Abstract
The characterization of soil microbial communities at different depths is essential to understand their impact on nutrient availability, soil fertility, plant growth and stress tolerance. We analyzed the microbial community at three depths (3 cm, 12 cm, and 30 cm) in the native fruit trees Annona cherimola (cherimoya) and Pouteria lucuma (lucuma), which provide fruits in vitamins, minerals, and antioxidants. We used PacBio-HiFi, a long-read high-throughput sequencing to explore the composition, diversity and putative functionality of rhizosphere bacterial communities at different soil depths. Bacterial diversity, encompassing various phyla, families, and genera, changed with depth. Notable differences were observed in the alpha diversity indices, especially the Shannon index. Beta diversity also varied based on plant type and depth. In cherimoya soils, positive correlations with Total Organic Carbon (TOC) and Cation Exchange Capacity (CEC) were observed, but negative ones with certain cations. In lucuma soils, indices like the Shannon index exhibited negative correlations with several metals and specific soil properties. We proposed that differences between the plant rhizosphere environments may explain the variance in their microbial diversity. This study provides insights into the microbial communities present at different soil depths, highlighting the prevalence of decomposer bacteria. Further research is necessary to elucidate their specific metabolic features and overall impact on crop growth and quality.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Introduction
Soil is an important ecosystem where microbial populations are established, which interact with plants through the roots, favoring their growth and protecting them from diseases caused by pathogens1. Successful microorganism utilization promotes soil health, water retention, carbon storage, root development, accessibility and recycling of nutrients, pollutant treatment, as well as biodiversity conservation2. The influence of plants on the microbial communities may not be readily apparent, making predictions difficult, as other factors come into play, such as the soil type3.
Superfoods such as cherimoya (Annona cherimola) have gained attention because of their anticancer bioactive chemicals and nutrients. Also, their family (Annonaceae) has medicinal use in the treatment of epilepsy, cardiac diseases, pathogenic infections, hemorrhage, ulcers, cancer, liver disorders, and other ailments4. Some results indicated that cherimoya juice is a potent antioxidant5. Similarly, lucuma (Pouteria lucuma) was an important part of the pre-Hispanic diet6,7. Because of its high dietary fiber, carotenoids, and sugar content, lucuma may be an alternative to refined sugars and artificial colorants in the dairy and bakery industry8. The cultivation of these plants has been reevaluated due to the demand for superfoods9,10.
The research in deep soils can reveal microorganisms with unique characteristics and adaptations to extreme conditions, opening a wide potential for biotechnology and the development of applications beneficial to society, such as new drugs, industrial enzymes, and bioplastics11,12. Also, examining changes in microbial properties across different soils and depths can enhance the understanding of long-term processes vital for agricultural ecosystem health. Previous studies have demonstrated that soil depth is a significant factor affecting microbial communities, especially alpha bacterial diversity, which exhibits its highest abundance in the upper 10 cm and tends to decrease by 20 to 40% in deeper layers13,14,15. The studies also note that microbial community composition is notably variable in the upper soil layers but maintains relative consistency in the deeper layers across different locations14,16,17,18.
Microbes via their enzymatic activities can alter soil physico chemical properties, organic matter, and potential soil respiration rate19. Also, the cycling of organic carbon and nitrogen, and their pools would be related to changes in the microbial community20. Other studies indicate that physico chemical soil properties, such as pH, carbon, electric conductivity, CEC, and soil texture were correlated with the differences between communities measured by UniFrac21. Differences in soil moisture, temperature, aeration, and substrate availability by soil depths may change the microbial communities. A linkage between tree species and microbial diversity reflects mutual feedback between plants and the soil22,23,24.
The comparison of the 16S rRNA gene sequences makes it possible to establish phylogenetic relationships between bacteria, facilitating their rapid identification25. The sequencing of the 16S rRNA gene is a widely used technique, with the V3-V4 regions being the most common. Miseq, Illumina's next-generation integrated sequencing instrument allows sequencing from 1 × 36 bp to 2 × 300 bp in length26. This is a limitation since the 16S region consists of 1550 bp in length. A new technology called PacBio-HiFi (Single-molecule, real-time sequencing developed by Pacific BioSciences) offers longer reads and faster executions27 and allows us to identify a higher proportion of phylum, class, family, genus, and species level than the V4 region-only dataset, expanding the microbial ecology studies28,29,30,31,32.
There is no information about the rhizosphere microbiome of these species, but it is important to understand their impact on the growth, development, and properties of these trees. The objective was to analyze how depth influences the diversity of microbes in the rhizosphere of fruit trees, how other soil properties impact microbial composition, and identify specific taxa associated with shallow or deep rhizosphere layers. Comprehension of soil microbial characteristics concerning alterations in soil depth has the potential to make significant contributions to the enduring well-being of agricultural soils or the identification of soil conditions that are not optimal.
Results
A total of 38,481 Operational taxonomic units (OTUs) were identified and assigned to 27 phyla, 71 classes, 178 orders, 382 families, and 955 genera. The rarefaction plot lines indicated the trend of the behavior of the curves, indicating that they tended to level off where the slope approached zero (Fig. S1).
Alpha diversity indices at different soil depths
Diversity was assessed using Shannon and Observed features indices, revealing variations across soil depth. Higher values of both indices were evident in samples closer to the surface.
In the ANOVA analysis, significant differences were observed for the Shannon and Observed features indices across different soil depths, but not among tree types. Subsequent post hoc Tukey's Honest Significant Difference (HSD) tests revealed significant differences (p < 0.05) between cherimoya samples at 3 cm and 30 cm depths in Shannon diversity index (Fig. 1a). Regarding the results of the Observed features test, a significant contrast was identified for the combinations of lucuma samples at a depth of 3 cm vs 30 cm (Fig. 1b).
Bacterial community composition changes with variations in soil depth
Principal coordinate analysis (PCoA) allowed us to represent differences or make comparisons between samples. The dissimilarity matrix was obtained with the Bray–Curtis method and the obtained coordinates were plotted with the help of PCoA to visualize and analyze patterns of dissimilarity in biological communities.
The beta diversity results (Fig. 2) illustrated the arrangement of the points of the Bray–Curtis dissimilarity matrix. It was observed that the lucuma samples at 30 cm replicates were grouped in a certain space, while the 3 cm and 12 cm samples overlapped. In the case of the cherimoya samples, there was an overlap between the 3 cm and 12 cm samples, but without a well-defined spacing pattern for the 30 cm samples. A PERMANOVA analysis using the Bray–Curtis distance as a measure of dissimilarity was performed to evaluate bacterial diversity and changes in composition between different tree types and soil depths. The results indicated that there was a significant difference in bacterial composition by plant type and no significant differences were observed as a function of soil depth (Table S5).
Taxonomic distribution in bacterial communities
Across all the samples, the relative abundance of bacterial phyla Proteobacteria, Acidobacteria, Planctomycetes, Actinobacteria, Firmicutes, and Gemmatimonadetes was 32, 21, 13, 11, 8 and 7%, respectively. The phyla with an abundance of less than 1% were Armatimonadetes, Candidatus Tectomicrobiota, Cyanobacteria, Deinococcus-Thermus, Fibrobacteres, and Verrucomicrobia. Fifteen phyla and the twenty most abundant families and genera were evaluated by each tree species. ANOVA and Tukey HSD statistical tests were performed to determine the significantly different samples (Fig. 3a).
In the case of lucuma, Proteobacteria, Acidobacteria, Planctomycetes, Actinobacteria, and Firmicutes were identified as the most abundant phyla (Fig. 3b).The ANOVA test indicated that the abundance of the phylum Acidobacteria is high at a depth of 3 cm and decreases as depth increases; while the phylum Actinobacteria and Planctomycetes increase in abundance at 30 cm depth. The most common families evaluated were: Alcaligenaceae, Bacillaceae, Gemmataceae, Gemmatimonadacea, Nitrospiraceae, and Pyrinomonadaceae, but significant distinctions were noted among the following four taxonomic categories: (i) Thermoanaerobaculaceae was the most abundant in shallow soils from 3 to 12 cm, while (ii) Burkholderiales, (iii) Hyphomicrobiaceae and (iv) Pirellulaceae were the most abundant in deep soils. The most abundant genera were Derxia, Bacillus, Gemmata, Gemmatimonas, and Nitrospira. Nevertheless, Pirellula and Thiobacter were significantly more abundant at a depth of 30 cm, whereas Thermoanaerobaculum had a predilection for growing in shallow areas (Table S1). For cherimoya, significant differences were observed in near-surface soils. It was observed that in soils 3 cm deep, there was a higher abundance of Armatimonadetes and the genus Holophaga (Table S2).
Soil physicochemical parameters related to diversity
Between lucuma and cherimoya trees, significant disparities were identified in parameters such as total nitrogen, available potassium, potassium exchangeable cation, calcium exchangeable cation, CEC, available phosphorus, B, Al, V, Cr, Cu, Zn, As, Mo, Ag, Cd, Sb, Ba, Pb, Na, Mg, and K content (Table S4).
The pH was neutral to basic (interval from 7.01 to 8.24), the lucuma soil was saline for the three depths (AVG: 4.44 dS m−1); although, for the cherimoya soil, up to the first 3 cm it was saline (AVG: 4.02 dS m−1), and for the other two depths slightly saline (AVG: 2.59 dS m−1). The P content was high for all three depths for both tree soils. The K content was medium for the lucuma soil at the depths of 3 and 12 cm, and low for the 30 cm depth. In contrast, for the cherimoya soil, it was high at depths of 3 and 12 cm, and medium at 30 cm. The organic matter content was very low at all three depths for both tree soils, resulting in low levels of carbon and nitrogen. The CEC was high for the lucuma soil at all depths for cherimoya soil at the depth of 3 cm, and medium for depths of 12 and 30 cm.
In the cherimoya soil, significant differences were detected in the total carbon, organic carbon, and available potassium contents between the content at 3 cm and that observed at 12 and 30 cm. Furthermore, significant differences were identified in potassium exchangeable cation, calcium exchangeable cation, and CEC between the content at 3 cm and that observed at 30 cm. Nonetheless, the total nitrogen content did not vary between the evaluated depths (Table S3). In the rhizospheric soil of the cherimoya crop, notable distinctions were noted. A negative correlation emerged between the total organic carbon (TOC) content and the se.ACE functional diversity index, whereas a positive correlation was evident between TOC and the Shannon and Inverse Simpson diversity indices. Similarly, a positive correlation was established between the total carbon (TC) content and the Shannon diversity index. Additionally, there was a negative correlation between Ka and se.ACE. Moreover, the Inverse Simpson index displayed a positive correlation with Ti and a negative correlation with Ni. Total nitrogen demonstrated positive correlation with phosphorus (Fig. 4a).
In the lucuma rhizospheric soil, only differences in calcium exchangeable cation were identified. No differences were observed in the contents of important parameters like total carbon, organic carbon, and total nitrogen content (Table S3). Pearson's correlation analysis on lucuma indicated that alpha diversity indices were significantly correlated with some of the soil physicochemical properties (Fig. 4). The indices of se.chao1, se.ACE, and Shannon were negatively correlated with several metals such as Ni, K, Al, Co, Fe, Na, Hg, Mg, Ba, Ti, Mn, Zn, and Be, as well as with sodium exchangeable cation, total organic carbon, and total carbon (Fig. 4a). On lucuma soil, total carbon (TC) demonstrated negative correlations with diversity indices, Observed, Chao1, ACE, and Fisher. Additionally, total organic carbon (TOC) exhibited negative correlations with Observed, Shannon, and Fisher indices. Total nitrogen (TN) exhibited a positive correlation with the Simpson index. Furthermore, phosphorus (Pa) displayed positive correlations with Simpson and Inverse Simpson indices. Beryllium (Be) exhibited negative correlations with Chao1, ACE, and Shannon indices. Moreover, the Chao1 index exhibited negative correlations with Mg2, K, Na, Ba, Ni, Ti, and Zn. The Shannon index displayed negative correlations with Ba, Cd, Cr, Cu, Pb, and Zn (Fig. 4b).
LEfSe analysis
The figure presents LEfSe analyses illustrating variations in OTU abundance between different soil depths. In Fig. 5a, corresponding to cherimoya soils, variations in OTU abundance are observed. Taxa such as Rubrobacteria and Holophagae were more enriched at a depth of 3 cm, while at 30 cm, OTUs from the phyla Proteobacteria and Nitrospirae predominate. In Fig. 5b, corresponding to lucuma soils, it is highlighted that taxa such as Acidobacteria, including classes Thermoanaerobaculia and Blastocatellia, as well as the family Pyrinomonadaceae, were enriched at a depht of 3 cm. At 30 cm, OTUs from the phylum Planctomycetes predominate. Additionally, Nitrospira also enriched at 3 cm, exhibits a significant presence, further illustrating the distribution patterns in these soil layers.
Discussion
The PacBio Hi-Fi system significantly improved sequencing quality with the introduction of circular consensus sequencing (CCS) protocols, producing accurate long high-fidelity (Hi-Fi) reads33. In this study, PacBio-HiFi sequencing technology was used for sequencing 16S rRNA gene, as it is more robust and of higher quality than technologies using short amplicons. This technology enhances the ability to achieve greater taxonomic annotation resolution, enabling the differentiation of closely related organisms at the species level or even within mild clones34,35,36. In addition, the detection of novel or rare variants within microbial communities is a distinct capability of long-read sequencing37. Nevertheless, there is insufficient experimental data on how new long-read technologies, particularly SMRT PacBio Hi-Fi, could be utilized to estimate the microbial composition in rhizosphere soils and extreme environments such as deep soils. On the other hand, PacBio Hi-Fi is more error-prone than the Illumina platform, and even though PacBio Hi-Fi is classified as third-generation sequencing (TGS), it represents a higher cost per base compared to short-read DNA metabarcoding.
A comparative analysis of the diversity of OTUs in two Peruvian superfruit trees, cherimoya and lucuma, and different soil depths was carried out. The results revealed significant differences in bacterial diversity between the different soil depths and trees evaluated. These findings suggest the existence of bacterial groups particularly associated, indicating specific adaptations or symbiotic relationships between these groups and trees38,39.
The alpha diversity was determined by Shannon and Observed features indices; both indicated that in surface layers the microbial diversity is higher in comparison to depth soil. The ANOVA test confirmed significant differences in microbial diversity with soil depth, aligning with other investigations that reported a decrease in microbial diversity at greater soil depths40,41. This decrease in richness and diversity of microbial communities has been observed across various ecosystems, including agricultural soils, reforested ecosystems, and eroded soils42,43. Beta diversity results demonstrated by the Bray–Curtis dissimilarity matrix revealed a clear separation between lucuma and cherimoya samples in the PCoA. Nonetheless, the replicates did not form distinct groups according to soil depth, except for the lucuma samples at 30 cm. Differences in microbial communities between the two soils may be due to species presence or relative abundances influenced by edaphic factors or root exudates, which provide nutrients and drive bacterial metabolic activities. Several studies indicate the direct effects that plants have on the composition and relative abundance of microbial diversity44,45,46.
There were no significant differences in the total carbon content and total organic carbon between crops; nevertheless, significant differences were observed at different depths in cherimoya soil. Carbon variations across the soil profile can be attributed to changes in labile carbon fractions. In other studies, the topsoil (0–10 cm and 10–20 cm) has been observed to harbor a higher concentration of SOC and plant roots compared to deeper soil layers. This distribution of SOC and roots can influence carbon dynamics across the soil profile47.
In a study, low stable carbon content reduced community richness. Nonetheless, high carbon sites with different depths exhibited a uniform diversity pattern48. The low nitrogen content of Peruvian coastal soils may contribute to the lack of significant differences between depths49. Nonetheless, nitrogen levels were higher in cherimoya soil, presumably due to the application of fertilizers (Table S4). Peruvian coastal soils also have a relatively high content of potassium, with glauconite-K being more enriched in these samples50. Therefore, it is expected that the crops do not have nutritional deficiencies.
Metals such as B, Zn, As, and Pb were determined to exceed the maximum allowable limit according to Canadian Environmental Quality Guidelines (CEQGs). In the case of copper and cadmium, higher concentrations were detected only in the cherimoya soil (Table S4). Variations in organic matter are related to the cation exchange capacity (CEC). Certain metal cations can stabilize SOC by facilitating the interaction between soil minerals and organic matter through cation bridging, and they can form complexes with organic molecules when their hydration shells are disrupted. The effective cation exchange capacity (CEC eff.) is a comprehensive measure that considers the available soil surfaces where metal cations are retained51. In this study, the effective CEC was not measured, as the neutral to basic pH condition allowed the assumption that there was no significant difference between the effective CEC and the total CEC.
A study evaluated the potential ecological risk index of heavy metals and suggested that the risk to soil bacterial community was more relevant to Cu than Cd52. The primary mechanism of copper toxicity is not oxidative stress, but rather the replacement of the Fe co-factor53. In contaminated soils, heavy metals such as Hg, Cr, Pb, Mn, and As can cause diseases and even death of organisms, although trace amounts can be beneficial54. The pH factor significantly impacts the dynamics of microbial communities; a slightly basic pH would reduce the availability of heavy metals in the soil, preventing harmful levels55. A negative correlation was identified between the Shannon index and the concentration of beryllium in lucuma soil, indicating a potential adverse impact on microbial diversity within this agricultural context. This finding corroborates previous studies that underscore the detrimental effects of beryllium on soil microbiota within soil–water systems56. Likewise, an inverse relationship was observed between concentrations of cadmium, lead, and zinc in cherimoya soil and the Shannon index, suggesting a decrease in microbial diversity. This concurs with prior research indicating the negative influence of these heavy metals on soil microbiome diversity. The notable decline in bacterial metabolic activity and the selective displacement of specific bacterial phyla further underscore the adverse effects of Cd, Pb, and Zn contamination on soil microbiota composition and diversity57. Additionally, the discovery of a negative correlation between copper concentration and the Shannon index implies a detrimental effect of copper on soil microbial diversity, consistent with previous findings illustrating the selective pressure exerted by copper on soil microbiota58. This association raises concerns regarding the potential adverse impacts of copper contamination on the health and functionality of soil microorganisms, pivotal for sustaining soil health and agricultural and natural ecosystem productivity59.
The bacterium Rubrobacter radiotolerans was identified in rhizospheric soils of cherimoya, suggesting a possible influence of heavy metal contamination on the microbial community in these soils60,61. The Rubrobacteraceae family exhibited significant differences in its abundance in the superficial soil of cherimoya plantations. The presence of this family was reported in soils evaluated at a depth of 0 to 15 cm62. Despite being a minority group of Acidobacteria, the abundance of the genus Holophaga was significant, especially in soils at a depth of 3 cm, highlighting its relevance in future research to better understand bacterial diversity in this environment63,64. The prevalence of Proteobacteria in the deeper layers of chirimoya rhizospheric soil at a 30 cm depth, as observed in our investigation, corroborates previous findings across diverse soil ecosystems65,66. This observation underscores the notable adaptation and persistence of Proteobacteria in agricultural settings67. Similarly, our LEfSe analysis identified Nitrospira as a distinctive biomarker in the 30 cm chirimoya rhizospheric soil. Moreover, the relative abundance of Nitrospirae exhibited a significant increase with soil depth, indicative of their competitive advantage in deeper soil layers68. These outcomes are consistent with those reported in a separate investigation conducted in a poplar plantation69. Nitrospira, a member of the bacterial phylum Nitrospirae, stands out as one of the most diverse and abundant groups of nitrite-oxidizing bacteria (NOB)70. In addition, Nitrospira was identified in 3 cm lucuma soil; Nevertheless, this reported discrepancy can be attributed to the significant variations in total nitrogen in soil type between these plant species (Table S4).
The presence of Thermoanaerobaculum aquaticum and Pyrinomonadaceae in the rhizospheric soil of the lucuma plant has significant implications for understanding biogeochemical processes in this unique environment71,72. Thermoanaerobaculum aquaticum known for its anaerobic reduction of iron and manganese oxides, probably plays a crucial role in soil nutrient availability and, consequently, in plant nutrition having been studied in superficial tundra soils73. Furthermore, the presence of Pyrinomonadaceae, adapted to nutrient-limiting conditions and capable of degrading complex organic compounds, underlines their fundamental role in nutrient cycling processes and organic matter decomposition of surface soil74.
Furthermore, the detection of Blastocatellia in lucuma rhizosphere soil demonstrates its adaptation to this different environment. A previous study reporting the prevalence of Blastocatellia in bulk soils underscores the critical need to understand microbial diversity in specific habitats such as the lucuma rhizosphere75,76. Additionally, the identification of Planctomycetes in the 30 cm rhizosphere of the lucuma plant77, known for their ability to degrade complex heteropolysaccharides, further emphasizes the intricate interplay between microbial communities and soil health. Their association with the degradation of complex organic compounds can profoundly influence rhizospheric microbial community dynamics, subsequently impacting nutrient availability and overall plant health78
Relative abundance analysis indicated that the phylum Acidobacteria was more common in superficial lucuma soils (3 cm) than in deep soils. On the other hand, Acidobacteria is a soil bacterial phylum that is not widely represented, but its members are ubiquitous and widely distributed in almost all ecosystems. Acidobacteria exhibit a remarkable ability to adapt to nutrient-deficient conditions and complex environments, allowing them to decompose complex substrates in tropical peatlands and other similar environments79. Burkholderiales, Hyphomicrobiaceae, and Pirellulaceae were identified as significantly more abundant in 30 cm lucuma soils. Hyphomicrobiaceae are key bacteria that play an important role in soil carbon and nitrogen cycling80. Studies have indicated that the relative abundance of Pirellulaceae is strongly correlated with certain soil pH levels81. Notably, it was intriguing to observe that Thiobacter was detected in significantly greater abundance at 30 cm of soil depth in lucuma compared to soils at different depths. This bacterium has also been documented in subterranean environments, such as caves and groundwater systems82. Moreover, Thiobacter exhibited remarkable resilience in heavy metal-rich soils, thanks to its ability to use sulfur compounds as an energy source and its capacity to endure adverse environmental conditions83. Also, a high concentration of metals was detected in the deepest soils.
PacBio sequencing, with its long reads, is advantageous for 16S rRNA gene metabarcoding, providing comprehensive taxonomic resolution84. Nonetheless, it produces fewer reads per sample compared to Illumina 2 × 300 bp, which generates more short, accurate reads. This lower read count can result in rarefaction curves not reaching a plateau, indicating incomplete microbial diversity coverage. Increasing sequencing depth or combining PacBio with Illumina can mitigate this issue85.
Methods
Field site and sampling
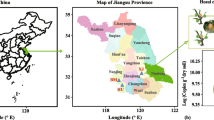
All tree specimens were roughly 15 years of age and had a history of fruit-bearing. Regardless of soil variation, all trees were subjected to standard irrigation practices at Sede Central Agricultural Research Station of INIA, maintaining constant agronomic management for five months from July to November 2022 ( moment of sampling). within La Molina district, Lima, Peru. This is situated at coordinates 12° 4′ 25″S and 76° 56′ 35″W and an elevation of 241 m about sea level (Fig. 6a). The region registers an average yearly temperature of 27 °C, an annual precipitation of 5 mm, and an average evaporation rate of 200 mm per year. At the experimental center, organic amendments, such as compost produced from crop residues and guinea pig manure, are primarily applied. The approximate amount is 1 t ha−1. Rhizosphere sampling involved taking three evenly spaced replicates from each soil depth level for each individual tree. These three subsamples were combined into a single replicate. Soil surrounding the roots with a diameter of less than 1 cm was collected at depths of 3 cm, 12 cm, and 30 cm for each plant (Fig. 6b), resulting in 10 samples for A. cherimola (Fig. 6c) and 12 samples for P. lucuma (Fig. 6d). After filtering through a 2 mm sieve, each soil was split into two segments: one was left to air-dry to assess soil biogeochemical properties, while the other was preserved at − 80 °C for DNA isolation. All the field experiments were approved by the Administration Bureau of the National Institute of Agrarian Innovation in Peru, following all the relevant guidelines.
(a) The geographical map of the location of the two plots: cherimoya and lucuma soil. (b) Reference image of soil sampling, (c) cherimoya tree, (d) and lucuma tree. Geographic map was built using software ArcGIS v10.8 (ESRI, California, USA)86.
Soil biogeochemical properties
The cherimoya soil was loamy, in contrast to the sandy loam soil where the lucuma trees were. The soil samples (1 kg) for these analyses were dried for 24 h at 40 °C and sieved 2 mm. The pH and electrical conductivity were measured using potentiometers pH7310 inoLab and Cond7310 inoLab (WTW™) in a 1:1: soil: water suspension (U.S.EPA SW-846-9045D)87 and considering a soil: water 1:1 ratio (ISO 11265:1994)88, respectively. Cation exchange capacity (CEC) was determined by saturation with ammonium acetate (pH 7.0). Available phosphorus was estimated by the modified Olsen method89, and available potassium was determined by extraction with ammonium acetate. Total nitrogen (TN) and total carbon (TC) were estimated by dry combustion (ISO 13878). Total organic carbon (TOC) was estimated after correcting for carbonates present in the sample (ISO 10694:1995 (E)), and total metals were estimated by microwave-assisted acid digestion of soils (EPA 3051A) and measured by Inductively Coupled Plasma-Mass Spectrometry (EPA 6020B).
Bacterial DNA extraction and sequencing
The total DNA of soil microorganisms was extracted from 0.5 g of each soil sample using a DNeasy PowerSoil Kit (QIAGEN Benelux B.V., Venlo, Netherlands), and each sample was run in triplicate. DNA concentration and quality were assessed using Qubit (Invitrogen, USA), a nucleic acid quantifier, and DNA integrity was checked using 1% agarose gel electrophoresis. DNA extraction of all samples was conducted on the same day. For bacterial identification purposes, the 16S rRNA gene PCR primers 27F/1492R with barcode on the forward primer were used in a 35-cycle PCR (5 cycles used on PCR products) using the HotStarTaq Plus Master Mix Kit (QIAGEN, USA) under the following conditions: 94 °C for 3 min, followed by 35 cycles of 94 °C for 30 s, 53 °C for 40 s and 72 °C for 90 s, after which a final elongation step at 72 °C for 5 min was performed. After amplification, PCR products were checked in 2% agarose gel to determine the success of amplification and the relative intensity of bands.
The complete sequence of 16S RNA was amplified by the PacBio platform, which has the advantage of providing more accurate species-level information. The SMRTbell libraries (Pacific Biosciences) were prepared following the manufacturer’s user guide, with indexing and library preparation executed according to the manufacturer’s instructions, applying the same PCR conditions as the first amplification, except the number of thermal cycles was reduced to 5. Sequencing was performed at the Molecular Research LP (MR DNA) (Shallowater, TX, USA) on the PacBio Sequel following the manufacturer’s guidelines. After completion of initial DNA sequencing, each library underwent a secondary analysis using PacBio’s CCS (circular consensus sequencing) algorithm. The CCS algorithm aligned the subreads individually from each template to generate consensus sequences, thereby correcting the stochastic errors generated in the initial analysis.
Bioinformatic and statistical analysis
Barcodes were removed from CCS sequencing data. The sequences were then oriented 5' to 3', and sequences with ambiguous base calls were removed. Later, sequences were denoised for the generation of OTUs at 3% divergence (97% similarity) using the software package Mothur v. 1.34.490, and chimeras were removed wih UCHIME91. Final OTUs were taxonomically classified using the 16S NCBI curated bacterial and Archaea RefSeq (http://www.ncbi.nlm.nih.gov) based on Blast92. searches (word size = 7; reward = 1; penalty = − 1; gap opening cost = 1; gap extension cost = 2).The data was normalized and filtered to remove low-quality sequences. Normalization uses the median of the sample sums to calculate a total normalization factor, which it uses to normalize the sample counts. Then, the result was multiplied by the total normalization factor. Normalization of the reads in each sample was performed using the Phyloseq package v4.0 in R software93. Rarefaction curve was calculated using species richness as a statistical estimator and plotted by MicrobiotaProcess packages in R software. The alpha diversity was determined via Shannon index and Observed features analysis. Two-way ANOVA was used to test the significant interaction between variables soil depth and type of plant. Tukey HSD was performed for multiple comparisons among different groups. Principal coordinate analysis (PCoA) using Bray–Curtis dissimilarity matrices was employed to measure beta-diversity. Through a permutational multivariate analysis of variance (PERMANOVA) with 999 permutations, soil depth and plant type were evaluated on the microbial community data using the adonis2 function of the R vegan package. Linear discriminant analysis effect size (LEfSe) was used to identify bacterial taxa that differed significantly between soil depth. All the analysis and graphics were generated by the following R packages: Phyloseq, ggplot2, and microbiome.
Data availability
The raw sequences have been deposited in the National Center for Biotechnology Information. The associated Bioproject and Biosample are PRJNA1059278, and SAMN39203878(9), respectively.
References
Yang, J., Kloepper, J. W. & Ryu, C.-M. Rhizosphere bacteria help plants tolerate abiotic stress. Trends Plant Sci. 14, 1–4. https://doi.org/10.1016/j.tplants.2008.10.004 (2009).
Suman, J. et al. Microbiome as a key player in sustainable agriculture and human health. Front. Soil Sci. 2, 821589. https://doi.org/10.3389/fsoil.2022.821589 (2022).
Fierer, N. Embracing the unknown: Disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 15, 579–590. https://doi.org/10.1038/nrmicro.2017.87 (2017).
Jamkhande, P. G., Ajgunde, B. R. & Jadge, D. R. Annona cherimola Mill. (Cherimoya): A review on its plant profile, nutritional values, traditional claims and ethnomedicinal properties. Orient Pharm. Exp. Med. 17, 189–201. https://doi.org/10.1007/s13596-017-0263-0 (2017).
Gupta-Elera, G., Garrett, A. R., Martinez, A., Robison, R. A. & O’Neill, K. L. The antioxidant properties of the cherimoya (Annona Cherimola) fruit. Food Res. Int. 44, 2205–2209. https://doi.org/10.1016/j.foodres.2010.10.038 (2011).
Padilla, I. M. G., Carmona, E., Westendorp, N. & Encina, C. L. Micropropagation and effects of mycorrhiza and soil bacteria on acclimatization and development of Lucumo (Pouteria lucuma R. and Pav.) Var. La Molina. In Vitro Cell. Dev. Biol. Plant 42, 193–196. https://doi.org/10.1079/IVP2006749 (2006).
Yahia, E.M. Postharvest Biology and Technology of Tropical and Subtropical Fruits: Fundamental Issues. ISBN 978-0-85709-362-2. https://doi.org/10.1533/9780857092762.437 (Woodhead Publishing in Food Science, Technology, and Nutrition; Elsevier Science, 2011).
García-Ríos, D., Aguilar-Galvez, A., Chirinos, R., Pedreschi, R. & Campos, D. Relevant physicochemical properties and metabolites with functional properties of two commercial varieties of Peruvian Pouteria lucuma. J. Food Process. Preserv. 44, e14479. https://doi.org/10.1111/jfpp.14479 (2020).
Durakova, A. et al. Storage studies of subtropical fruit Lucuma in powdered form. Bulgarian J. Agric. Sci. 25, 1287–1292. https://doi.org/10.5555/20203350829 (2019).
Rosa, O. L. et al. Nutritional and antioxidant profile of the Physalis fruit grown in three Andean regions of Peru. Roczniki Państwowego Zakładu Higieny https://doi.org/10.32394/rpzh.2023.0247 (2023).
Frey, B. et al. Deep soil layers of drought-exposed forests harbor poorly known bacterial and fungal communities. Front. Microbiol. https://doi.org/10.3389/fmicb.2021.674160 (2021).
Schlatter, D. C., Kahl, K., Carlson, B., Huggins, D. R. & Paulitz, T. Soil acidification modifies soil depth-microbiome relationships in a no-till wheat cropping system. Soil Biol. Biochem. 149, 107939. https://doi.org/10.1016/j.soilbio.2020.107939 (2020).
Hao, J. et al. The effects of soil depth on the structure of microbial communities in agricultural soils in Iowa (United States). Appl. Environ. Microbiol. 87, e02673-e2720. https://doi.org/10.1128/AEM.02673-20 (2021).
Eilers, K. G., Debenport, S., Anderson, S. & Fierer, N. Digging deeper to find unique microbial communities: The strong effect of depth on the structure of bacterial and archaeal communities in soil. Soil Biol. Biochem. 50, 58–65. https://doi.org/10.1016/j.soilbio.2012.03.011 (2012).
Fierer, N., Schimel, J. P. & Holden, P. A. Variations in microbial community composition through two soil depth profiles. Soil Biol. Biochem. 35, 167–176. https://doi.org/10.1016/S0038-0717(02)00251-1 (2003).
Naylor, D., McClure, R. & Jansson, J. Trends in microbial community composition and by soil depth. Microorganisms 10, 540. https://doi.org/10.3390/microorganisms10030540 (2022).
Chu, H. et al. Bacterial community dissimilarity between the surface and subsurface soils equals horizontal differences over several kilometers in the Western Tibetan Plateau. Environ. Microbiol. 18, 1523–1533. https://doi.org/10.1111/1462-2920.13236 (2016).
Gu, Y. et al. Long-term fertilization structures bacterial and archaeal communities along soil depth gradient in a paddy soil. Front. Microbiol. https://doi.org/10.3389/fmicb.2017.01516 (2017).
Naz, M. et al. The soil PH and heavy metals revealed their impact on soil microbial community. J. Environ. Manag. 321, 115770. https://doi.org/10.1016/j.jenvman.2022.115770 (2022).
Cookson, W. R. et al. The contribution of soil organic matter fractions to carbon and nitrogen mineralization and microbial community size and structure. Soil Biol. Biochem. 37, 1726–1737. https://doi.org/10.1016/j.soilbio.2005.02.007 (2005).
Andrew, D. R. et al. Abiotic factors shape microbial diversity in Sonoran desert soils. Appl. Environ. Microbiol. 78, 7527–7537. https://doi.org/10.1128/AEM.01459-12 (2012).
Scheibe, A. et al. Effects of tree identity dominate over tree diversity on the soil microbial community structure. Soil Biol. Biochem. https://doi.org/10.1016/j.soilbio.2014.11.020 (2015).
Thoms, C., Gattinger, A., Jacob, M., Thomas, F. M. & Gleixner, G. Direct and indirect effects of tree diversity drive soil microbial diversity in temperate deciduous forest. Soil Biol. Biochem. 42, 1558–1565. https://doi.org/10.1016/j.soilbio.2010.05.030 (2010).
Wu, H. et al. Linkage between tree species richness and soil microbial diversity improves phosphorus bioavailability. Funct. Ecol. 33, 1549–1560. https://doi.org/10.1111/1365-2435.13355 (2019).
Sobolev, D. & Begonia, M. F. T. Effects of heavy metal contamination upon soil microbes: Lead-induced changes in general and denitrifying microbial communities as evidenced by molecular markers. Int. J. Environ. Res. Public Health 5, 450–456. https://doi.org/10.3390/ijerph5050450 (2008).
Rodicio, M. D. R. & Mendoza, M. D. C. Identificación bacteriana mediante secuenciación del ARNr 16S: fundamento, metodología y aplicaciones en microbiología clínica. Enferm Infecc Microbiol. Clin. 22, 238–245. https://doi.org/10.1016/S0213-005X(04)73073-6 (2004).
Ravi, R. K., Walton, K. & Khosroheidari, M. MiSeq: A next generation sequencing platform for genomic analysis. Methods Mol. Biol. 1706, 223–232. https://doi.org/10.1007/978-1-4939-7471-9_12 (2018).
Rhoads, A. & Au, K. F. PacBio sequencing and its applications. Genomics Proteomics Bioinform. 13, 278–289. https://doi.org/10.1016/j.gpb.2015.08.002 (2015).
Wang, S. et al. Microbial richness of marine biofilms revealed by sequencing full-length 16S rRNA genes. Genes 13, 1050. https://doi.org/10.3390/genes13061050 (2022).
Notario, E. et al. Amplicon-based microbiome profiling: From second- to third-generation sequencing for higher taxonomic resolution. Genes 14, 1567. https://doi.org/10.3390/genes14081567 (2023).
Zhang, J., Su, L., Wang, Y. & Deng, S. Improved high-throughput sequencing of the human oral microbiome: From illumina to PacBio. Can. J. Infect. Dis. Med. Microbiol. 2020, e6678872. https://doi.org/10.1155/2020/6678872 (2020).
Ji, B., Zhang, X., Zhang, S., Song, H. & Kong, Z. Insights into the bacterial species and communities of a full-scale anaerobic/anoxic/oxic wastewater treatment plant by using third-generation sequencing. J. Biosci. Bioeng. 128, 744–750. https://doi.org/10.1016/j.jbiosc.2019.06.007 (2019).
Wenger, A. M. et al. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat. Biotechnol. 37, 1155–1162. https://doi.org/10.1038/s41587-019-0217-9 (2019).
Wang, S. et al. Microbial richness of marine biofilms revealed by sequencing full-length 16S rRNA genes. Genes 13(6), 1050. https://doi.org/10.3390/genes13061050 (2022).
Notario, E. et al. Amplicon-based microbiome profiling: From second- to third-generation sequencing for higher taxonomic resolution. Genes 14(8), 1567. https://doi.org/10.3390/genes14081567 (2023).
Zhang, J., Su, L., Wang, Y. & Deng, S. Improved high-throughput sequencing of the human oral microbiome: From illumina to PacBio. Can. J. Infect. Dis. Med. Microbiol. 2020, 6678872. https://doi.org/10.1155/2020/6678872 (2020).
Tedersoo, L., Albertsen, M., Anslan, S. & Callahan, B. Perspectives and benefits of high-throughput long-read sequencing in microbial ecology. Appl. Environ. Microbiol. 87(17), e0062621. https://doi.org/10.1128/AEM.00626-21 (2021).
Paul, E. & Frey, S. Soil Microbiology, Ecology and Biochemistry ISBN 978-0-12-823415-0. https://doi.org/10.1016/C2011-0-05497-2 (Elsevier, 2023).
Jacoby, R., Peukert, M., Succurro, A., Koprivova, A. & Kopriva, S. The role of soil microorganisms in plant mineral nutrition—current knowledge and future directions. Front. Plant Sci. https://doi.org/10.3389/fpls.2017.01617 (2017).
Kibblewhite, M. G., Ritz, K. & Swift, M. J. Soil health in agricultural systems. Philos. Trans. R. Soc. Lond. B Biol. Sci. 363, 685–701. https://doi.org/10.1098/rstb.2007.2178 (2008).
Paterson, E., Gebbing, T., Abel, C., Sim, A. & Telfer, G. Rhizodeposition shapes rhizosphere microbial community structure in organic soil. New Phytol. 173, 600–610. https://doi.org/10.1111/j.1469-8137.2006.01931.x (2007).
Jiao, S. et al. Soil microbiomes with distinct assemblies through vertical soil profiles drive the cycling of multiple nutrients in reforested ecosystems. Microbiome 6, 146. https://doi.org/10.1186/s40168-018-0526-0 (2018).
de Vries, F. T. et al. Changes in root-exudate-induced respiration reveal a novel mechanism through which drought affects ecosystem carbon cycling. New Phytol. 224, 132–145. https://doi.org/10.1111/nph.16001 (2019).
Lei, S. et al. Analysis of the community composition and bacterial diversity of the rhizosphere microbiome across different plant taxa. Microbiologyopen 8, e00762. https://doi.org/10.1002/mbo3.762 (2018).
Hu, J., Miller, G. & Shi, W. Abundance, diversity, and composition of root-associated microbial communities varied with tall fescue cultivars under water deficit. Front. Microbiol. https://doi.org/10.3389/fmicb.2022.1078836 (2023).
Prescott, C.E., Frouz, J., Grayston, S.J., Quideau, S.A. & Straker, J. Chapter 13—Rehabilitating forest soils after disturbance. In Developments in Soil Science; Global Change and Forest Soils, Vol. 36, 309–343. https://doi.org/10.1016/B978-0-444-63998-1.00013-6 (Elsevier, 2019).
Li, L. et al. Rhizobacterial compositions and their relationships with soil properties and medicinal bioactive ingredients in Cinnamomum Migao. Front. Microbiol. https://doi.org/10.3389/fmicb.2023.1078886 (2023).
Rangjaroen, C., Rerkasem, B., Teaumroong, N., Noisangiam, R. & Lumyong, S. Promoting plant growth in a commercial rice cultivar by endophytic diazotrophic bacteria isolated from rice landraces. Ann. Microbiol. 65, 253–266. https://doi.org/10.1007/s13213-014-0857-4 (2015).
Zhou, J. et al. Spatial and resource factors influencing high microbial diversity in soil. Appl. Environ. Microbiol. 68, 326–334. https://doi.org/10.1128/AEM.68.1.326-334.2002 (2002).
Morales, R. A., Zorogastúa, P., De Mendiburu, F. & Quiroz, R. Producción mecanizada de maíz, camote y yuca en la Costa Desértica Peruana: Estimación de la huella de carbono y propuestas de mitigación. Ecología Aplicada 17, 13–21. https://doi.org/10.21704/rea.v17i1.1169 (2018).
Wood, J. L., Tang, C. & Franks, A. E. Competitive traits are more important than stress-tolerance traits in a cadmium-contaminated rhizosphere: A role for trait theory in microbial ecology. Front. Microbiol. https://doi.org/10.3389/fmicb.2018.00121 (2018).
Solly, E. F. et al. A critical evaluation of the relationship between the effective cation exchange capacity and soil organic carbon content in swiss forest soils. Front. For. Glob. Change 3, 98. https://doi.org/10.3389/ffgc.2020.00098 (2020).
Chen, J. et al. The toxic factor of copper should be adjusted during the ecological risk assessment for soil bacterial community. Ecol. Indic. 111, 106072. https://doi.org/10.1016/j.ecolind.2020.106072 (2020).
Shuaib, M. et al. Variation and succession of microbial communities under the conditions of persistent heavy metal and their survival mechanism. Microb. Pathog. 150, 104713. https://doi.org/10.1016/j.micpath.2020.104713 (2021).
Solioz, M. Copper and bacteria: Evolution, homeostasis and toxicity. SpringerBriefs in Molecular Science ISBN 978-3-319-94438-8. https://doi.org/10.1007/978-3-319-94439-5. (Springer, 2018).
Islam, M. R., Sanderson, P., Johansen, M. P., Payne, T. E. & Naidu, R. Environmental chemistry response of beryllium to diverse soil-solution conditions at a waste disposal site. Environ. Sci. Process. Impacts 25, 94–109. https://doi.org/10.1039/D2EM00313A (2023).
Fajardo, C. et al. Pb, Cd, and Zn soil contamination: Monitoring functional and structural impacts on the microbiome. Appl. Soil Ecol. 135, 56–64. https://doi.org/10.1016/j.apsoil.2018.10.022 (2019).
Zhang, X. et al. Structure and diversity of fungal communities in long-term copper-contaminated agricultural soil. Sci. Total Environ. 806, 151302. https://doi.org/10.1016/j.scitotenv.2021.151302 (2022).
Fagnano, M. et al. Copper accumulation in agricultural soils: Risks for the food chain and soil microbial populations. Sci. Total Environ. 734, 139434. https://doi.org/10.1016/j.scitotenv.2020.139434 (2020).
Al-Najjar, M. A. & Albokari, M. M. Shifts in microbial community composition in tannery-contaminated soil in response to increased gamma radiation. Ann. Microbiol. 69, 1567–1577. https://doi.org/10.1007/s13213-019-01541-z (2019).
Meier, D. V., Imminger, S., Gillor, O. & Woebken, D. Distribution of mixotrophy and desiccation survival mechanisms across microbial genomes in an arid biological soil crust community. mSystems 6, e00786-e820. https://doi.org/10.1128/mSystems.00786-20 (2021).
Da Rocha, U. N. Ecology of Acidobacteria and Verrucomicrobia in the plant-soil ecosystem. Univ. Lib. Groningen. 10, 326–339 (2010).
Joo, H. K., Park, Y. W., Jang, Y. Y. & Lee, J. Y. Structural analysis of glutamine synthetase from Helicobacter pylori. Sci. Rep. 8, 11657. https://doi.org/10.1038/s41598-018-30191-5 (2018).
Roy, A. B. 1 The hydrolysis of sulfate esters. In The Enzymes Vol. 5 (ed. Boyer, P. D.) 1–19 (Academic Press, 1971). https://doi.org/10.1016/S1874-6047(08)60084-0.
Hao, J. et al. The effects of soil depth on the structure of microbial communities in agricultural soils in Iowa. Appl. Environ. Microbiol. 87, e02673-e2720. https://doi.org/10.1128/AEM.02673-20 (2021).
Li, C., Yan, K., Tang, L., Jia, Z. & Li, Y. Change in deep soil microbial communities due to long-term fertilization. Soil Biol. Biochem. 75, 264–272. https://doi.org/10.1016/j.soilbio.2014.04.023 (2014).
Zhao, F. Z. et al. Change in soil bacterial community during secondary succession depend on plant and soil characteristics. Catena. 173, 246–252. https://doi.org/10.1016/j.catena.2018.10.024 (2019).
Daims, H., Nielsen, J. L., Nielsen, P. H., Schleifer, K.-H. & Wagner, M. In situ characterization of nitrospira-like nitrite-oxidizing bacteria active in wastewater treatment plants. Appl. Environ. Microbiol. 67, 5273–5284. https://doi.org/10.1128/AEM.67.11.5273-5284.2001 (2001).
Feng, H., Guo, J., Wang, W., Song, X. & Yu, S. Soil depth determines the composition and diversity of bacterial and archaeal communities in a poplar plantation. Forests 10, 550. https://doi.org/10.3390/f10070550 (2019).
Dlamini, S. P., Akanmu, A. O. & Babalola, O. O. Rhizospheric microorganisms: The gateway to a sustainable plant health. Front. Sustain. Food Syst. 6, 925802. https://doi.org/10.3389/fsufs.2022.925802 (2022).
Tschoeke, D. A. et al. New bacterial and archaeal lineages discovered in organic rich sediments of a large tropical Bay. Mar. Genomics 54, 100789. https://doi.org/10.1038/s41396-021-00992-0 (2020).
Losey, N. A. et al. Thermoanaerobaculum aquaticum gen. nov., sp. nov., the first cultivated member of Acidobacteria subdivision 23, isolated from a hot spring. Int. J. Syst. Evol. Microbiol. 63, 4149–4157. https://doi.org/10.1099/ijs.0.051425-0 (2013).
Pessi, I. S. et al. In-depth characterization of denitrifier communities across different soil ecosystems in the tundra. Environ. Microbiome 17, 30. https://doi.org/10.1186/s40793-022-00424-2 (2022).
Huijun, L. I., Weijian, Z. & Weijian, W. U. Effects of sea rice on soil chemical properties and microbial community structure in coastal solonchaks. J. Agric. Sci. Technol. https://doi.org/10.13304/j.nykjdb.2022.0294 (2023).
Fernandez-Gomez, B. et al. Bacterial communities associated to Chilean altiplanic native plants from the Andean grasslands soils. Sci. Rep. 9, 1042. https://doi.org/10.1093/femsec/fiac079 (2019).
Riksen, V. S. & Korobova, L. N. Biodiversity of the microbiome as an indicator of phytomeliorative soil transformation. In IOP Conf. Ser. Earth Environ. Sci. 1154. https://doi.org/10.1088/1755-1315/1154/1/012017 (2023).
Liu, J. L. et al. Characteristics of bulk and rhizosphere soil microbial community in an ancient Platycladus orientalis forest. Appl. Soil Ecol. 132, 91–98. https://doi.org/10.1016/j.apsoil.2018.08.014 (2018).
Wang, X. et al. Stable-isotope probing identifies uncultured planctomycetes as primary degraders of a complex heteropolysaccharide in soil. Appl. Environ. Microbiol. 81, 4607–4615. https://doi.org/10.1128/aem.00055-15 (2015).
Hermans, S. M. et al. Bacteria as emerging indicators of soil condition. Appl. Environ. Microbiol. 83, e02826-e2916. https://doi.org/10.1128/AEM.02826-16 (2017).
Hirayama, H., Takai, K., Inagaki, F., Nealson, K. H. & Horikoshi, K. Thiobacter subterraneus gen. nov., sp. nov., an obligately chemolithoautotrophic, thermophilic, sulfur-oxidizing bacterium from a subsurface hot aquifer. Int. J. Syst. Evol. Microbiol. 55, 467–472. https://doi.org/10.1099/ijs.0.63389-0 (2005).
Chen, Z. et al. Impact of different sulfur sources on the structure and function of sulfur autotrophic denitrification bacteria. Sci. Rep. 13, 19404. https://doi.org/10.1038/s41598-023-46829-y (2023).
Mhete, M., Eze, P. N., Rahube, T. O. & Akinyemi, F. O. Soil properties influence bacterial abundance and diversity under different land-use regimes in semi-arid environments. Sci. Afr. 7, e00246. https://doi.org/10.1016/j.sciaf.2019.e00246 (2020).
Sessitsch, A., Weilharter, A., Gerzabek, M. H., Kirchmann, H. & Kandeler, E. Microbial population structures in soil particle size fractions of a long-term fertilizer field experiment. Appl. Environ. Microbiol. 67, 4215–4224. https://doi.org/10.1128/AEM.67.9.4215-4224.2001 (2001).
Schloss, P. D., Jenior, M. L., Koumpouras, C. C., Westcott, S. L. & Highlander, S. K. Sequencing 16S rRNA gene fragments using the PacBio SMRT DNA sequencing system. PeerJ 4, e1869 (2016).
Cook, R. et al. The long and short of it: Benchmarking viromics using Illumina, Nanopore and PacBio sequencing technologies. Microb. Genomics 10(2), 001198 (2024).
ESRI ArcGIS Desktop: Release 10.2.2 (Environmental Systems Research Institute, 2022).
US Environmental Protection Agency Method 9045D Soil and Waste PH. (2004).
International Standard Organisation (ISO). Soil Quality: Determination of the Specific Electrical -Conductivity. (1996). https://www.iso.org/standard/19243.html (Accessed 10 May 2023).
Secretaría de Medio Ambiente y Recursos Naturales (SEMARNAT). Norma Oficial Mexicana NOM-021-RECNAT-2000. http://www.ordenjuridico.gob.mx/Documentos/Federal/wo69255.pdf (Accessed 10 May 2023).
Schloss, P. D. et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. https://doi.org/10.1128/aem.01541-09 (2009).
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C. & Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. https://doi.org/10.1093/bioinformatics/btr381 (2011).
Camacho, C. et al. BLAST+: Architecture and applications. BMC Bioinform. 10, 421. https://doi.org/10.1186/1471-2105-10-421 (2009).
R Core Team. R: A Language and Environment for Statistical Computing (R Fundation for Statistical Computing, 2020).
Acknowledgements
We thank Ivan Ucharima, Rosa Poccomucha, and Linda Arteaga for supporting the logistic activities in our laboratory. Also, W.E.P and J.C. thank INIA Project CUI N° 2487112; and C.I.A. thanks Vicerrectorado de Investigación of UNTRM.
Author information
Authors and Affiliations
Contributions
Conceptualization, R.E., T.P., J.C., W.E.P. and E.A.V; METHODOLOGY, R.E., T.P, W.E.P.; software, R.E. and T.P.; validation, R.E and Y.R.; formal analysis, R.E., T.P. Y.R. and W.E.P.; investigation, R.E., Y.R., W.E.P., C.I.A. and T.P..; resources, J.C., E.A.V., and R.E.; data curation, R.E., T.P. and W.E.P.; writing—original draft, R.E.,T.P., and W.E.P; writing—review and editing, R.E., Y.R. and C.I.A.; supervision, E.A.V., and C.I.A.; project administration, C.I.A.; funding acquisition, C.I.A. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Estrada, R., Porras, T., Romero, Y. et al. Soil depth and physicochemical properties influence microbial dynamics in the rhizosphere of two Peruvian superfood trees, cherimoya and lucuma, as shown by PacBio-HiFi sequencing. Sci Rep 14, 19508 (2024). https://doi.org/10.1038/s41598-024-69945-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-69945-9
- Springer Nature Limited