Abstract
This study aimed to determine the sequence type (ST) of Bartonella henselae infecting small Indian mongooses from Saint Kitts via multi-locus sequence typing (MLST). This investigation used stored EDTA blood (n = 22) samples from mongooses previously identified as positive for B. henselae. Chocolate agar plates were enriched with Bartonella alpha-Proteobacteria growth medium (BAPGM) to culture and isolate Bartonella from the blood samples. To perform MLST, DNA was extracted and purified from isolates followed by amplification by conventional PCR (300–500 bp) for eight genes (16S rDNA, batR, gltA, groEL, ftsZ, nlpD, ribC, and rpoB). Bartonella henselae STs were deposited in the PubMLST repository. Out of 22 B. henselae-positive blood samples, isolates were obtained from 12 mongooses (54.5%; 12/22). Each mongoose was infected with one ST. The studied mongoose population was infected with sequence types ST2, ST3, ST8, and a novel ST represented by ST38. Bartonella henselae ST2, ST3 and ST8 infecting mongooses are known to circulate in humans and cats, with ST2 and ST8 associated with Cat Scratch Disease (bartonellosis) in humans. The results presented herein denote the circulation of B. henselae STs with zoonotic potential in mongooses with risk of B. henselae transmission to humans.
Similar content being viewed by others
Introduction
The genus Bartonella belongs to the family Bartonellaceae of the sub-class Alpha-2-Proteobacteria. It comprises facultative intracellular Gram-negative, haemotropic, slow-growing, vector-borne bacteria (Chomel et al.1). Over the last 20 years, the number of Bartonella species identified from a wide range of mammals has increased considerably1. Noteworthy, Bartonella bacteria are a seemingly ubiquitous genus with their DNA demonstrated in a plethora of species throughout the animal kingdom, including, but not limited to, domestic2 and wildlife hosts3,4,5. Among the species known or suspected to be pathogenic for humans, Bartonella henselae is the genus’s primary zoonotic agent, and cats are often natural asymptomatic reservoirs6. Though the domestic cat serves as a primary host for B. henselae, this is the species most found in wild carnivores7.
The small Indian mongoose (Urva auropunctata) is a highly invasive, terrestrial carnivorous wild mammal that belongs to the Herpestidae family, Feliformia superfamily, of ‘cat-like’ animal species, widespread in most Caribbean islands and listed under the 100 worst invasive species (IUCN 2000). Mongooses were also recognized as reservoirs of Bartonellae bacteria, with Bartonella henselae being detected in specimens from Japan8 and Grenada9. Our research group recently identified10 that B. henselae is also prevalent in mongooses and cat fleas (Ctenocephalides felis) collected from them on the Island of Saint Kitts, Wes Indies.
In humans B. henselae is responsible for Cat Scratch Disease (CDS). Although CSD is usually a benign self-limiting disease that develops as fever and lymphadenopathy, more recently, B. henselae has been found in association with several unspecific health conditions. These include but are not limited to, central nervous system syndromes, fever of unknown origin (FUO), hepatosplenic disease, negative blood culture endocarditis, neuroretinitis, and musculoskeletal and cutaneous lesions11.
Although B. henselae is a zoonotic pathogen, the worldwide distribution of its sequence types (STs) is not homogeneous. Multi-locus sequence typing (MLST) has shown that some of the STs of B. henselae are only found in the feline population, representing a group of strains less pathogenic to humans12. Until now, 37 STs of B. henselae have been identified13. Among the B. henselae strains described in humans, the strains classified into ST1 are causative for most cases of CSD14. Other zoonosis-associated strains belong to ST2, ST5, and ST8. It is postulated that those STs could possess additional virulence factors capable of coding for a more efficient transmission from cats to humans or, alternatively, better survival of the pathogen in the human host15. Conversely, most feline STs belong to ST4, ST6, and ST716. ST5 was the only ST described in fleas so far17.
Batonella henselae STs can vary according to geographic distribution; most isolates of B. henselae are obtained from cats in Asia (Japan and the Philippines)14, Israel, North America15, Central America (Guatemala)18 and South America (Argentina)19 were the zoonotic type ST1. Conversely, there is little description of zoonotic strains of B. henselae in the cat populations of Europe, (France, Germany, Italy, the Netherlands, and the United Kingdom) and Australia15.
In regions such as the Caribbean, where mongooses and domestic cats have direct interactions and share the same ectoparasite, Ctenocephalides felis9, which is the vector for the bacteria20,21 it is possible that mongooses can serve as potential reservoirs of infection with B. henselae STs that are found in cats but are also STs pathogenic for humans. To date, all B. henselae STs deposited in PubMLST belong to cats, humans, or fleas (https://pubmlst.org/organisms/bartonella-henselae). We are not aware of studies involving wildlife. To that end, the aim of this study was to determine the Sequence Type (ST) of B. henselae infecting small Indian mongooses from Saint Kitts via MLST analysis.
Results
Culture and isolation of Bartonella henselae from mongooses’ blood
Out of 22 mongooses’ blood positive for B. henselae infection, Bartonella-like colonies were obtained from 12 blood samples and confirmed to be Bartonella bacteria via qPCR after the first passage. Accordingly, Bartonella isolates (after six passages of subculturing the bacteria in six consecutive plates) were obtained from 12 mongooses (54.5%; 12/22), with a total of 22 pure isolates, five isolates from different plates obtained from one mongoose; a total two isolates from six mongooses, and one isolate obtained from five mongooses.
Genotyping via multi-locus sequence typing (MLST) analysis of Bartonella henselae
All obtained isolates were confirmed to be B. henselae by gltA sequencing and Blastn analyses (with 99–100% of query cover and 99.86–100% identity to B. henselae to GenBank deposited sequences: KY913627.1; KY913626.1; MN107415.1; MN107415.3). MLST based on 16S rRNA, batR, ftsZ, gltA, groEL, nlpD, ribC, and rpoB genes was achieved for all 22 isolates, resulting in the presence of three previously described STs (2, 3, and 8) and novel sequence type represented by ST38. All the mongooses from which more than one colony was obtained were infected with just one sequence type. Of the 12 mongooses, 33% (4/12) were bacteremic with ST3, followed by 25% (3/12) infected with ST8, 25% (3/12) with ST2, and the less prevalent ST (16%; 2/12) was the newly described ST38 (Table 1).
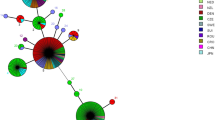
The new ST38 was closely related and originated from ST2. While this ST was only found in mongooses, the other STs (STs 2, 3, and 8) described in mongooses have been reported in humans and cats. Of the B. henselae STs detected in mongooses, ST8 is the most geographically dispersed worldwide, followed by ST2 and ST3 that were only shared with a few countries and ST38 only described in St. Kitts. The relatedness of the B. henselae STs found in the present study with those detected worldwide was assessed by GrapeTree analyses (Fig. 1).
Relatedness of the Bartonella henselae sequence types (STs) detected in small Indian mongooses from Saint Kitts in the present study by (A) sequence type; (B) sample source; (C) geographical distribution. Mongooses’ samples in (B) are identified as mongooses. The parenthesis in each colored coded column represents the numbers of samples per ST (A), per source (B) and per country (C).
Discussion
To date, 37 B. henselae sequence types have been described, with 28 unique STs from cats, eight shared between cats and humans, and one described in cats, humans, and fleas (https://pubmlst.org/organisms/bartonella-henselae)22. To the best of the authors’ knowledge, this is the first investigation exploring STs of B. henselae circulating in a wildlife species, and the first study within the Caribbean that included B. henselae genotyping. This approach has already been used for genotyping B. henselae in cats and/or humans from North America (USA), South America (Argentina, Brazil)13,19,23 Central America (Guatemala)18 Europe (United Kingdom, Germany, Italy, France, Croatia, Spain, Germany and Czech Republic)15,16,17,24,25 Asia (Japan, the Philippines, Turkey, Israel)14,15,26, and Oceania (Australia, New Zealand)15,27.
Upon initially conducting the MLST, purified isolates following three culture passages were used. Regrettably, incomplete gene amplification from the 8 loci led to an inability to obtain a comprehensive MLST profile. Consequently, a new attempt was made after six culture passages, akin to the authors’ approach with the cat blood samples utilized for Whole Genome Sequencing in previous experiments28. On the contrary, another previous research revealed that using isolation in chocolate agar followed by MLST in cats across up to three passages was enough for a complete MSLT profile23. The authors are unsure if the inability to retrieve all genes after only three passages was due to the host species (mongooses). Considering this, the authors advocate for the implementation of these multiple culture passages, especially for the MLST on mammals different from their preferred host, as the new standard protocol.
In our study, ST2, ST3, ST8, and the newly described ST38 were detected in mongooses’ blood. Previously, ST2, ST3, and ST8 were described in humans and cats, with ST8 being the most geographically dispersed from the three known STs detected in our study, with reports in European countries, New Zealand, and Argentina. Otherwise, ST1, ST5, and ST6 have a worldwide distribution, as they are present on three continents (America, Europe, and Oceania)15 but were not detected in the B. henselae isolates from the Caribbean mongooses. Whether this is because those STs do not infect mongooses or they do not circulate within the Caribbean region, is yet to be determined. ST8 is seemingly the most frequent ST infecting humans, along with ST1, ST2 (detected in this study) and ST5. Bartonella henselae strains belonging to ST1 may be more likely to be associated with zoonosis in Australia, Japan, and potentially the United States, however, this association does not necessarily apply elsewhere16. Other less frequent STs in humans include ST3 (detected in this study), ST4, ST6, ST7, and ST922. In Spain, all three STs found in our study (ST2, ST3, ST8) were associated with human infection17, while ST2 also infected humans in Australia22. On the other hand, ST2 and ST8 have been encountered infecting humans with symptoms of Cat Scratch Disease (CSD) in the UK16 and ST2 in Romania22. Investigations within the Caribbean should survey the STs circulating and causing disease in humans.
A new allelic combination described as B. henselae ST3822 was found in the mongooses from St. Kitts. This novel ST38 originated from ST2 that was previously detected in cats and/or humans from Australia, Spain, and the UK22. ST38 represents a clonal evolution as described previously24. Bartonella henselae displays specific adaptations of a complex facultative intracellular lifestyle that enables the colonisation of distinct mammalian reservoir hosts. This remarkable host adaptability has a multifactorial basis and is thought to be driven by horizontal gene transfer (HGT). Highly efficient HGT is described for Bartonellae and could thus drive evolution of this genus, expected to occur through the recurrent transmission bottlenecks during the complex infection cycle of these pathogens in their mammalian reservoir hosts and arthropod vectors29. Ctenocephalides felis fleas, vectors of B. henselae among cats, may have an important role in the generation of the genetic diversity of this agent. Homologous recombination among genotypes of the same or different species of Bartonella can occur in the vector intestine30,31. Future investigations should aim to explore if ST38 is only circulating on a specific host (mongooses) or specific geographical area (St. Kitts).
Only single ST infections were documented in mongooses from this study in St Kitts. Although possible in cats, co-infection with more than one B. henselae STs are infrequently described. Three studies have described co-infections so far. ST1 and ST5 were detected in the blood of one cat in Brazil13; ST5 and ST7 in a cat, and ST6 and ST8 in another cat from the UK16 and B. henselae type I (Houston I) and Type II (Marseille) STs were documented in experimental co-transmission of the two B. henselae, to Specific Pathogen Free (SPF) kittens by the natural vector, the cat flea32. The latter study32 hypothesized that co-infection with ST1 and ST2 resulted in in vivo competition between the two STs, with a selective advantage of ST2 over ST1 during the 2 years of persistent bloodstream infection. It is possible that the absence of co-infections observed in mongooses is due to competition between STs or that co-infection does not occur in this animal species. Future longitudinal studies should explore those hypotheses.
The results presented herein denote the circulation of B. henselae STs with zoonotic potential in mongooses (ST2, ST3, ST8), with a possible risk of B. henselae infection in humans. Encounters between mongooses and humans throughout the Caribbean islands are known to be frequent, and direct human exposure to this wild mammal was described before in Puerto Rico, resulting in a bite33. The small Indian mongoose is a highly invasive, terrestrial wild mammal34 widespread in most Caribbean islands. Being a member of the Herpestidae family, the mongooses fall under Feliformia (NCBI: txid48418), a suborder of carnivores which also includes animals such felines, hyenas, and civets. Bartonella henselae is described in several wildlife species from the suborder Feliformia, being identified in wild cats, civets, and mongooses8. Wildlife provides blood meals for vector growth and reproduction, serves as pathogen reservoirs, and can disperse vectors and pathogens35; as such, it is important to identify B. henselae STs in wildlife, including but not limited to mongooses. By 2050, invasive mongoose populations are likely to expand their geographic distribution across island and continental ecosystems beyond the Caribbean including North America and Europe35. When hosts disperse vectors and their associated pathogens into new, and suitable environments, a rise in vector-borne diseases may follow36. The recognition of the STs infecting mongooses within the Caribbean for the first time provides the foundation for preventative measures to be applied, which might include a selection of an appropriate strain for the development and/or improvement of local diagnostic tests37 and future vaccine development32.
Conclusions
This study investigated B. henselae sequence types (STs) circulating in a wildlife species and was the first study within the Caribbean that incorporated B. henselae genotyping. Only single ST infections were observed in mongooses from Saint Kitts. The sequence types ST2, ST3, ST8 and a novel ST represented by ST38, were identified in mongooses’ blood isolates. Bartonella henselae ST2, ST3 and ST8 infecting mongooses are known to circulate in humans and cats, with ST2 and ST8 associated with Cat Scratch Disease in humans. The results presented herein denote the circulation of B. henselae STs with zoonotic potential in mongooses with possible risk of B. henselae transmission to humans within the Caribbean, since encounters between mongooses and humans throughout the islands are known to be frequent. The novel ST38 originated from ST2 and represents a clonal evolution of Bartonellae in a certain geographical region and/or host, and future investigations should aim to explore its geographic dispersion and other reservoirs.
Methods
Mongooses samples origin
The research was conducted with mongooses’ stored blood samples. Animals were captured in Saint Kitts (17.3578° N, 62.7830° W) between August 2019 and January 2020 and bled by venipuncture of the cranial vena cava and released in the same trapping location. All methods were carried out in accordance with relevant guidelines and regulations and reported in accordance with ARRIVE guidelines and extensive information on the sampling procedure can be found in10. The use of mongooses’ samples for research was approved by the RUSVM Institutional Animal Care and Use Committee (IACUC) (TSU7.24.19). Hence, this study used PCR EDTA blood (n = 22) stored samples (− 80 °C) from mongooses previously identified as positive for B. henselae10 via DNA sequence analyses (nBLAST), phylogenetic inference and haplotype diversity.
Culture and isolation of Bartonella henselae from mongooses’ blood
Culture and isolation from 22 EDTA blood samples from B. henselae-positive mongooses was attempted. Accordingly, the samples were diluted 1:2 in Schneider’s Insect medium and 2.0 μg/mL amphotericin B to enhance the Bartonella isolation and reduce fungal contamination, respectively, as previously recommended38. Briefly, 100 µL of each diluted sample was directly plated on chocolate agar and incubated at 37 °C with a 10% CO2 atmosphere. After growth in the first plate, any small, round, and Bartonella-like colony was: (1) diluted in 300 µL Bartonella alpha-Proteobacteria growth medium (BAPGM)39 and sub-cultured with a loop for re-isolation in two chocolate agar plates, each subdivided into three and identified as sub-colonies A, B, C and sub-colonies D, E F; (2) diluted in 200 µL of PBS for DNA extraction by a thermal protocol (i.e. 95 °C for 12 min). DNA was collected from the supernatant after centrifugation at 4 °C at 8500 rpm for 5 min. and submitted to qPCR for Bartonella spp.40. Only colonies molecularly confirmed as Bartonella spp. were subsequently re-inoculated in BAPGM enriched agar chocolate plates, with a total of six passages, until pure isolated cultures were obtained (up to eight weeks). DNA was extracted from the pure isolated colonies using the Illustra Tissue and Cells genomic Prep Mini Spin Kit, following the manufacturer’s recommendations. Extracted DNA was submitted to Bartonella sp. conventional (c)PCR for gltA41 for sequencing and further confirmation that the pure isolates corresponded to B. henselae.
Genotyping via multi-locus sequence typing (MLST) analysis of Bartonella henselae
To identify B. henselae sequence types (STs), DNA obtained from all B. henselae blood isolates after six passages were amplified by cPCR (300–500 bp products) for eight genes (16S rDNA, batR, gltA, groEL, ftsZ, nlpD, ribC, and rpoB), to perform MLST, according to a previously described protocol15,27. Each reaction mixture comprised 12.5 µL of 2 × PCR master mix (Promega®, Madison, WI, USA), 0.5 µL of a 20 ρmol/μL solution of both forward and reverse primers (Table 2), 10.5 µL sterile, distilled water, and 1 µL DNA extracted from the B. henselae colony.
As positive and negative controls for the cPCR, DNA extracted from blood of a B. henselae naturally infected cat42 and nuclease-free water (Promega®, Madison, WI, USA) were used, respectively. The thermic profile consisted of 96 °C for 5 min followed by 40 cycles of 96 °C for 10 s, 55 °C for 10 s and 72 °C for 50 s, with a final extension step of 72 °C for 10 min15,27. A T100TM Thermal Cycler (Bio-Rad, Hercules, CA, USA) was used for all the cPCR reactions. The PCR products were stained with SYBR® Safe DNA gel stain (Thermo ScientificTM, USA) and run in a 1.5% agarose gel electrophoresis. The positive bands at expected sizes were purified by enzymatic reaction using Exo-CIP™ Rapid PCR Cleanup Kit (New England Biolab inc., Ipswich, MA, USA), according to the manufacturer’s instructions. Sequencing of the purified DNA was performed in Macrogen (Seoul, Korea). To obtain consensus sequences, forward and reverse sequences were processed in Geneious Prime 7.1. Percentages of identity were obtained using BLASTn43.
Bartonella henselae obtained consensus sequences had their strain identified (ST), according to the online database of B. henselae MLST and were deposited in the PubMLST repository (https://pubmlst.org/organisms/bartonella-henselae)22, under the numbers: 476-494. Bartonella henselae STs found in this study were compared to other worldwide B. henselae STs from humans, domestic cats, and fleas, deposited at the MLST bank (https://pubmlst.org/organisms/bartonella-henselae). A Graphtree was constructed using the pubMLST tool22 to represent (A) the frequency of each B. henselae ST available on pubMLST; (B) the sample source and (C) their distribution by country.
Ethics approval
This study’s Animal capture and handling were approved by the University of Montreal's Animal Use Ethics Committee (CÉUA 19-Rech-1993 and 19-Rech-1945), and the use of the samples was endorsed by the Ross University School of Veterinary Medicine (RUSVM) Institutional Animal Care and Use Committee (IACUC #TSU7.24.19).
Data availability
The datasets generated and analyzed during the current study are available in the PubMLST (https://pubmlst.org/organisms/bartonella-henselae) repository submission number: 476-494.
References
Chomel, B. B. et al. Ecological fitness and strategies of adaptation of Bartonella species to their hosts and vectors. Vet. Res. 40, 29 (2009).
Álvarez-Fernández, A., Breitschwerdt, E. B. & Solano-Gallego, L. Bartonella infections in cats and dogs including zoonotic aspects. Parasit. Vectors. 11, 1–21 (2018).
Maggi, R. G. et al. Bartonella henselae in captive and hunter-harvested Beluga (Delphinapterus Leucas). J. Wildl. Dis. 44, 871–877 (2008).
Jones, D. C. et al. Distribution, diversity, and host specificity of Bartonella in rodents from the Southeastern United States. Am. J. Trop. Med. Hyg. 57, 578–588 (1997).
Veikkolainen, V., Vesterinen, E. J., Lilley, T. M. & Pulliainen, A. T. Bats as reservoir hosts of human bacterial pathogen, Bartonella mayotimonensis. Emerg. Infect. Dis. https://doi.org/10.3201/eid2006.130956 (2014).
Mogollon-Pasapera, E., Otvos, L., Giordano, A. & Cassone, M. Bartonella: Emerging pathogen or emerging awareness?. Int. J. Infect. Dis.. 13, 3–8 (2009).
Kosoy, M. & Goodrich, I. Comparative ecology of Bartonella and Brucella infections in wild carnivores. Front. Vet. Sci. 5, 1–32 (2019).
Sato, S. et al. Small indian mongooses and masked palm civets serve as new reservoirs of Bartonella henselae and potential sources of infection for humans. Clin. Microbiol. Infect. 19, 1181–1187 (2013).
Jaffe, D. A. et al. Bartonella henselae in small Indian mongooses (Herpestes auropunctatus) from Grenada, West Indies. Vet. Microbiol. 2018(216), 119–122 (2017).
Mau, A. et al. Molecular survey and genetic diversity of Bartonella spp. in small Indian Mongooses (Urva auropunctata) and their fleas on Saint Kitts, West Indies. Microorganisms 9, 1350 (2021).
Cheslock, M. A. & Embers, M. E. Human bartonellosis: An underappreciated public health problem?. Trop. Med. Infect. Dis. 69, 4 (2019).
Arvand, M. & Viezens, J. Evaluation of pulsed-field gel electrophoresis and multi-locus sequence typing for the analysis of clonal relatedness among Bartonella henselae isolates. Int. J. Med. Microbiol. 297, 255–262 (2007).
Dias, C. M. et al. Multi-locus sequencing typing of Bartonella henselae isolates reveals coinfection with different variants in domestic cats from Midwestern Brazil. Acta Trop. 237, 106742 (2023).
Boulouis, H.-J., Chao-chin, C., Henn, J. B., Kasten, R. W. & Chomel, B. B. Factors associated with the rapid emergence of zoonotic Bartonella infections. Vet. Res. 36, 383–410 (2005).
Arvand, M., Feil, E. J., Giladi, M., Boulouis, H. & Viezens, J. Multi-locus sequence typing of Bartonella henselae isolates from three continents reveals hypervirulent and feline-associated clones. PLoS One. 2, e1346 (2007).
Chaloner, G. L., Harrison, T. G., Coyne, K. P., Aanensen, D. M. & Birtles, R. J. Multilocus sequence typing of Bartonella henselae in the United Kingdom indicates that only a few, uncommon sequence types are associated with zoonotic disease. J. Clin. Microbiol. 49, 2132–2137 (2011).
Gil, H. et al. Distribution of Bartonella henselae variants in patients reservoir hosts and vectors in Spain. PLoS One. 8, 7 (2013).
Bai, Y. et al. Coexistence of Bartonella henselae and B. clarridgeiae in populations of cats and their fleas in Guatemala. J. Vector Ecol. 40, 327–332 (2015).
Cicuttin, G. L. et al. Bartonella spp. in cats from Buenos Aires, Argentina. Vet. Microbiol. 168, 225–228 (2014).
Foil, L. et al. Experimental infection of domestic cats with Bartonella henselae by inoculation of ctenocephalides fells (Siphonaptera: Pulicidae) feces. J. Med. Entomol. 35, 625 (1998).
Chomel, B. B. et al. Experimental transmission of Bartonella henselae by the cat flea. J. Clin. Microbiol. 34, 1952 (1996).
Jolley, K. A., Bray, J. E. & Maiden, M. C. J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 3, 124 (2018).
Furquim, M. E. et al. Genetic diversity and multilocus sequence typing analysis of Bartonella henselae in domestic cats from Southeastern Brazil. Acta Trop. 222, 106037 (2021).
Mietze, A. et al. Combined MLST and AFLP typing of Bartonella henselae isolated from cats reveals new sequence types and suggests clonal evolution. Vet. Microbiol. 148, 238–245 (2011).
Stepanić, M. et al. First isolation and genotyping of Bartonella henselae from a cat living with a patient with cat scratch disease in Southeast Europe. BMC Infect. Dis. 2019, 19 (2019).
Can, H. et al. Genetic characterization of Bartonella henselae samples isolated from stray cats by multi-locus sequence typing. BMC Vet. Res. 2023, 19 (2023).
Iredell, J. et al. Characterization of the natural population of Bartonella henselae by multilocus sequence typing. Society 41, 5071–5079 (2003).
Sepulveda, P. et al. Draft genomes of 16 Bartonella henselae strains from cats in Valdivia, Chile. Microbiol. Resour. Announc. 12, 11 (2023).
Québatte, M. & Dehio, C. Bartonella gene transfer agent: Evolution, function, and proposed role in host adaptation. Cell. Microbiol. 21, 11 (2019).
Guy, L. et al. A genome-wide study of recombination rate variation in Bartonella henselae. BMC Evol. Biol. 12, 65 (2012).
Kosoy, M., Mckee, C., Albayrak, L. & Fofanov, Y. Genotyping of Bartonella bacteria and their animal hosts: Current status and perspectives. Parasitology. 145, 1–20 (2017).
Huwyler, C. et al. Dynamics of co-infection with Bartonella henselae genotypes I and II in naturally infected cats: Implications for feline vaccine development. Microb. Ecol. 74, 474–484 (2017).
Styczynski, A., Tran, C. & Dirlikov, E. Human rabies—Puerto Rico, 2015. Infect. Control Hosp. Epidemiol. 17, 53–80 (1996).
Nellis, D. W. & Everard, C. O. R. The biology of the mongoose in the Caribbean. Stud. Fauna Curaçao Caribbean Islands 64, 1–162 (1983).
Louppe, V., Leroy, B., Herrel, A. & Veron, G. The globally invasive small Indian mongoose Urva auropunctata is likely to spread with climate change. Sci. Rep. 10, 7461 (2020).
Lodge, D. M. et al. Biological invasions: Recommendations for US policy and management. Ecol. Appl. 16, 2035–2054 (2006).
Tsuneoka, H. et al. The utility of a country-specific Bartonella henselae antigen in an IgM-indirect fluorescent antibody assay for the improved diagnosis of cat scratch disease. Diagn. Microbiol. Infect. Dis. 87, 22–24 (2017).
Kosoy, M. Y. et al. Distribution, diversity, and host specificity of Bartonella in rodents from the Southeastern United States. Am. J. Trop. Med. Hyg. 57, 578–588 (1997).
Maggi, R. G., Duncan, A. W. & Breitschwerdt, E. B. Novel chemically modified liquid medium that will support the growth of seven Bartonella species. J. Clin. Microbiol. 43, 2651–2655 (2005).
Andre, M. R. et al. Assessment of a quantitative 5’ nuclease real-time polymerase chain reaction using the nicotinamide adenine dinucleotide dehydrogenase gamma subunit (nuoG) for Bartonella species in domiciled and stray cats in Brazil. J. Feline Med. Surg. 10986, 1–9 (2015).
Billeter, S. A., Gundi, V. A. K. B., Rood, M. P. & Kosoy, M. Y. Molecular detection and identification of Bartonella species in Xenopsylla cheopis fleas (Siphonaptera: Pulicidae) collected from Rattus norvegicus rats in Los Angeles, California. Appl. Environ. Microbiol. 77, 7850–7852 (2011).
Muller, A. et al. Prevalence, hematological findings and genetic diversity of Bartonella spp. in domestic cats from Valdivia, Southern Chile. Parasitology 144, 773–782 (2017).
Basic, A. S. & Tool, L. A. S. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Acknowledgements
We wish to thank A. Conan, A. R. Berentsen, P. A. Leighton, A. Allibert, M. J. Rivera and J.-P. Viau for their invaluable help with mongoose capture and handling.
Funding
This research was founded by the Center for Conservation Medicine and Ecosystem Health, Ross University School of Veterinary Medicine. Foundation Grant No 43015-2023.
Author information
Authors and Affiliations
Contributions
AM: Conceptualization, Methodology, Project administration, Funding acquisition, Formal Analysis, Validation Writing–original draft; RM: Conceptualization, Funding acquisition, Methodology, Writing – review & editing PS: Methodology, Software, Writing – review & editing; AM (Alex Mau): Methodology, Writing – review & editing; CS: Methodology, Writing – review & editing; AC: Methodology, Writing – review & editing; IB: Validation, Methodology, Writing – review & editing; PB: Methodology, Writing – review & editing; EB: Conceptualization, Funding acquisition, Methodology, Writing – review & editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Muller, A., Maggi, R., Sepulveda-Garcia, P. et al. Sequence typing of Bartonella henselae in small Indian mongooses (Urva auropunctata). Sci Rep 14, 18654 (2024). https://doi.org/10.1038/s41598-024-69909-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-69909-z
- Springer Nature Limited