Abstract
When IL-1 receptor antagonist (IL-1rn) is knocked out, mice have shown strain background dependent and major QTL regulated susceptibility to spontaneously inflammatory arthritis disease (SAD). The impact on bone properties resulting from the interactions of IL-1rn, genomic background strains, and the QTL locus, is unknown. Bone properties in the four specifically bred mouse strains with mutation of IL-1rn and variations in genomic components were investigated with high-resolution MicroCT and genomic analytical tools. Two congenic mouse strains were also measured to evaluate the influence on bone properties by a QTL in the region in chromosome 1. Our results reveal that several bone phenotypes, including bone mineral density (BMD), bone volume, tibial length, and cortical thickness of the tibia are different between wild type and IL-1rn knockout mice in both Balb/c and DBA/1 backgrounds, but IL-1rn knockout affects BMD differently between the two mouse strains. The absence of IL-1rn decreases BMD in Balb/c mice but increases BMD in DBA/1−/− mice compared to their respective wild type counterparts. A QTL transferred from the Balb/c genetic background which affects arthritis in congenic strains appears to also regulate BMD. While several genes, including Ctsg and Prg2, may affect BMD, Ifi202b is the most favored candidate gene for regulating BMD as well as SAD. In conclusion, the previously mentioned bone phenotypes are each influenced in different ways by the loss of IL-1ra when considered in mice from varying genomic backgrounds.
Similar content being viewed by others
Introduction
Our previous research has reported on the association between arthritis and bone mineral density (BMD)1. As a wide-spectrum inflammatory cytokine, interleukin-1 (IL-1) is implicated in many different diseases such as diabetes, leukemia, and inflammatory bowel disease1,2,3,4. It has also been reported that the inflammatory reaction in the synovium results in the production of IL-1, which together with other cytokines, has a systemic effect leading to generalized bone loss1. There is literature linking variations in the IL-1rn gene with early postmenopausal bone loss at the spine, as well as variations in forearm BMD in humans5,6. Studies in animal models indicate that the overexpression of human IL-1 in mice leads to bone loss5. These data suggest that IL-1 plays an important role in the regulation of BMD.
The interleukin-1 receptor antagonist (IL-1ra), encoded by the IL-1rn gene, is structurally related to and competes with IL-1 for the occupancy of the IL-1 cell receptors. It acts as a competitive inhibitor, preventing the triggering of signal transduction. Studies in two strains of mice developed with IL-1rn knockouts show that spontaneous inflammatory arthritis disease (SAD) is dependent not only on IL-1ra deficiency but also on other unidentified genetic factors7,8,9. It can be observed that while IL-1rn knockout mice in the Balb/c strain spontaneously develop autoimmunity and joint-specific inflammation, IL-1rn knockout mice in the DBA/1 strain do not10. However, the effect of the IL-1rn mutation on bone phenotypes of the different genomic backgrounds has not been investigated. Furthermore, the relation between the SAD and BMD under the IL-1rn gene-mouse strain genomic background interaction is not known.
Such phenotypic variation caused by the interaction between IL-1ra and the genomic background in mice may provide insight to the variation in the relationship between IL-1rn and BMD in the human population9,10,11. Due to the heterogeneity of the human population, it is difficult to separate genomic components specific to either bone loss or arthritis. However, phenotypes from mouse strains homozygous for different genomic components may provide a link between individual phenotype and genomic components. Thus, the mouse model provides an opportunity to investigate the relationship between IL-1rn and the genomic background which leads to variation in bone phenotypes, such as bone volume, tibial length, and cortical thickness of the tibia. This research observes the bone phenotypes under the effect of the IL-1ra deficiency in mice of different strain backgrounds and determine whether the effects on bone phenotype are influenced by a QTL contributing to SAD.
Materials and methods
Mouse strains
The mouse models for this study include six strains: four control stains (wild type and mutant) and two congenic strains derived from them. In previous studies, we identified a QTL region on chromosome 1 which regulates arthritis severity and incidence in Balb/c and DBA/1 strains by analyzing the F2 population from crossing DBA/1 IL-1rn−/− and Balb/c IL-1rn−/− mice1,8,9. In the IL-1rn−/− mutant model, the disease status between the Balb/c and DBA/1 is opposite to that in the collagen-induced arthritis (CIA) model. In CIA model, mice of DBA/1 are susceptible to arthritis while mice of Balb/c resist to the disease. When the IL-1rn is mutated, Babl/c is susceptible while the DBA/1 is resistant. All strains of mice in this study were kept and bred in the same animal facility at Memphis Veterans Affairs Medical Center. All experimental protocols and mouse husbandry were performed in accordance with the National Institutes of Health’s Guide for the Care and Use of Laboratory Animals and approved by the VAMC Institutional Animal Care and Use Committee. All methods are reported in accordance with ARRIVE guidelines.
Control strains
BALB/c IL-1rn−/− served as a disease control via IL-1rn knockout. This strain has been observed to develop arthritis and hind joint specific inflammation with an occurrence approaching 100% at 6 weeks of age11.
DBA/1 IL-1rn−/− served as a healthy control via their resistance to developing SAD. DBA/1 IL-1rn−/− does not develop arthritis and hind joint specific inflammation at 6 weeks of age11.
Balb/c wild type without any mutations served as a non-disease control.
DBA/1 wild type without any mutations as a non-disease control.
Congenic strains
Homozygous spontaneous disease models, both deficient in IL-1ra production (Fig. 1), have been self-crossed for 8 generations. These mice developed SAD with a much less severity8,9.
BALB.D1-1 IL-1rn−/−—Represents mice from a Balb/c background with IL-1rn knockout and a genomic QTL region from chromosome 1 of the DBA/1 genetic background.
DBA.B1 IL-1rn−/−—Represents mice from a DBA/1 background with IL-1rn knockout and a genomic QTL region from chromosome 1 of the Balb/c genetic background.
MicroCT analysis
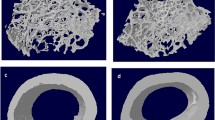
To quantitatively assess the structural changes associated with genetic variation, morphometric and architectural indices were determined from microtomographic examinations. We harvested 4 months old mice’s hindlimbs (males and females, n = 6–14) of Balb/c wildtype, DBA/1 wildtype, and IL-1rn knockout mice. Standard carbon dioxide (CO2) inhalation followed by cervical dislocation procedures were used for the euthanasia of the mice at the animal facility of VAMC. Whole bone BMD and midshaft BMD (the middle 100 slides in the tibia) were measured. Tibial thickness, tibial length, and cortical bone volume (BV) were also collected. All measurements and analyses were performed using the software included with the SCANCO µCT40 instrument. The bone samples were placed in an 8 mm diameter sample holder in 70% ethanol and immobilized with plastic foam. A μCT 40 desktop scanner (Scanco Medical, Southeastern, PA) was used to scan the samples at 11.5 μm resolution, at an energy level of 55 keV, with 2000 projections, an integration time of 300 ms, and an intensity of 145 μA. Morphometric and architectural parameters of bones were assessed, and realistic 3D visual models were constructed for each bone sample by selecting the volumes of interest (VOI). 3D trabecular parameters were evaluated using a fixed Gaussian filter and a threshold of 220 for the cancellous bone and 260 for the cortical envelope (Fig. 2). During the development and application of the micro CT in the studies of animal models, Scanco Medical has ensured the equipment is designed to make the adjustment according to the length of the tibia. Measurement of the lengths of the tibia was done from ankle to knee using calipers from the images of the microCT. In addition, we have the same internal control for every measurement to ensure the same paraments are in the different bones.
RNA isolation and microarray
Spleens were quickly frozen in liquid nitrogen and stored at − 80 °C. Total RNA was extracted using TRIzol Reagent (Invitrogen) according to the manufacturer’s instructions and RNA quality was quantified was determined by Agilent’s Bioanalyzer. The ratio of 28S/18S was above 1·8 for all the samples8. Spleen Microarray data have been reported in a previously published article and some of the genes have been validated by Realtime PCR7. RNA extraction and microarray methods were described in previous publications7,8.
Realtime PCR
Taqman Real-Time PCR reactions were carried out with an ABI 7900 Real Time PCR System (software: Sequence Detection System 2.3 (ABI)) using ABI’s standard protocol for RT-PCR and PCR. Samples were normalized to GAPDH mRNA expression levels. Data for treatment groups are shown as ΔΔCT normalized to an endogenous reference (vehicle).
Rat genome database
The Rat Genome Database (http://rgd.mcw.edu) is a model organism database and a website that could provide comprehensive human, rat, and mouse data, which helps to integrate phenotypic measurements of strains used as disease models. It can also analyze the QTL region for one phenotype12.
GeneNetwork analysis
Genenetowrk is an interactive tool that enables a user to readily reconstruct genetic networks based on genome expression data without having intimate knowledge of the mathematical models10,13. Using GeneNetwork (www.genenetwork.org), we chose the spleen dataset (UTHSC Affy MoGene 1.0 ST Spleen (Dec10) RMA) and (UCLA GSE27483 BXD Bone Femur ILM Mouse WG-6 v1, v1.1 (Jan13) RSN) to analysis the correlation. Pearson’s and Spearman’s Rank correlation have been used to validate the relationship among targeted genes in the potential pathway. More detail on the steps followed can be found in our previous study13.
Statistical analysis
Statistical analyses were performed in R and Excel, and some figures were gathered with Paint14,15. A significance level of 0.05 was selected. To statistically quantify differences in microCT-measured bone parameters between strains, adjustment by regression was first performed to control for the effect of sex. Analysis of variance (ANOVA) and two-sample t-tests were employed, and all relevant p-values are two-sided. 95% confidence intervals were constructed using quantiles from the t distribution. The Bonferroni correction was used to control the family-wise Type I error rate during multiple comparisons. Both t and p values are produced in the ANOVA. The t and p values represent the probability that the null hypothesis – that both (or all) populations are the same—is no and true. The bigger the t value is, the lower the probability of a null hypothesis is true; the small the p value is, the lesser the probability of the hypothesis is true. All distributions were found to be fairly normal through quantile–quantile plots and assumptions of homoscedasticity validated through Levene’s test.
Age of the mice
We select our measurements of bone properties of the mice at age of 4 months. Mouse bodies grow mature about three months, while the bone mineral density reaches the maximum at the time about 4 months. Previous majority of the studies on bone mineral density have chosen the mice at 4 months15. Accordingly, our study at 4 months is at the optimal time point as well as in agreement with other studies.
Results
Isolating the effect of strain on the bone parameters
Analysis of variance was performed to test the main effects of strain, sex, and the interaction of strain and sex on the bone parameters of interest. Table 1 displays our ANOVA results from the comparison of all animals for the effect of strains, sex, and their interactions. For all bone parameters, the effect of strain was highly significant (p < 0.001), indicating that the parameters differ significantly across the different strains. The effect of sex was significant only for bone volume (p < 0.001), below-knee BMD (p = 0.004), and cortical thickness (p = 0.003), indicating that these three parameters differ significantly across male and female mice. None of the strain-sex interactions were significant, indicating that the patterns of differences in the bone parameters across strains did not vary significantly between male and female mice.
To expand upon ANOVA’s significant main effects of sex, male mice had significantly higher below-knee BMD (t = 5.345, p < 0.001), bone volume (t = 2.708, p = 0.010), and cortical thickness (t = 2.692, p = 0.010) than female mice. For our analyses, in order to isolate the effect of strain while controlling for sex, all bone parameters of interest were regressed over sex, and the residuals were treated as the adjusted responses. Plots of the adjusted responses are displayed in Fig. 3.
Bone volume, tibial length, cortical thickness, and SMI in wild type and knockout mice
We first looked at the mice’s bone volume, tibial length, midshaft tibia cortical thickness, and structure model index (Supplementary Figure S1). A summary of our data is presented in Table 2.
Comparisons across strains are shown in Fig. 3. After adjusting for sex, tibial lengths were significantly lower in the IL-1rn−/− strains of both the Balb/c (t = − 6.765, p < 0.001) and DBA/1 (t = − 7.685, p < 0.001) backgrounds than their wild type counterparts. Similarly, below knee bone volume was significantly lower in IL-1rn−/− mice of both the Balb/c (t = − 6.772, p < 0.001) and DBA/1 (t = − 3.459, p = 0.020) backgrounds than their wild type counterparts. Cortical thickness was significantly lower in Balb/c IL-1rn−/− mice (t = − 5.837, p < 0.001) but not in DBA/1 IL-1rn−/− mice (t = 1.186, p = 0.810). Finally, SMI was significantly reduced in Balb/c IL-1rn−/− mice (t = − 4.610, p < 0.001) but not in DBA/1 IL-1rn−/− mice (t = 0.219, p = 1.000). This evidence suggests that bone phenotype is affected by the regulation of gene expression under the loss of IL-1ra, and it interacts with genetic background. IL-1rn knockout decreases below-knee bone volume and tibia length in both DBA/1 and Balb/c mice, but it reduces SMI and cortical thickness only in Balb/c mice.
Bone mineral density in wild type and knockout mice
Next, we analyzed the mice’s below-knee (foot and lower leg) bone mineral density and midshaft tibial bone mineral density. A summary of our data is presented in Table 3.
Comparisons across strains are shown in Fig. 3. After adjusting for sex and compared to the wild type strains, below-knee BMD was significantly higher in the DBA/1 IL-1rn−/− strain (t = 4.279, p = 0.007); however, below-knee BMD was insignificantly lower in the Balb/c IL-1rn−/− strain (t = -2.302, p = 0.112). Because the below-knee measurement contained the ankle which developed arthritis, as shown in Fig. 1, we then obtained a BMD measurement taken from 100 slides in the middle of the tibia to validate the results by excluding interference from arthritis in the ankle. In this case, both Balb/c and DBA/1 strains manifested a significant change in BMD. When IL-1rn is knocked out, the tibial BMD of Balb/c mice decreased (t = − 4.096, p = 0.002) while the tibial BMD of DBA/1 mice increased (t = 3.027, p = 0.047).
Congenic mice’s bone parameters and QTL region regulating BMD in DBA/1 strains
We further tested the congenic strains of Balb/c and DBA/1 mice created by transferring a QTL region on chromosome 1 from one strain to another. A summary of our data for the congenic strains is presented in Table 4. Again, comparisons across strains are shown in Fig. 3.
There were no statistically significant differences between the Balb.D1 IL-1rn−/− congenic strain with a QTL region from the DBA/1 background and the Balb/c IL-1rn−/− strain. Compared to the Balb/c WT strain, Balb.D1 IL-1rn−/− mice had significantly lower tibial lengths (t = 3.800, p = 0.015), below knee bone volume (t = 5.997, p < 0.001), SMI (t = 4.705, p = 0.002), cortical thickness (t = 6.181, p < 0.001), and midshaft tibial BMD (t = 3.977, p = 0.006) after adjusting for sex. In short, the congenic Balb.D1 IL-1rn−/− strain is essentially phenotypically identical to the Balb/c IL-1rn−/− strain. All significant comparisons between the Balb/c IL-1rn−/− strain and the Balb/c WT strain were held for comparisons of the Balb.D1 IL-1rn−/− strain versus the Balb/c WT strain.
On the other hand, the congenic DBA.B1-1 IL-1rn−/− differed noticeably in phenotype from the DBA/1 IL-1rn−/− strain, seeing a significant reduction in midshaft tibia cortical thickness (t = − 4.835, p < 0.001), SMI (t = − 3.332, p = 0.009), below-knee BMD (t = − 10.545, p < 0.001), and midshaft tibial BMD (t = − 5.186, p < 0.001). This suggests that the QTL region transferred from the Balb/c genetic background in DBA.B1 IL-1rn−/− mice may have the ability to regulate BMD in multiple positions of the bone. However, an examination of the other bone parameters measured reveals that tibial length was significantly greater in the DBA.B1-1 IL-1rn−/− strain than in the DBA/1 IL-1rn−/− strain (t = 9.244, p < 0.001) whereas below-knee bone volume remained similar, suggesting that DBA.B1 IL-1rn−/− may affect differently on different bone properties.
Identification of the gene regulating BMD in DBA/1 strains
Using the RGD database, we sorted the QTL region in each chromosome that regulated BMD. In a previous study, we found that the Ifi200 gene cluster contained the favorite candidate gene for SAD resistance in the congenic strain16. Supplementary Figure S2 shows that the location of the BMD-regulating QTL region in chromosome 1 is similar to the position of the arthritis-regulating QTL region in our mice. After performing real-time PCR, we believe the expression levels of Ifi202b can explain the discrepancies in BMD among the strains (Fig. 4).
The relative expression levels of Ifi202b in Balb/c WT mice are distinct from that of Balb/c IL-1rn−/− mice and congenic Balb.D1-1 IL-1rn−/− mice. Additionally, the relative expression levels of Ifi202b in DBA/1 IL-1rn−/− mice is distinct from that of DBA/1 WT mice and congenic DBA.B1 IL-1rn−/− mice. These patterns of Ifi202b expression match the patterns of differences in BMD (Fig. 3e,f). 50 likely candidate genes related to BMD have been identified, and Ifi202b is one of the candidate genes currently without any experimentally validated function in bone18. Overall, this evidence suggests that Ifi202b is perhaps the most favored BMD-regulating gene in chromosome 1.
Whole genome expression levels in wild type and knockout mice
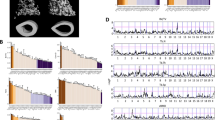
We compared spleen microarray data between DBA/1 WT and DBA/1 IL-1rn−/− mice and compiled a list of the genes that demonstrated greater than a twofold change. The same method was then applied to Balb/c WT and Balb/c IL-1rn−/− mice. Comparing the two lists allowed us to acquire a list of genes that are affected by IL-1rn knockout in both Balb/c and DBA/1 strains. These genes are listed in Supplementary Figure S3. Figure 5 shows the expression levels of Ctsg and Prg2 in wild type and knockout mice of the DBA/1 and Balb/c backgrounds. When IL-1rn is knocked out, the expression level of Ctsg and Prg2 is increased in both Balb/c and DBA/1 strains. Cathepsin G (Ctsg) is a bactericidal serine protease that has antimicrobial properties and can regulate inflammation17. Additionally, Ctsg also activates osteoclast precursors by stimulating the expression of RANKL, enhancing osteolysis18.
Ctsg and Prg2 expression levels in wild type and IL-1rn−/− mice of both Balb/c and DBA/1 genetic backgrounds. The scale on y-axis in (A) and (B) are gene relative expression level unit from microarray analysis. We consider the r value between positive or negative 0.35 and 0.7 as there is a negative or positive correlation. When the r values are more than positive or negative 0.7, we consider that a significant negative or positive correlation exists. From (A) to (F) are based on gene expression data from the spleen. From (G) to (L) are based on the gene expression data from bone femurs. (A) Ctsg expression level is higher in IL-1rn−/− mice under both Balb/c and DBA/1 genetic backgrounds than their wild types. (B) Prg2 expression level is higher in IL-1rn−/− mice of both Balb/c and DBA/1 genetic backgrounds than that of their wild types. (C) Gene network of interleukin family and Ctsg and Prg2. (D) Positive correlation between the express levels of Il1rn and IL1r2. (E) Positive correlation between the Ctsg and Prg2 with significant positive linear correlation r = 0.734, p < 0.001. (G–L) (F). Positive correlation between IL1r2 and Crsg. The figures G to L demonstrate the similar gene network (G) and gene–gene positive correlations (H–L).
GeneNetwork analysis
Using the GeneNetwork database, from “UTHSC Affy MoGene 1.0 ST Spleen (Dec10) RMA”, we found that there is a significant positive linear correlation (r = 0.734, p < 0.001) between Ctsg and Prg2. Ctsg is also significantly correlated with IL-1r2 (r = 0.618, p < 0.001). From “UCLA GSE27483 BXD Bone Femur ILM Mouse WG-6 v1, v1.1 (Jan13) RSN”, we also found that Ctsg and Prg2 are always significantly, moderately, and positively associated (r > 0.5, p < 0.001) with the IL-1 family genes, implying that there are some connections among Ctsg, Prg2, and the IL-1 families.
Discussion
This study investigates the effect of the interaction of different genomic backgrounds with the same mutation, IL-1rn knockout, on phenotypic variation of bone. Knockout has been used as an essential method to study the function of genes, but no study has focused on the effects on bone properties based on the absence of IL-1rn. Our study answered the question of how IL-1rn knockouts affect BMD, and the answer is that the effect of IL-1rn knockout is genome/strain dependent.
In the previous study, we found that a deletion of IL-1rn was associated with arthritis and led to different effects in Balb/c IL-1rn−/− and DBA/1 IL-1rn−/− mouse backgrounds1,8,9,10,11. Such a difference is possibly caused by the major histocompatibility complex (MHC)19. Subsequently, we also identified a region on chromosome 1 that regulates arthritis, and then we bred two different congenic strains, Balb.D1-1 IL-1rn−/− and DBA.B1 IL-1rn−/− mice1. Balb.D1-1 IL-1rn−/− mice can develop arthritis, but the onset of the disease is less severe than that of Balb/c IL-1rn−/−, because this congenic strain carried a QTL region from DBA/1. DBA.B1 IL-1rn−/− mice carried a QTL region from the Balb/c genetic background, so their SAD went from non-onset to onset, but the onset is also much milder11.
When IL-1rn was knocked out, reduced below knee bone volume and shortened tibias were observed in both Balb/c IL-1rn−/− and DBA/1 IL-1rn−/− mice, illustrating the association between IL-1rn and these two bone properties under different genomic backgrounds. Based on previous experience, we chose to use 6 to 14 mice per group for experiments, which is sufficient to show differences, if any7,8. Accordingly, we consider this result to be reliable. Significant thinning of tibial thickness was observed in Balb/c IL-1rn−/− mice, while there was no change in DBA/1 IL-1rn−/− mice. The differences in the bone phenotype between Balb/c and DBA/1 mice indicate that no individual phenotype in one mouse strain by a knockout of a gene does not necessarily mean the gene does not cause an abnormal phenotype in a different mouse strain.
These findings also support that the absence of IL-1rn decreases bone volume and tibia length in both Balb/c IL-1rn−/− and DBA/1 IL-1rn−/− strains compared with wild type mice. IL-1rn impacts BMD differently in Balb/c and DBA/1 mouse strains. The absence of IL-1rn decreases BMD in Balb/c IL-1rn−/− mice and increases it in BMD in DBA/1 IL-1rn−/− mice relative to their wild type mice. In both Balb/c and DBA/1 strains, the increased expression of Ctsg may be the key factor in regulating bone volume and bone length. The QTL in DBA.B1 IL-1rn−/− which affects arthritis in congenic strains also regulates BMD, and Ifi202b may be the most favored candidate gene for BMD. The translational significance for the human population is that a mutation may have completely different phenotypic effects on different populations. Gene flexibility, the different function, or degrees of influences of the same gene when it is under different genomic backgrounds, will be an issue of worthy attention both in humans and mice.
Spleen, as an organ important to immune system, secretes multiple molecules that affect bone and immunological responses12,22,23,24. The immune function of Ifi202b has been previously reported23,24. However, recently, its potential regulatory role in bone properties is also reported12,25. Considering the fact that this gene is located within the QTL and its expression is differentially associated with both BMD and inflammatory arthritis in these mouse stains, it is possible that Ifi202b affect the susceptibility to inflammatory arthritis and variation in BMD in these mouse strains. It is necessary to further confirm its function and molecular pathway in the immunological and bone properties. In addition, at present it is not clear whether it has the same effect on other mouse strains.
The decreased BMD in Balb/c mice may suggest that a negative effect of the IL-1rn knockout under the genomic background of Balb/c. The increased BMD in DBA/1−/− mice hints no negative effect rather the DBA/1 benefits from the knockout of IL-1rn. This phenomenon is similar to the phenotypes of inflammation arthritis in both strains. In CIA model, DBA/1 suscept to arthritis while Balb/c resist. However, when the IL-1rn is knocked out, Balb/c becomes susceptible, and DBA/1 becomes resistant to the inflammation arthritis.
Limitation of the study
One question is whether and how arthritis disease affected bone properties. While we were not able to clearly distinguish between the effects of arthritis disease and the bone properties, the effect on the backgrounds of both strains suggests that indeed the IL-1rn−/− and its interaction with the QTL on chromosome 1 do affect the bone properties independently from that of the effect on arthritis. An early study on the human population has indicated that the IL-1RN affects the bone loss in women20. A meta-analysis found that polymorphisms of interleukin-1 (IL-1) confer susceptibility to ankylosing spondylitis (AS) in Europeans but not in Asians21, suggesting a potential genome dependent function of genes in the interleukin-1family. Future studies are necessary to analyze the interaction between the arthritis and bone properties.
We realized that one limitation in our study is that we only adjusted the data with the length of the bone. Although in general, the length of the bone may positively relate to the body size, there could be variation because of the amount of fat. Our assumption was that because these mice are homozygous strains and grow under the same environment, there should not be much difference within the same strain. However, we still can not rule out the possibility of variation in fat and body sizes. In this regard, readers should be cautious when use our data.
There is a difference in the size of the QTL between arthritis and BMD. Because the QTL for arthritis was done in F2 and several congenic strains8,9,10,11, we were able to reduce the size of the genomic region. while for the BMD study, we only based on these six mouse strains. We can not rule out the possibility that the real location of genes of arthritis and BMD might be different, or they may not be the same gene.
Our study used an unequal number of male and female mice, which could introduce bias in the results. As such, the bone parameters, such as bone volume, BMD, and cortical thickness, are affected by sex in our study. This limitation suggests the need for caution when interpreting the data. Future studies should aim to use a balanced number of male and female mice to ensure that the observed effects are not confounded by sex differences.
Data availability
The authors declare that the data will be available without restrictions by contacting corresponding author: Weikuan Gu.
References
Liu, W. et al. Gene expression profiling studies using microarray in osteoarthritis: Genes in common and different conditions. Arch. Immunol. Ther. Exp. (Warsz) 68(5), 28 (2020).
Fardellone, P., Salawati, E., Le Monnier, L. & Goëb, V. Bone loss, osteoporosis, and fractures in patients with rheumatoid arthritis: A review. J. Clin. Med. 9(10), 3361 (2020).
Zerbini, C. A. F. et al. Biologic therapies and bone loss in rheumatoid arthritis. Osteoporos. Int. 28(2), 429–446 (2017).
Galindo Zavala, R. et al. Low bone mineral density in juvenile idiopathic arthritis: Prevalence and related factors. An. Pediatr. (Barc) 87(4), 218–225 (2017).
Makhdoom, A. et al. Bone mineral density level by dual energy X-ray absorptiometry in rheumatoid arthritis. J. Pak. Med. Assoc. 67(1), 15–19 (2017).
Aoki, Y. et al. Overexpression of the human interleukin 1a gene causes osteopenia in mice. J. Rheumatol. 32(2), 320–324 (2005).
Ivanova, J. T., Boyanov, M. A. & Toshev, A. K. Polymorphisms of the human IL-1 receptor antagonist gene and forearm bone mineral density in postmenopausal women. Indian J. Endocrinol. Metab. 16(4), 580–584 (2012).
Jiao, Y. et al. Identifying a major locus that regulates spontaneous arthritis in IL-1ra-deficient mice and analysis of potential candidates. Genet. Res. (Camb) 93(2), 95–103 (2011).
Deng, N. et al. Genomic locus on chromosome 1 regulates susceptibility to spontaneous arthritis in mice deficiency of IL-1RA. BMC Immunol. 15, 57 (2014).
Liu, X. et al. Decreased expression levels of Ifi genes is associated to the increased resistance to spontaneous arthritis disease in mice deficiency of IL-1RA. BMC Immunol. 17(1), 25 (2016).
Zhou, F., He, X., Iwakura, Y., Horai, R. & Stuart, J. M. Arthritis in mice that are deficient in interleukin-1 receptor antagonist is dependent on genetic background. Arthritis Rheum. 52(12), 3731–3738 (2005).
Lu, L. et al. Genetic dissection of femoral and tibial microarchitecture. JBMR Plus 3(12), e10241 (2019).
Cao, Y. et al. Anakinra as an interleukin 1 receptor antagonist, complicated genetics and molecular impacts–from the point of view of mouse genomics. Int. Immunopharmacol. 13(1), 28–36 (2012).
Han, K. O. et al. Nonassociation of interleukin-1 receptor antagonist genotypes with bone mineral density, bone turnover status, and estrogen responsiveness in Korean postmenopausal women. Bone 31, 612–615 (2002).
Wang, L. et al. Trps1 differentially modulates the bone mineral density between male and female mice and its polymorphism associates with BMD differently between women and men. PLoS ONE 9(1), e84485 (2014).
Tian, C. et al. Ifi204 as the most favored candidate gene that regulates susceptibility to spontaneous arthritis in mice deficient in IL-1ra. Gene Rep. 12, 21–29. https://doi.org/10.1016/j.genrep.2018.05.006 (2018).
Pérez-Is, L. et al. The N125S polymorphism in the cathepsin G gene (rs45567233) is associated with susceptibility to osteomyelitis in a Spanish population. PLoS ONE 14(10), e0220022. https://doi.org/10.1371/journal.pone.0220022 (2019).
Wilson, T. J., Nannuru, K. C. & Singh, R. K. Cathepsin G-mediated activation of pro-matrix metalloproteinase 9 at the tumor-bone interface promotes transforming growth factor-beta signaling and bone destruction. Mol. Cancer Res. 7(8), 1224–1233. https://doi.org/10.1158/1541-7786.MCR-09-0028 (2009).
Daser, A., Mitchison, H., Mitchison, A. & Müller, B. Non-classical-MHC genetics of immunological disease in man and mouse. The key role of pro-inflammatory cytokine genes. Cytokine 8(8), 593–7 (1996).
Keen, R. W., Woodford-Richens, K. L., Lanchbury, J. S. & Spector, T. D. Allelic variation at the interleukin-1 receptor antagonist gene is associated with early postmenopausal bone loss at the spine. Bone. 23(4), 367–371. https://doi.org/10.1016/s8756-3282(98)00109-4 (1998).
Lea, W. I. & Lee, Y. H. The associations between interleukin-1 polymorphisms and susceptibility to ankylosing spondylitis: A meta-analysis. Jt. Bone Spine 79(4), 370–374. https://doi.org/10.1016/j.jbspin.2011.06.010 (2012).
Ludlow, L. E. et al. The role of p202 in regulating hematopoietic cell proliferation and differentiation. J. Interferon Cytokine Res. 28(1), 5–11 (2008).
Hueber, W., Zeng, D., Strober, S. & Utz, P. J. Interferon-alpha-inducible proteins are novel autoantigens in murine lupus. Arthritis Rheum. 50(10), 3239–3249 (2004).
Cao, Y. et al. Sex differences in correlation with gene expression levels between Ifi200 family genes and four sets of immune disease-relevant genes. J. Immunol. Res. 28(2018), 1290814 (2018).
Zhang, L., Wang, C., Zhang, X. & Li, H. The interferon-inducible protein p202 promotes osteogenesis in mouse bone marrow stromal cells. Biosci. Rep. 38(3), BSR0171618 (2018).
Acknowledgements
The study was partially supported by a merit grant (I01 BX000671 to Weikuan Gu, 1I01RX004283 to H. Cho) from the Department of Veterans Affairs and the Veterans Administration Medical Center in Memphis, National Institutes of Health (R01 AR51190 to WG; R01 AR50785 to JS). The contents of the article are solely the responsibility of the authors and do not necessarily represent the official views of the funding agencies.
Author information
Authors and Affiliations
Contributions
Conception and design: W.G., Y.J., J.S., Karen Hasty. Data collection: W.D., C.T., A.B., Y.J., X.L., Y.C. Data organization and analysis: W.D., C.T., Z.G.L., J.M., M.B., W.G. Data interpretation, W.G., W.D., D.B., K.L., E.A.F., L.K.M., H.J.C., K.H., J.S. Drafting the manuscript: W.D., C.T., G.L., Y.J., W.G. Review and/or revision of the manuscript: All authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Dong, W., Tian, C., Li, Z.G. et al. Deficiency of interleukin-1 receptor antagonist in mice differentially affects bone properties under different genomic backgrounds. Sci Rep 14, 19889 (2024). https://doi.org/10.1038/s41598-024-70454-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-70454-y
- Springer Nature Limited