores, whereas the mutation of fiuA affects it by 40% after 3 h of incubation, indicating that iron import primarily occurs through FiuA and less through FpvB. Such a difference was not observed for iron import via DFOG through the TBDTs FoxA and FpvB.
Altogether, the data show that RHODO-Fe is imported into bacterial cells by FpvB, FiuA, and another unidentified TBDT. Coprogen-Fe, TAFC-Fe, fusigen-Fe, ferrirhodin-Fe, and ferrirubin-Fe are imported solely by FpvB. DFOG-Fe is imported by FpvB and FoxA. Ferricrocin-Fe and ferrichrycin-Fe are imported by FpvB and FiuA.
RHODO, ferricrocin and ferrichrysin induce the transcription and expression of fiuA and DFOG of foxA
The transcriptional regulation of genes encoding TBDTs is primarily controlled by the Ferric uptake regulator (Fur) protein in most bacteria55,56,57,58. Fur is a global regulatory protein that responds to iron availability in the bacterial cells. When iron is abundant, Fur binds to iron and forms a complex that can attach to specific DNA sequences called Fur boxes, which are located in the promoter regions of genes encoding TBDTs. This binding represses the transcription of these genes, preventing the synthesis of TBDTs. Conversely, under low iron conditions, the Fur-iron complex dissociates, leading to derepression of TBDT genes transcription. This allows the bacteria to express TBDTs, facilitating the uptake of iron from the environment to meet cellular needs. In addition, it is well known that certain genes encoding TBDTs have their transcription regulated by the presence of their corresponding iron-loaded xenosiderophores in the environment of P. aeruginosa, in iron deficiency growth conditions28,31,50,52,59. These transcriptional regulations necessitating the presence of iron-loaded xenosiderophores involve sigma and anti-sigma factors, two-component systems, and transcriptional regulators of the AraC family60,61,62,63,64,65,66. The transcription of fiuA and foxA is regulated by Fur and by the sigma and anti-sigma factors FiuI/FiuR and FoxI/FoxR respectively63. For the fpvB gene, only regulation by Fur is present according to genome analysis67.
To verify if the transcription of fiuA, foxA, and fpvB could be regulated by certain siderophores tested in this study, a qRT-PCR study was conducted (Fig. 4). We examined the transcription of three genes fiuA, foxA, and fpvB, as well as fptA and fpvB, because often the induction of the transcription of a gene encoding TBDTs goes hand in hand with the repression of the transcription of fptA, the gene encoding for the TBDT involved in iron import by pyochelin, and more rarely, repression of fpvA, the gene encoding for iron import by pyoverdine28,31,50,52,59. This study was conducted on the strain not producing pyoverdine and pyochelin (∆pvdF∆pchA), in the presence of 10 µM of one of the tested xenosiderophores, and after 8 h of culture under iron restricted conditions (the preculture of ∆pvdF∆pchA was already carried out under iron restricted conditions).
Modulation of TBDT gene transcription by RHODO, coprogen, DFOG, TAFC, fusigen, ferricrocin, ferrichrysin, ferrirhodin, or ferrirubin. P. aeruginosa ∆pvdF∆pchA cells were grown for 8 h in the presence or absence of 10 µM of one of the xenosiderophores. The transcription of fpvA, fpvB, fptA, foxA and fiuA was followed by qRT-PCR53. fpvA and fpvB encodes the ferri-pyoverdine TBDT, fptA the ferri-pyochelin TBDT, foxA the ferri-nocardamine TBDT and fiuA the ferri-ferrichrome TBDT24.
No significant induction of the transcription of the genes fpvB, foxA, and fiuA was observed in the presence of coprogen, TAFC, fusigen, ferrirhodin and ferrirubin (Fig. 4): log2 fold change (FC) around 1. However, in the presence of 10 µM DFOG, we observed a strong induction of foxA transcription (log2 (FC) = 5,9). In the presence of 10 µM ferricrocin, and ferrichrysin, an induction of fiuA transcription was observed (log2 (FC) of 3,5 and 2,5 respectively) as well as in the presence of 10 µM RHODO but in a lesser extent (log2 (FC) = 1,4). A strong repression of fptA transcription was observed with TAFC, and a repression of both fptA and fpvA transcription was observed with ferrirhodin and ferrirubin. All the other xenosiderophores had no effect on the transcription of fpvA and fptA.
A differential proteomic approach was used to confirm the RT-qPCR data for ferricrocin and ferrichrysin, which induce the transcription of fiuA (Fig. 5). Additionally, this approach aimed to determine if, in the case of RHODO, we could identify another TBDT besides FoxA or FiuA whose expression is induced by the presence of this molecule. The proteomic approach could not be carried out with DFOG and the other xenosiderophores because we had no more of these molecules available. In the case of RHODO, aside from FiuA (log2 fold change of 3.72), no other TBDT was identified with induced expression in the presence of this molecule. Moreover, the presence of RHODO had no effect on the expression levels of the various proteins in the pyoverdine and pyochelin pathways. Ferricrocin and ferrichrysin both induced only the expression of fiuA, with log2 fold changes of 7.75 and 6.09 respectively, confirming the RT-qPCR data. Regarding the expression levels of the proteins in the pyochelin and pyoverdine iron uptake pathways, only an induction of FpvR expression was observed in the presence of ferrichrysin. No other effects were seen. The molecular mechanism by which this xenosiderophore modulates FpvR expression, an anti-sigma factor involved in regulating the transcription of various genes in the pyoverdine pathways, remains unclear.
Analysis of changes in the expression of proteins involved in iron-uptake pathways in P. aeruginosa cells grown under iron-limited conditions in the presence of RHODO, ferricrocin or ferrichrysin. A. Proteomic analyses were performed on the pyoverdine- and pyochelin-deficient P. aeruginosa strains (∆pvdF∆pchA) grown overnight in CAA supplemented, or not, with 10 µM RHODO, ferricrocin or ferrichrysin. Average values measured in the presence of 10 µM RHODO, ferricrocin or ferrichrysin are plotted against average values measured in the absence of any supplementation with xenosiderophores. Median values represent the median of the relative intensity of each protein, normalized against all proteins detected by shotgun analysis (n = 3). The proteins of the pyochelin pathway are represented by orange dots, those of the pyoverdine pathway by green dots, and the TBDTs by blue dots. B. Heat maps of various TBDTs and proteins involved in the pyochelin and pyoverdine iron uptake pathways: the darker the shade of blue, the higher the expression of the protein; the darker the shade of red, the more the expression of the protein is repressed. NS, not significant.
In conclusion, RT-qPCR data showed an induction of foxA transcription in the presence of DFOG and of fiuA transcription in the presence of ferricrocin and ferrichrysin, which was confirmed by proteomic analysis. RHODO also seems to induce the transcription of fiuA according to proteomic data, but with lower efficiency than ferricrocin and ferrichrysin. This leads to the conclusion that all xenosiderophores utilizing the TBDT FiuA and FoxA are probably able to induce its transcription and expression with varying efficiencies. These regulations certainly involve the sigma and anti-sigma factors FiuI/FiuR and FoxI/FoxR for fiuA and foxA respectively.
Discussion
According to the nature of the chemical groups that chelate iron, siderophores can be classified into four classes: hydroxamates, catechols, carboxylic acids, and mixed siderophores, which contain at least two chelating groups of different natures10,68. Regarding hydroxamate-type siderophores, this group can be further subdivided into five subtypes (rhodotorulic acid, coprogen, fusarinine, ferrioxamine, and ferrichrome type siderophores) based on their chemical structure, with the chelating functions being exclusively hydroxamates in all five subtypes35. In this study, we demonstrated that P. aeruginosa can utilize siderophores from each of these five subtypes to access iron.
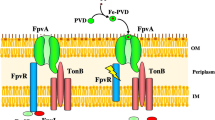
The import of iron by these nine xenosiderophores into P. aeruginosa cells occurs via four different TBDTs: FiuA, FoxA, FpvB and another unidentified TBDT that we were unable to identify (Fig. 6). Coprogen, triacetylfusarinine C, fusigen, ferrirhodin, and ferrirubin siderophores import iron into P. aeruginosa cells exclusively via FpvB. Iron complexed by DFOG is imported as previously shown for desferioxamine B-Fe (DFOB)29 by two TBDTs, FpvB and FoxA. As previously shown for ferrichrome-Fe29, ferricrocin-Fe and ferrichrysin-Fe complexes, enter the bacteria through FpvB and FiuA. Finally, rhodotorulic acid-Fe complexes are imported by FpvB, FiuA, and another unidentified TBDT.
FiuA, FpvB, FoxA and FpvA transport selectivity. FiuA is the TBDT of the ferric forms of ferrichrome, ferricrocin, ferrichrysin and RHODO; FpvB of the ferric forms of ferrichrome, ferricrocin, ferrichrysin, RHODO, TAFC, fusigen, coprogen, ferrirhodin, ferrirubin, pyoverdine, DFOB and DFOG; FoxA of the ferric forms of nocardamine, bisucaberin, DFOB and DFOG; and at last FpvA of the ferric form of pyoverdine. In pink are highlighted the differences in the chemical structures of ferrichrome, ferricrocin, ferrichrysin, ferrirhodin and ferrirubin.
These data also confirm that FpvB, FiuA, and FoxA have distinct selectivities for siderophore-Fe uptake. FpvB can import the ferric complexes of all nine hydroxamate-type xenosiderophores tested, indicating a broad uptake and certainly binding selectivity. Initially described as an alternative TBDT for iron uptake by the siderophore pyoverdine, with FpvA being the primary transporter, FpvB has more recently been identified as a TBDT involved in the uptake of iron by ferrichrome and DFOB29,69. The same authors determined the affinity of FpvB for iron-chelated ferrichrome, DFOB, and pyoverdine, showing that FpvB has a higher affinity for iron-chelated ferrichrome and DFOB than for pyoverdine-Fe29. In the case of DFOB-Fe, they observed a two-site binding model, which was not seen for the other two siderophores29. Additionally, a structural model of FpvB generated by AlphaFold2, combined with docking studies and site-directed mutagenesis experiments conducted by the same group, suggests that all three ligands bind to a similar hydrophobic pocket in FpvB29. Since FpvB imports iron via all nine hydroxamate-type siderophores tested in the present study without exception, it is likely that this transporter is capable of recognizing and transporting many other hydroxamate-type siderophores found in nature.
Our data show that FiuA specifically recognizes and transports ferrichrome-type xenosiderophores: ferrichrome as already previously described27,29, ferrichrysin, and ferricrocin. The cyclic hexapeptide structure of these compounds appears crucial for recognition and uptake by FiuA, as this TBDT exclusively transports xenosiderophores with such a structure. A certain variability in the sequence of this hexacyclic structure is tolerated, as the three cyclic hexapeptide sequences Gly-Gly-Gly-Orn-Orn-Orn (ferrichrome), Gly-Gly-Ser-Orn-Orn-Orn (ferrichrocin), and Gly-Ser-Ser-Orn-Orn-Orn (ferrichrycin) can all be recognized and transported by FiuA when complexed with iron. The three-dimensional X-ray structure of the cyclic iron(III)-containing hexapeptide ferrichrysin shows similarities with that of the related ferrichrome, which probably explains why both compounds can be recognized by FiuA12. In both structures, the iron(III) ions are coordinated by three hydroxamate groups in an octahedral cis-Λ arrangement12. However, if the three acylated δ-N-hydroxyornithyl residues are modified or bulky, as seen in ferrirubin or ferrirhodin, it affects recognition by FiuA without impacting binding to FpvB. FiuA also recognizes ferric forms of RHODO, which likely have a distinct 3D structure compared to the ferric forms of ferrichrome, ferrichrysine, and ferricrocin, and may bind to FiuA differently.
FoxA transports iron chelated by DFOG, DFOB, and nocardamine (also known as DFOE), which are hydroxamate xenosiderophores of the desferrioxamine family, as well as bisucaberin28,30,30. Although DFOG and DFOB are linear tris-hydroxamate siderophores with the same tris-hydroxamate backbone as the cyclic nocardamine, this structural difference does not seem to affect recognition and transport by FoxA, because these compounds form a similar iron-binding shell as shown by the FoxA X-ray structures solved in complex with DFOB-Fe and nocardamine-Fe28,70. These structures show that NOCA-Fe and DFOB-Fe perfectly superimpose in the FoxA binding site, with the Fe atoms located in exactly the same position in both complexes, suggesting that DFOG could have a similar binding mode. Fluorescence titration experiments showed that the binding affinities for DFOB-Fe and nocardamine-Fe to FoxA are of the same order of magnitude. The dissociation constant (Kd) of nocardamine-Fe for FoxA is 178 ± 16 nM, and that of DFOB-Fe is 100 ± 10 nM, both forming 1:1 siderophore-Fe stoichiometry complexes with28,70. Additionally, the same team that solved the structures of FoxA also demonstrated, through structural and biophysical interaction studies, that ferri-DFOB binds to the transporter via a two-step TonB-binding mechanism70. The X-ray structure of the FoxA-bisucaberin-Fe complex has also been solved, showing that the bisucaberin-Fe complex is distinct from the two other siderophore-Fe complexes because it forms a 3:2 rather than a 1:1 siderophore-iron complex30. The X-ray structure also reveals that bisucaberin-Fe mimics the binding conformation of nocardamine-Fe but forms different hydrogen bonds with the transporter30. Chan et al. also demonstrated that extracellular loop 8 of FoxA plays a role in the binding and uptake of all three ligands30. It is also important to note that the thiopeptide antibiotic thiocillin, which has a different chemical structure than DFOB, nocardamine, and bisucaberin, also exploits FoxA for uptake. Molecular docking approaches suggest that thiocillin has a binding mode similar to that of nocardamine-Fe29,30. Thiocillin uses FoxA to cross the outer membrane and inhibit the growth of P. aeruginosa71. Our data show that the ferric forms of fusigen and coprogen are only recognized and transported by FpvB and not by FoxA. These compounds have unsaturations in their carbon chains, which can cause rigidities in their structures, affecting their interaction with FoxA.
The efficiency of 55Fe uptake varies among the different xenosiderophores tested, between TAFC importing 50 pmol·mL−1·OD600 nm−1 and ferricrocin importing 360 pmol·mL−1·OD600 nm−1. The lowest amount of 55Fe imported is observed for the xenosiderophores using only FpvB for transport: TAFC, fusigen, ferrirubin, and coprogen (between 50 to 100 pmol·mL−1·OD600 nm−1). The highest amount of 55Fe imported was observed for ferricrocin and ferrichrysin, which use both FpvB and FiuA (360 and 250 pmol·mL−1·OD600 nm−1 respectively). It should be noted that this difference in the amount of 55Fe imported into P. aeruginosa cells, depending on the xenosiderophore used, does not affect bacterial growth (Figs. 2, 3). At this stage, it is not possible to conclude whether FiuA has a higher uptake rate of ferri-siderophore complexes compared to FpvB and FoxA, or if the observed differences in the amount of imported 55Fe are due to varying expression levels of these transporters. However, it is important to note that, according to genome annotation, fpvB transcription is solely regulated by Fur, whereas the transcription of fiuA and foxA is regulated by Fur and sigma/anti-sigma factors (FiuI/FiuR and FoxI/FoxR, respectively)63,65. Through RT-qPCR and proteomics approaches, we observed an induction of foxA transcription in the presence of DFOG and an induction of fiuA transcription and expression in the presence of ferricrocin, ferrichrysin, and RHODO. These regulations likely involve the sigma and anti-sigma factors FiuI/FiuR for fiuA and FoxI/FoxR for foxA (for more details concerning the molecular mechanisms involved, see 63,64,65,72). It can be hypothesized that this positive regulation by sigma and anti-sigma factors results in a higher number of FoxA and FiuA transporters compared to FpvB in the outer membranes of P. aeruginosa. However, this remains a hypothesis and needs to be verified with additional studies quantifying the TBDTs in the membranes of P. aeruginosa.
This screening of nine different xenosiderophores already provides a good overview of the transport selectivities among FpvB, FiuA, and FoxA. These data also significantly extend the list of xenosiderophores that P. aeruginosa can utilize: 29 xenosiderophores are currently listed as usable by P. aeruginosa (Table S5 in Supplemental Materials). The broad transport selectivity of FpvB highlights the importance of obtaining the X-ray structure of this transporter to gain insight into the recognition and binding mechanisms between FpvB and various hydroxamate siderophores. With more than 40 structures of TBDTs currently described in the literature, elucidating the structure of FpvB with different siderophores bound to its binding site(s) would help understand why this transporter, unlike other TBDTs, recognizes so many different siderophores. Given the low specificity of FpvB’s recognition capabilities, this transporter could potentially be used to import siderophore-antibiotic conjugates. One of the major challenges in developing new antibiotics against Gram-negative bacteria is ensuring that these drugs can enter the bacterium to reach their biological targets. Due to the low binding and uptake selectivity of FpvB for hydroxamate-type siderophores, this transporter could serve as an entry point for siderophore-conjugated antibiotics (for reviews concerning this strategy, see Ref. 73,74,75,76,77). In addition, FpvB sequence is highly conserved in the various isolates of P. aeruginosa that have been fully sequenced: 99–100% sequence identity for 270 different isolates67, indicating that siderophore-antibiotic conjugates targeting this transports could be used against many isolates of P. aeruginosa.
Pseudomonas aeruginosa is a common opportunistic pathogen that can cause infections in immunocompromised individuals, particularly those with cystic fibrosis, burns, or compromised immune systems78,79. Fungi, including Candida species and Aspergillus species, are also known to cause opportunistic infections in similar patient populations80. Co-infections involving P. aeruginosa and fungi can occur in various clinical settings and often lead to more severe disease outcomes compared to infections with either pathogen alone81,82,83. The interactions between P. aeruginosa and fungi in co-infections are complex and can involve competition for nutrients, production of virulence factors, and modulation of host immune responses. Both P. aeruginosa and fungi produce and use siderophores to acquire iron from host tissues, competing for these essential resources. P. aeruginosa produces pyoverdine and pyochelin84,85,86. Aspergillus species, including Aspergillus fumigatus, produce siderophores such as ferricrocin and fusarinine C to acquire iron from the environment. In the context of co-infections, the production and utilization of siderophores by P. aeruginosa and fungi can influence the dynamics of microbial growth and virulence87. Moreover, using a neutropenic mouse model of microbial gastrointestinal colonization and dissemination, Lopez-Medina and collaborators demonstrated that the fungus Candida albicans inhibits the virulence of the bacterium P. aeruginosa by suppressing the expression of pyochelin and pyoverdine genes, which are critical for iron acquisition and virulence88. Furthermore, deletion of the fiuA gene in P. aeruginosa was shown to affect biofilm formation under sublethal carbenicillin stress conditions and to reduce the production of elastase, a major virulence determinant89. Understanding the interactions between P. aeruginosa, fungi, and siderophores in co-infections is crucial for developing strategies to prevent and treat infections caused by these pathogens and to combat multidrug-resistant infections in clinical settings.
P. aeruginosa is also known to coexist with various Streptomyces species and fungi in different environments, like soil and rhizosphere ecosystems, with synergistic or competitive interactions depending on the specific strains and environmental conditions90,91,92. One significant aspect of these interactions involves the production and utilization of siderophores. Each organism produces siderophores to scavenge iron from the surroundings, thereby reducing the availability of iron for the others. In some cases, as demonstrated in the present work for P. aeruginosa and in previous studies by Jurkevitch et al. on P. aeruginosa and P. putida32,93, Pseudomonas and fungi or Streptomyces may engage in cooperative interactions regarding siderophore production and utilization. Indeed, the siderophores produced by fungi and Streptomyces can be used by P. aeruginosa, leading to cooperative relationships where both partners benefit from increased iron acquisition. These interactions between P. aeruginosa, P. putida, fungi, Streptomyces, and the siderophores they produce are dynamic and can vary depending on factors such as environmental conditions, nutrient availability, and the specific microbial species involved. Understanding these interactions is important not only for elucidating microbial ecology but also for potential applications in agriculture, bioremediation, and medical microbiology.
In conclusion, we have demonstrated that P. aeruginosa can utilize the nine hydroxamate siderophores tested to access iron, with the TBDTs involved being FpvB, FiuA, FoxA and another unidentified TBDT. These TBDTs exhibit distinct transport selectivities: FpvB transports and imports ferric complexes of all nine siderophores tested, FiuA predominantly recognizes siderophores of the ferrichrome subtype, and FoxA recognizes those of the DFOB family. The expression of FiuA and FoxA is regulated by the absence of iron and the presence of the siderophores they transport, unlike FpvB, which appears to be consistently expressed at the same levels under iron-restricted conditions.
These findings (i) demonstrate that hydroxamate siderophores must play a key role in the dynamics of fungal-bacterial interactions, (ii) provide insights into the ecological balance of microbial communities, and (iii) may have implications for various fields, including agriculture and medicine, where iron availability plays a critical role in microbial pathogenesis and nutrient cycling. For example, our data suggest that FpvB could be an interesting transporter for facilitating the uptake of antibiotics via siderophore vectorization due to its broad binding and transport selectivity. Consequently, understanding the biochemistry and physiological roles of hydroxamate siderophores offers valuable insights into microbial iron homeostasis and opens avenues for the development of innovative strategies to combat bacterial infections.
Methods
Chemicals
The siderophores rhodotoluric acid (RHODO), coprogen, Desferrioxamine G (DFOG), Triacetylfusarinine C (TAFC), fusigen, ferricrocin, ferrichrysin, ferrirhodin and ferrirubin were obtained from Biophore Research Products. The 55FeCl3 was purchased from Perkin Elmer Life and Analytical Science (Waltham, MA, USA) and the protonophore CCCP (carbonyl cyanide m-chlorophenylhydrazone) was purchased from Sigma-Aldrich (Saint-Louis, MO, USA).
Bacterial strains and growth conditions
The P. aeruginosa strains used in this study are listed in Table S2 (Supplemental Materials). For all experiments in iron restricted conditions, the strains were first grown overnight in LB medium with shaking (220 rpm) at 30°C. Bacteria were then washed and resuspended in iron-restricted casamino acid (CAA) medium composed of 5 g l-1 low-iron CAA (Difco, Franklin Lakes, NJ, USA), 1.46 g l-1 K2HPO4 3H2O, and 0.25 g.l-1 MgSO4 7H2O.
Plasmids and strains construction
Enzymes were obtained from ThermoFisher Scientific and Eurofins Genomics and oligonucleotides used in this study are listed in Table S3 (Supplemental Materials). Escherichia coli TOP10 strain (Invitrogen, MS, USA) was used as the host strain for the plasmids. Plasmid construction and mutation in the chromosomal genome of P. aeruginosa were carried out as detailed before94. Briefly, 1400 bp of flanking sequences of the genes fiuA, foxA or fpvB were inserted into the suicide vector pEXG2 using a blunt-end ligation strategy. The plasmids were sequenced and then used to generate chromosomal mutations into P. aeruginosa strain. All deletion mutants were confirmed by Polymerase Chain Reaction (PCR) and sequencing.
Growth assays in iron-restricted conditions
To monitor growth in microplates in the presence of hydroxamate siderophores, P. aeruginosa strains were first grown as described above. After an overnight culture in CAA medium at 30°C, bacteria cultures were then washed and resuspended in CAA medium at an optical density of 0,01 at 600 nm in 96-well plates (Greiner, U-bottom microplate, Frickenhausen, Germany) and in the presence or absence of 10 µM RHODO, coprogen, TAFC, fusigen, DFOG, ferricrocin, ferrichrysin, ferrirhodin or ferrirubin50,51,52,94. Plates were incubated with shaking at 30°C, in a microplate reader (Infinite M200, Tecan). The bacterial growth was monitored every 30 min for 24 h by measuring the optical density at 600 nm. The data for each measurement represents the mean of three biological replicates.
Iron uptake assay
As for the growth assay, P. aeruginosa strains were first grown overnight in LB broth, followed by a second overnight in CAA medium, all at 30°C. Bacterial cultures were then washed and resuspended in CAA medium at an optical density of 0,1 at 600 nm in the presence of 10 µM of one of the siderophores RHODO, coprogen, TAFC, fusigen, DFOG, ferricrocin, ferrichrysin, ferrirhodin or ferrirubin to induce the expression of the corresponding TBDT24,50,60,61,65. Siderophore-55Fe complexes and 55Fe uptake kinetics were carried out as described previously95. Briefly, the uptake of 55Fe into P. aeruginosa cells was monitored for 3 h at a concentration of xenosiderophore-55Fe complexes of 500 nM, and in the presence or absence of 200 µM CCCP, a proton motive force inhibitor96. Xenosiderophore-55Fe complexes were prepared with the siderophore:iron (mol:mol) ratio of 20:1 for coprogen, TAFC, ferricrocin, ferrirhodin, ferrichrysin et ferrirubin, 30:1 for DFOG and 40:1 for RHODO and fusigen to avoid any 55Fe precipitation because not chelated by the xenosiderophores.
Quantitative real-time PCR
To monitor the expression of genes of interest, P. aeruginosa ∆pvdF∆pchA cells were grown as described above. After an overnight culture at 30°C in CAA medium, bacterial cultures were washed and resuspended in CAA medium at an optical density of 0,1 at 600 nm in the absence or presence of 10 µM of RHODO, coprogen, TAFC, fusigen, DFOG, ferricrocin, ferrichrysin, ferrirhodin or ferrirubin. After 8 h of incubation with shaking (220 rpm) at 30°C, 2.5 X 108 cells were harvested from each culture and 2 volumes of RNAprotect Bacteria Reagent (Qiagen, Hilden, Germany) was added. The RNeasy Plus Mini Kit (Qiagen) was used to extract total RNA from the pellet following the manufacturer instructions. 1 µg of RNA was reverse transcribed using the iScript cDNA Synthesis Kit (Biorad, Hercules, CA, USA). Expression of specific genes was measured in a CFX Opus 96 device (Biorad) using the iTaq Universal SYBR Green Supermix (Biorad) and the appropriate primers (Table S4). The transcript levels were normalized using those of uvrD and rpsl. The data are expressed as a log2 ratio (fold-change) relative to the reference condition and are collected from three biological replicates.
Proteomic analysis
P. aeruginosa ∆pvdF∆pchA cells were grown as for quantitative real-time PCR experiment in the absence or presence of 10 µM of RHODO, ferricrocin or ferrichrysin. After 8 h of incubation at 30°C, 5 × 108 cells were collected and used for proteomic analysis. Each cell-culture condition was prepared in biological triplicates and the peptide mixture of every sample was prepared for injection as described previously31.
Ferricrocin treated and untreated samples were prepared and analyzed on a Qexactive Plus (Thermo-Fisher Scientific) mass spectrometer as described in50.
The RHODO and ferrichrysin treated and untreated samples were injected on a reversed phase nanoElute 2 coupled to a TIMS-TOF Pro 2 mass spectrometer (Bruker Daltonik Gmbh) using a DDA-PASEF acquisition strategy. Peptides were separated on the integrated emitter column IonOpticks Aurora Elite (25 cm × 75μM, 1.7μm particle size and 120 Å pore size; AUR3–15075C18-CSI) with 70 min gradients.
Proteins were identified via comparison to the Uniprot, P. aeruginosa database (v 2020_04, strain PAO1, 5564 forward protein sequences) using Mascot algorithm (version 2.8, Matrix Science) with a decoy strategy. Mass error was set to 20 ppm for precursor ions and 30 ppm for fragment ions. Oxidation (M), carbamidomethylation FCR and its untreated controls (C) and acetylation (protein Nter) were considered as variable modifications. Data were then imported into Proline v2.0 software97, proteins were validated with a Mascot pretty rank equal to 1, a Mascot score threshold set at 25 and 1% FDR (False Discovery Rate) on both peptide spectrum matches (PSM score) and protein sets (Protein Set score) and MS1 eXtracted Ion Chromatograms (XIC) were used to quantify each protein. An exhaustive map alignment followed by a median ratio normalization of the intensities was computed on Proline. A 50s cross assignment was carried out within groups only.
For the statistical analyses of the data, Prostar 1.26.4 was used. Partially observed values (POV) were imputed with structured least square adaptative regression (SLSA) method while values missing in the entire condition (MEC) were imputed with det quantile 1%. A LIMMA statistical test with a log (FC) threshold of 1 and a Benjamini–Hochberg correction were used to generate log2(FC) and adjusted p-values. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD052483, Token: BLPXzP6mEmEX.
Data availability
Proteomic data are available via ProteomeXchange with identifier PXD052483. Project accession: PXD052483 Token: BLPXzP6mEmEX.
References
Hider, R. Siderophore mediated absorption of iron. Struct. Bond. 58, 28 (1984).
Dev, S. & Babitt, J. L. Overview of iron metabolism in health and disease. Hemodial. Int. 21(Suppl 1), S6–S20 (2017).
Kraemer, S. M. Iron oxide dissolution and solubility in the presence of siderophores. Aquat. Sci. 66, 3–18 (2004).
Ellermann, M. & Arthur, J. C. Siderophore-mediated iron acquisition and modulation of host-bacterial interactions. Free Radic. Biol. Med. 105, 68–78 (2017).
Schalk, I. J., Mislin, G. L. A. & Brillet, K. Structure, function and binding selectivity and stereoselectivity of siderophore-iron outer membrane transporters. Curr. Top. Membr. 69, 37–66 (2012).
Celia, H. et al. Structural insight into the role of the Ton complex in energy transduction. Nature 538, 60–65 (2016).
Celia, H. et al. Cryo-EM structure of the bacterial Ton motor subcomplex ExbB-ExbD provides information on structure and stoichiometry. Commun. Biol. 2, 358 (2019).
Celia, H., Noinaj, N. & Buchanan, S. K. Structure and stoichiometry of the ton molecular motor. Int. J. Mol. Sci. 21, 375 (2020).
Schalk, I. J. & Guillon, L. Fate of ferrisiderophores after import across bacterial outer membranes: different iron release strategies are observed in the cytoplasm or periplasm depending on the siderophore pathways. Amino Acids 44, 1267–1277 (2013).
Boukhalfa, H. & Crumbliss, A. L. Chemical aspects of siderophore mediated iron transport. Biometals 15, 325–339 (2002).
Zalkin, A., Forrester, J. D. & Templeton, D. H. Crystal and molecular structure of ferrichrome A. Science 146, 261–263 (1964).
Norrestam, R., Stensland, B. & Brändén, C. I. On the conformation of cyclic iron-containing hexapeptides: the crystal and molecular structure of ferrichrysin. J. Mol. Biol. 99, 501–506 (1975).
Raymond, K. N., Dertz, E. A. & Kim, S. S. Enterobactin: an archetype for microbial iron transport. Proc. Natl. Acad. Sci. U.S.A. 100, 3584–3588 (2003).
Konetschny-Rapp, S., Jung, G., Meiwes, J. & Zähner, H. Staphyloferrin A: A structurally new siderophore from Staphylococci. Eur. J. Biochem. 191, 65–74 (1990).
Welz, D. & Braun, V. Ferric citrate transport of Escherichia coli: functional regions of the FecR transmembrane regulatory protein. J. Bacteriol. 180, 2387–2394 (1998).
Silva, A. M., Kong, X., Parkin, M. C., Cammack, R. & Hider, R. C. Iron(III) citrate speciation in aqueous solution. Dalton Trans. 40, 8616–8625 (2009).
Demange, P. et al. Bacterial siderophores: structure and NMR assigment of pyoverdins PaA, siderophores of Pseudomonas aeruginosa ATCC 15692. Biol. Metals 3, 155–170 (1990).
Gibson, F. & Magrath, D. I. The isolation and characterization of a hydroxamic acid (aerobactin) formed by Aerobacter aerogenes 62–1. Biochim. Biophys. Acta Gen. Subj. 192, 175–184 (1969).
Lee, B. H. & Miller, M. J. Natural ferric ionophores: total synthesis of schizokinen, schizokinen A, and arthrobactin. J. Org. Chem. 48, 24–31 (1983).
Mullis, K. B., Pollack, J. R. & Neilands, J. B. Structure of schizokinen, An iron-transport compound from Bacillus megaterium. Biochemistry 10, 4894 (1971).
Winkelmann, G. Structures and functions of fungal siderophores containing hydroxamate and complexone type iron binding ligands. Mycol. Res. 96, 529–534 (1992).
Renshaw, J. C. et al. Fungal siderophores: structures, functions and applications. Mycol. Res. 106, 1123–1142 (2002).
Winkelmann, G. Ecology of siderophores with special reference to the fungi. Biometals 20, 379–392 (2007).
Schalk, I. J. & Perraud, Q. Pseudomonas aeruginosa and its multiple strategies to access iron. Environ. Microbiol. 25, 811–831 (2022).
Cornelis, P. & Bodilis, J. A survey of TonB-dependent receptors in fluorescent pseudomonads. Environ. Microbiol. Rep. 1, 256–262 (2009).
Schalk, I. J. & Cunrath, O. An overview of the biological metal uptake pathways in Pseudomonas aeruginosa. Environ. Microbiol. 18, 3227–3246 (2016).
Hannauer, M., Barda, Y., Mislin, G. L., Shanzer, A. & Schalk, I. J. The ferrichrome uptake pathway in Pseudomonas aeruginosa involves an iron release mechanism with acylation of the siderophore and a recycling of the modified desferrichrome. J. Bacteriol. 192, 1212–1220 (2010).
Normant, V. et al. Nocardamine-Dependent Iron Uptake in Pseudomonas aeruginosa: Exclusive Involvement of the FoxA Outer Membrane Transporter. ACS Chem. Biol. 15, 2741–2751 (2020).
Chan, D. C. K. & Burrows, L. L. Pseudomonas aeruginosa FpvB Is a High-Affinity Transporter for Xenosiderophores Ferrichrome and Ferrioxamine B. mBio 14, e0314922 (2023).
Chan, D. C. K. et al. Interactions of TonB-dependent transporter FoxA with siderophores and antibiotics that affect binding, uptake, and signal transduction. Proc. Natl. Acad. Sci. U.S.A. 120, e2221253120 (2023).
Will, V. et al. Siderophore specificities of the Pseudomonas aeruginosa TonB-dependent transporters ChtA and ActA. FEBS Lett 597, 2963–2974 (2023).
Jurkevitch, E., Hadar, Y. & Chen, Y. Differential siderophore utilization and iron uptake by soil and rhizosphere bacteria. Appl. Environ. Microbiol. 58, 119–124 (1992).
Winkelmann, G. Structural and stereochemical aspects of iron transport in fungi. Biotechnol. Adv. 8, 207–231 (1990).
Neilands, J. B. Siderophores: Structure and function of microbial iron transport compounds. J. Biol. Chem. 270, 26723–26726 (1995).
Van Der Helm, D. & Winkelmann, G. Hydroxamates and Polycarboxylates as Iron Transport Agents (Siderophores) in Fungi. Metal Ions in Fungi (CRC Press, Boca Raton, 1994).
Carrano, C. J. & Raymond, K. N. Coordination chemistry of microbial iron transport compounds: Rhodotorulic acid and iron uptake in Rhodotorula pilimanae. J. Bacteriol. 136, 69–74 (1978).
Calvente, V., de Orellano, M. E., Sansone, G., Benuzzi, D. & Sanz de Tosetti, M. I. Effect of nitrogen source and pH on siderophore production by Rhodotorula strains and their application to biocontrol of phytopathogenic moulds. J. Ind. Microbiol. Biotechnol. 26, 226–229 (2001).
Wong, G. B., Kappel, M. J., Raymond, K. N., Matzanke, B. & Winkelmann, G. Coordination chemistry of microbial iron transport compounds. 24. Characterization of coprogen and ferricrocin two ferric hydroxamate siderophores. J. Am. Chem. Soc. 105, 810–815 (1983).
Hossain, M. B., Eng-Wilmot, D. L., Loghry, R. A. & Van der Helm, D. Circular dichroism, crystal structure, and absolute configuration of the siderophore ferric N, N’, N"-triacetylfusarinine, FeC39H57N6O15. J. Am. Chem. Soc. 102, 5766–5773 (1980).
Aguiar, M. et al. Uptake of the Siderophore Triacetylfusarinine C, but Not Fusarinine C, Is Crucial for Virulence of Aspergillus fumigatus. mBio 13, e0219222 (2022).
Diekmann, H. & Zähner, H. Konstitution von Fusigen und dessen Abbau zu Δ2-Anhydromevalonsäurelacton. Eur. J. Biochem. 3, 213–218 (1967).
Haselwandter, K. et al. Isolation and identification of hydroxamate siderophores of ericoid mycorrhizal fungi. Biometals 5, 51–56 (1992).
Haselwandter, K. et al. Linear fusigen as the major hydroxamate siderophore of the ectomycorrhizal Basidiomycota Laccaria laccata and Laccaria bicolor. Biometals 26, 969–979 (2013).
Reissbrodt, R., Rabsch, W., Chapeaurouge, A., Jung, G. & Winkelmann, G. Isolation and identification of ferrioxamine G and E in Hafnia alvei. Biol. Metals 3, 54–60 (1990).
Jalal, M. A. et al. Extracellular siderophores from Aspergillus ochraceous. J. Bact. 158, 683–688 (1984).
Fidelis, K., Hossain, M. B., Jalal, M. A. & van der Helm, D. Structure and molecular mechanics of ferrirhodin. Acta. Crystallogr. C 46(Pt 9), 1612–1617 (1990).
Haselwandter, K. & Winkelmann, G. Ferricrocin–an ectomycorrhizal siderophore of Cenococcum geophilum. Biometals 15, 73–77 (2002).
Konetschny-Rapp, S., Jung, G., Huschka, H.-G. & Winkelmann, G. Isolation and identification of the principal siderophore of the plant pathogenic fungus Botrytis cinerea. Biol. Metals 1, 90–98 (1988).
Munawar, A., Marshall, J. W., Cox, R. J., Bailey, A. M. & Lazarus, C. M. Isolation and characterisation of a ferrirhodin synthetase gene from the sugarcane pathogen Fusarium sacchari. Chembiochem 14, 388–394 (2013).
Perraud, Q. et al. Phenotypic adaption of Pseudomonas aeruginosa by hacking siderophores produced by other microorganisms. Mol. Cell Proteomics 19, 589–607 (2020).
Perraud, Q. et al. Opportunistic use of catecholamine neurotransmitters as siderophores to access iron by Pseudomonas aeruginosa. Environ. Microbiol. 24, 878–893 (2022).
Fritsch, S. et al. Uptake mechanisms and regulatory responses to MECAM- and DOTAM-Based artificial Siderophores and Their Antibiotic Conjugates in Pseudomonas aeruginosa. ACS Infect. Dis. 8, 1134–1146 (2022).
Gasser, V. et al. Catechol siderophores repress the pyochelin pathway and activate the enterobactin pathway in Pseudomonas aeruginosa: an opportunity for siderophore-antibiotic conjugates development. Environ. Microbiol. 18, 819–832 (2016).
Cunrath, O., Geoffroy, V. A. & Schalk, I. J. Metallome of Pseudomonas aeruginosa: a role for siderophores. Environ. Microbiol. 18, 3258–3267 (2016).
Coy, M. & Neilands, J. B. Structural dynamics and functional domains of the fur protein. Biochemistry 30, 8201–8210 (1991).
Hassett, D. J. et al. Ferric uptake regulator (Fur) mutants of Pseudomonas aeruginosa demonstrate defective siderophore-mediated iron uptake, altered aerobic growth, and decreased superoxide dismutase and catalase activities. J. Bacteriol. 178, 3996–4003 (1996).
Fillat, M. F. The FUR (ferric uptake regulator) superfamily: diversity and versatility of key transcriptional regulators. Arch. Biochem. Biophys. 546, 41–52 (2014).
Nader, S. et al. New insights into the tetrameric family of the Fur metalloregulators. Biometals 32, 501–519 (2019).
Perraud, Q. et al. Phenotypic Adaptation of Pseudomonas aeruginosa in the Presence of Siderophore-Antibiotic Conjugates during Epithelial Cell Infection. Microorganisms 8, 1820 (2020).
Heinrichs, D. E. & Poole, K. PchR, a regulator of ferripyochelin receptor gene (fptA) expression in Pseudomonas aeruginosa, functions both as an activator and as a repressor. J. Bacteriol 178, 2586–2592 (1996).
Dean, C. R., Neshat, S. & Poole, K. PfeR, an enterobactin-responsive activator of ferric enterobactin receptor gene expression in Pseudomonas aeruginosa. J. Bacteriol. 178, 5361–5369 (1996).
Michel, L., Gonzalez, N., Jagdeep, S., Nguyen-Ngoc, T. & Reimmann, C. PchR-box recognition by the AraC-type regulator PchR of Pseudomonas aeruginosa requires the siderophore pyochelin as an effector. Mol. Microbiol. 58, 495–509 (2005).
Llamas, M. A. et al. The heterologous siderophores ferrioxamine B and ferrichrome activate signaling pathways in Pseudomonas aeruginosa. J. Bacteriol. 188, 1882–1891 (2006).
Llamas, M. A. et al. Characterization of five novel Pseudomonas aeruginosa cell-surface signalling systems. Mol. Microbiol. 67, 458–472 (2008).
Llamas, M. A., Imperi, F., Visca, P. & Lamont, I. L. Cell-surface signaling in Pseudomonas: stress responses, iron transport, and pathogenicity. FEMS Microbiol. Rev. 38, 569–597 (2014).
Luscher, A. et al. Plant-derived catechols are substrates of TonB-dependent transporters and sensitize Pseudomonas aeruginosa to siderophore-drug conjugates. MBio 22 (2022).
Winsor, G. L. et al. Pseudomonas genome database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res. 39, D596-600 (2011).
Hider, R. C. & Kong, X. Chemistry and biology of siderophores. Nat. Prod. Rep. 27, 637–657 (2011).
Ghysels, B. et al. FpvB, an alternative type I ferripyoverdine receptor of Pseudomonas aeruginosa. Microbiology 150, 1671–1680 (2004).
Josts, I., Veith, K. & Tidow, H. Ternary structure of the outer membrane transporter FoxA with resolved signalling domain provides insights into TonB-mediated siderophore uptake. eLife 8, e48528 (2019).
Chan, D. C. K. & Burrows, L. L. Thiocillin and micrococcin exploit the ferrioxamine receptor of Pseudomonas aeruginosa for uptake. J. Antimicrob. Chemother. 76, 2029–2039 (2021).
Bastiaansen, K. C., van Ulsen, P., Wijtmans, M., Bitter, W. & Llamas, M. A. Self-cleavage of the Pseudomonas aeruginosa cell-surface signaling anti-sigma factor FoxR occurs through an N-O acyl rearrangement. J. Biol. Chem. 290, 12237–12246 (2015).
Mislin, G. L. A. & Schalk, I. J. Siderophore-dependent iron uptake systems as gates for antibiotic Trojan horse strategies against Pseudomonas aeruginosa. Metallomics 6, 408–420 (2014).
Negash, K. H., Norris, J. K. S. & Hodgkinson, J. T. Siderophore-antibiotic conjugate design: New drugs for bad bugs?. Molecules 24, E3314 (2019).
Pham, T.-N., Loupias, P., Dassonville-Klimpt, A. & Sonnet, P. Drug delivery systems designed to overcome antimicrobial resistance. Med. Res. Rev. 39, 2343–2396 (2019).
Al Shaer, D., Al Musaimi, O., de la Torre, B. G. & Albericio, F. Hydroxamate siderophores: Natural occurrence, chemical synthesis, iron binding affinity and use as Trojan horses against pathogens. Eur. J. Med. Chem. 208, 112791 (2020).
Southwell, J. W., Black, C. M. & Duhme-Klair, A.-K. Experimental methods for evaluating the bacterial uptake of Trojan Horse antibacterials. ChemMedChem 16, 1063–1076 (2021).
Faure, E., Kwong, K. & Nguyen, D. Pseudomonas aeruginosa in chronic lung infections: How to adapt within the host?. Front. Immunol. 9, 2416 (2018).
Wood, S. J., Kuzel, T. M. & Shafikhani, S. H. Pseudomonas aeruginosa: Infections, animal modeling, and therapeutics. Cells 12, 199 (2023).
Kelly, B. T., Pennington, K. M. & Limper, A. H. Advances in the diagnosis of fungal pneumonias. Expert. Rev. Respir. Med. 14, 703–714 (2020).
Méar, J.-B. et al. Candida albicans and Pseudomonas aeruginosa interactions: more than an opportunistic criminal association?. Med. Mal. Infect. 43, 146–151 (2013).
Oliveira, M., Cunha, E., Tavares, L. & Serrano, I. P. aeruginosa interactions with other microbes in biofilms during co-infection. AIMS Microbiol. 9, 612–646 (2023).
Kahl, L. J. et al. Interkingdom interactions between Pseudomonas aeruginosa and Candida albicans affect clinical outcomes and antimicrobial responses. Curr. Opin. Microbiol. 75, 102368 (2023).
Sass, G. et al. Studies of Pseudomonas aeruginosa Mutants Indicate Pyoverdine as the Central Factor in Inhibition of Aspergillus fumigatus Biofilm. J. Bacteriol. 200, e00345-e417 (2017).
Sass, G. et al. Aspergillus-Pseudomonas interaction, relevant to competition in airways. Med. Mycol. 57, S228–S232 (2019).
Schalk, I. J., Rigouin, C. & Godet, J. An overview of siderophore biosynthesis among fluorescent Pseudomonads and new insights into their complex cellular organization. Environ. Microbiol. 22, 1447–1466 (2020).
Schrettl, M. et al. Distinct roles for intra- and extracellular siderophores during Aspergillus fumigatus infection. PLoS Pathog. 3, 1195–1207 (2007).
Lopez-Medina, E. et al. Candida albicans Inhibits Pseudomonas aeruginosa virulence through suppression of Pyochelin and Pyoverdine Biosynthesis. PLoS Pathog. 11, e1005129 (2015).
Lee, K. et al. The ferrichrome receptor A as a new target for Pseudomonas aeruginosa virulence attenuation. FEMS Microbiol. Lett. 363, fnw104 (2016).
Berg, G., Eberl, L. & Hartmann, A. The rhizosphere as a reservoir for opportunistic human pathogenic bacteria. Environ. Microbiol. 7, 1673–1685 (2005).
Braga, R. M., Dourado, M. N. & Araújo, W. L. Microbial interactions: ecology in a molecular perspective. Braz. J. Microbiol. 47, 86–98 (2016).
Babalola, O. O. et al. Rhizosphere microbiome cooperations: Strategies for sustainable crop production. Curr. Microbiol. 78, 1069–1085 (2021).
Jurkevitch, E., Hadar, Y., Chen, Y., Libman, J. & Shanzer, A. Iron uptake and molecular recognition in Pseudomonas putida: receptor mapping with ferrichrome and its biomimetic analogs. J. Bacteriol. 174, 78–83 (1992).
Gasser, V. et al. The esterase PfeE, the Achilles’ Heel in the battle for iron between Pseudomonas aeruginosa and Escherichia coli. Int. J. Mol. Sci. 22, 2814 (2021).
Hoegy, F. & Schalk, I. J. Monitoring iron uptake by siderophores. Methods Mol. Biol. 1149, 337–346 (2014).
Clément, E., Mesini, P. J., Pattus, F., Abdallah, M. A. & Schalk, I. J. The binding mechanism of pyoverdin with the outer membrane receptor FpvA in Pseudomonas aeruginosa is dependent on its iron-loaded status. Biochemistry 43, 7954–7965 (2004).
Bouyssié, D. et al. Proline: an efficient and user-friendly software suite for large-scale proteomics. Bioinformatics 36, 3148–3155 (2020).
Acknowledgements
Authors acknowledge the Centre National de la Recherche Scientifique (CNRS) for general financial support. This work was also supported by a CNRS grant (80/Prime 2020), a grant from the Agence Nationale de la Recherche (ANR, grant number: ANR-22-CE44-0024-01), and a grant from the associations Vaincre la Mucoviscidose and Gregory Lemarchal (grant number: RF20230503252). We also acknowledge the Interdisciplinary Thematic Institute (ITI) InnoVec (Innovative Vectorization of Biomolecules, IdEx, ANR-10-IDEX-0002). The mass spectrometry instrumentation at the IBMC was funded by the Interdisciplinary Thematic Institute IMCBio+ , as part of the ITI 2021-2028 program of the University of Strasbourg, CNRS and Inserm, IdEx Unistra (ANR-10-IDEX-0002), SFRI-STRAT’US (ANR-20-SFRI-0012), and EUR IMCBio (ANR-17-EURE-0023) under the framework of the French Investments of the France 2030 Program as well as by EquipEx I2MC (ANR-11-EQPX-0022). Further funding supports were from the CPER 2021-2027 (ImaProGen Project) and the Strasbourg eurometropole. VW received a fellowship from the Ministry of Research and Education. We thank Anne Bonneau for providing the strain ∆pvdF∆pchA∆fpvB.
Author information
Authors and Affiliations
Contributions
VW, CF, and VN conducted the 55Fe uptake assays, qRT-PCR, and bacterial growth assays. VW also conceptualized the experiments, prepared the manuscript figures, and participated in writing, review, and editing. FV generated some of the mutants used in the study. LK and JC performed the proteomic experiments and analyses, and contributed to writing, review, and editing. IJS conceptualized and supervised the biological experiments, contributed to writing, review, and editing the manuscript, and was responsible for funding acquisition and project administration. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Will, V., Frey, C., Normant, V. et al. The role of FoxA, FiuA, and FpvB in iron acquisition via hydroxamate-type siderophores in Pseudomonas aeruginosa. Sci Rep 14, 18795 (2024). https://doi.org/10.1038/s41598-024-69152-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-69152-6
- Springer Nature Limited