Abstract
Recent advancements in cancer therapy have led to the development of novel nanoparticle-based drug delivery systems aimed at enhancing the efficacy of chemotherapeutic agents. This study focuses on evaluating aprepitant-loaded PLGA and Eudragit RS 100 nanoparticles for their potential antiangiogenic effects. Characterization studies revealed that aprepitant-loaded nanoparticles exhibited particle sizes ranging from 208.50 to 238.67 nm, with monodisperse distributions (PDI < 0.7) and stable zeta potentials (between − 5.0 and − 15.0 mV). Encapsulation efficiencies exceeding 99% were achieved, highlighting the efficacy of PLGA and Eudragit RS 100 as carriers for aprepitant. Cellular uptake studies demonstrated enhanced internalization of aprepitant-loaded nanoparticles by HUVEC cells compared to free aprepitant, as confirmed by fluorescence microscopy. Furthermore, cytotoxicity assays revealed significant dose-dependent effects of aprepitant-loaded nanoparticles on HUVEC cell viability, with IC50 values at 24 h of 11.9 µg/mL for Eudragit RS 100 and 94.3 µg/mL for PLGA formulations. Importantly, these nanoparticles effectively inhibited HUVEC cell migration and invasion induced by M2c supernatant, as evidenced by real-time cell analysis and gene expression studies. Moreover, aprepitant-loaded nanoparticles downregulated VEGFA and VEGFB gene expressions and reduced VEGFR-2 protein levels in HUVEC cells, highlighting their potential as antiangiogenic agents. Overall, this research underscores the promise of nanoparticle-based aprepitant formulations in targeted cancer therapy, offering enhanced therapeutic outcomes through improved drug delivery and efficacy against angiogenesis.
Similar content being viewed by others
Introduction
Modern chemotherapy has a long history, with advances accelerating in recent years. Effective pharmacological therapy has traditionally relied on using sufficiently large dosages of a cytotoxic agent, which kills rapidly dividing cells and significantly shrinks the tumor. This strategy is not always effective, though, as the majority of such drugs do not specifically target cancer cells and frequently have negative side effects1. Targeted medicines with cancer cell selectivity have lately been introduced into clinical practice. The majority of these are cytostatic in nature, affecting cell signaling and replication but do not result in an immediate reduction in tumor burden2. At low doses and on cells resistant to apoptosis, cytotoxic compounds can produce stasis, but cytostatic actions typically result in cell death in any cell that is not in stationary phase3. These two mechanisms of action, however, have distinct impacts on the underlying cell population dynamics. For these reasons, as well as a lack of selectivity and substantial side effects, cytostatics are today's medications but not tomorrow's, and new anticancer strategies must be developed. A good alternative is aprepitant, a neurokinin-1 receptor (NK-1R) antagonist with anticancer and antiemetic activity that is safe to use and has minimal side effects. In this context, nanoparticles were synthesized using the nanoprecipitation method, chosen for its ability to enhance safety, and therapeutic efficacy4,5,6.
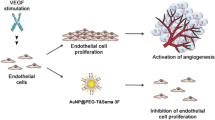
Peptides secreted by tumors are of interest because they have the potential to facilitate the diagnosis of tumors and to be targeted in therapy. Furthermore, data suggests that these peptides may contribute to cancer progression. Substance P (SP) is a neuroactive peptide that was discovered in the nervous system and is thought to function as a neurotransmitter/neuromodulator7. SP has been demonstrated to promote tumor mass growth and activation of tumor cell migration (required for invasion and metastasis) via its mitogenic capabilities and stimulation of endothelial cell growth (angiogenesis) by binding to the NK-1R88. Tumors must rapidly establish a new vascular network to support the high growth rate of cancer cells, a process known as angiogenesis, which is defined as the formation of new blood vessels from pre-existing vessels. Tumor cells secrete large amounts of pro-angiogenic substances, which lead to the creation of an abnormal vascular network characterized by disordered, immature, and permeable blood vessels, resulting in tumors that are poorly perfused. The hypoxic microenvironment formed by poor tumor perfusion may favor the selection of more invasive and aggressive tumor cells while also inhibiting immune cells' tumor-killing function. Pharmacological agents with anti-angiogenic effects that impair vascular nutrition by blocking vascular endothelial growth factor (VEGF) signaling and deprive the tumor of nutrients and oxygen are important9. Based on whether they have anti- or pro-inflammatory properties, macrophages are divided into the M1 and M2 (M2a, -b, -c, and -d) subgroups10. Macrophages change into M2a and M2c-like phenotypes in the microenvironment of tumors and tumor vessels, leading to epithelial-mesenchymal transition, cell invasion, and tumor growth with a poor prognosis11. Co-culture of human umbilical vein endothelial cells (HUVEC) and human monocyte THP-1 cells differentiated into M2 macrophages, which are abundant in the tumor microenvironment and constitute the majority of the tumor mass, is frequently used to model angiogenesis in the tumor microenvironment12.
Because of its weak basic compound, aprepitant, a non-peptide NK-1R antagonist with notable antiemetic and antitumor effects, has poor water solubility between pH 2 and 1013. Due to its poor solubility, it is difficult to administer as a systemic and effective therapy using conventional formulations containing aprepitant. Therefore, efforts to increase the solubility and dissolution rate of aprepitant are currently ongoing. Recently, attention has been focused on nanoparticle carrier systems to overcome such problems. Nanoparticles are solid colloidal drug carrier systems, 10–1000 nm in size, prepared with natural or synthetic polymer and ceramic or inorganic element. The active substance to be loaded in the nanoparticle system may be dissolved or dispersed in the nanoparticle, or may be attached to its surface or covalently bound. The nanocapsule is the matrix system in which the active ingredient is physically dispersed equally in the polymer, whereas the nanosphere is the system in which the active component is enclosed by the polymer membrane14,15.
Therefore, aprepitant-loaded PLGA and Eudragit RS 100-based drug delivery systems that have a greater impact than the current formulation were developed, and their efficacy were compared with the original formulation on angiogenesis.
Materials and methods
Chemical compounds
Eudragit RS 100 was a kind gift from Evonik Industries AG (Essen, Germany). Resomer® RG 502 H [Poly (d,l-lactide-co-glycolide), acid-terminated, lactide:glycolide 50:50, Mw: 7.000–17.000] and Span® 60 were purchased from Sigma-Aldrich (St. Louis, MO, USA). Pluronic® F-68 was purchased from Alfa-Aesar (Kandel, Germany). All other chemicals used were of analytical grade.
Preparation of nanoparticles
The nanoprecipitation process was used to create PLGA and Eudragit RS 100 based nanoparticles with a few modifications16,17,18,19. Nanoparticles made of PLGA were created as follows. First, PLGA (60 mg) and Span® 60 (20 mg) were dissolved in 4 mL of acetone. Following that, 3 mL of this solution was added dropwise at a rate of 5 mL h−1 into Pluronic® F-68 aqueous solution (10 mL, 0.5%, w/v) under magnetic stirring. The next step was to allow the acetone to evaporate for 4 h at room temperature while magnetic stirring. To gather the nanoparticles, the resulting aqueous dispersion was centrifuged. 5 mL of distilled water was added after the nanoparticles were collected to wash the particles. The nanoparticles dispersed in water were again subjected to the above-mentioned centrifugation process. To wash the nanoparticles, this procedure was repeated twice. To generate aprepitant-loaded PLGA-based nanoparticles, the process began by adding 6 mg of aprepitant to the organic phase. Then, 3 mL of such solution with drugs were added dropwise at rate 5 mL h−1 into Pluronic® F-68 aqueous solution (10 mL, 0.5%, w/v) under magnetic stirring. The next step was to allow the acetone to evaporate for 4 h at room temperature while magnetic stirring. To gather the nanoparticles, the resulting aqueous dispersion was centrifuged (11,000 rpm, 45 min, 4 °C). To wash the nanoparticles, this procedure was repeated twice.
Following are the steps used to prepare nanoparticles based on Eudragit RS 100. First, Span® 60 (20 mg) and Eudragit RS 100 (60 mg) were dissolved in 4 mL of ethanol. Following that, 3 mL of this solution was added dropwise at a rate of 5 mL h−1 into Pluronic® F-68 aqueous solution (10 mL, 0.5%, w/v) under magnetic stirring. Then, ethanol was given 4 h to evaporate at room temperature while being stirred magnetically. To gather the nanoparticles, the resulting aqueous dispersion was centrifuged. (11,000 rpm, 45 min, 4 °C). After the nanoparticles were collected, 5 mL of distilled water was added to wash the particles. The previously mentioned centrifugation technique was once more applied to the nanoparticles that had been dispersed in water (Table 1). To wash the nanoparticles, this procedure was repeated twice.
For aprepitant loaded Eudragit RS 100-based nanoparticles preparation, briefly, the procedure started by adding 6 mg aprepitant to organic phase. Following that, 3 mL of this solution was added dropwise at a rate of 5 mL h−1 into Pluronic® F-68 aqueous solution (10 mL, 0.5%, w/v) under magnetic stirring. Ethanol was then allowed to evaporate at room temperature under magnetic stirring for 4 h. The resulting aqueous dispersion was centrifuged to collect the nanoparticles (11,000 rpm, 45 min, 4 °C). To wash the nanoparticles, this procedure was repeated twice.
Similar to this, it was possible to dye the nanoparticles with the fluorophore Nile red prior to cellular uptake studies by adding 100 L of a 10 mM acetonic solution to the PLGA or Eudragit RS 100 organic solution while carefully following the instructions for making PLGA or Eudragit RS 100 based nanoparticles20.
Characterization of nanoparticles
Particle size, polydispersity index and zeta potential
Dynamic light scattering technique (DLS) was used to assess the particle size and polydispersity index (PDI) of nanoparticle formulations based on PLGA and Eudragit RS 100 on the Zetasizer Nano (Zetasizer Nano ZS, Malvern Instruments, Malvern, UK). By dispersing the formulation in distilled water, the produced nanoparticles' PDI and particle size were evaluated. The same instrument was used to measure the zeta potential values in a disposable folded capillary zeta cell, which was diluted with distilled water and kept at room temperature of 25 °C. All samples were measured in triplicate for statistical analysis, and the average values and standard deviation of the measurements were computed21,22.
UV–visible spectrophotometric method
Prior to the drug content investigation, a precise and sensitive UV spectrophotometric method for aprepitant was developed23. UV method was performed on a UV/Vis Recording Spectrophotometer UV-160A (Shimadzu) at 272 nm and using quartz cell. UV spectrophotometric method used for aprepitant in this study was validated for linearity, precision and accuracy. For standard solution, accurately weighed 100 mg of aprepitant was transferred to 100 mL volumetric flask and dissolved in ethanol: acetone (1:1). The calibration curve was obtained with 6 concentrations of the standard solution (2.0–10.0 μg mL−1). The precision of this study was determined by repeatability (intra-day) and intermediate precision (inter-day). By testing samples on the same day, at the same concentration, repeatability was assessed. The intermediate precision was studied by comparing the assays on different days (3 days). Six sample solutions for each concentration (4.0 μg mL−1, 5.0 μg mL−1, 6.0 μg mL−1) were prepared and analyzed. The recovery of known concentrations of a aprepitant reference standard given to the samples at the start of the procedure served to determine the accuracy 24. For this aim, an accurately weighed 10 mg of aprepitant was transferred to 100 mL volumetric flask and dissolved in ethanol: acetone (1:1) (final concentration of 100 μg mL−1). From this solution, solutions at concentrations of 4.0, 5.0 and 6.0 μg mL−1 were prepared for accuracy study. All solutions were prepared in triplicate and analyzed24,25.
Encapsulation efficiency
By carefully managing the extraction of aprepitant from nanoparticles, the encapsulation efficiency (EE%) of PLGA and Eudragit RS 100 based nanoparticles was assessed. 5 mg of precisely weighed nanoparticles were dissolved in a 1:1 mixture of ethanol and acetone. The final samples were filtered through 0.45 μm membrane filters and analyzed using UV/Visible spectrophotometer (Shimadzu UV VIS 160) at 272 nm. Three times each experiment was run to ensure statistical validity. Calculations were made to determine the measurements' average values and standard deviation. The EE % of nanoparticles was calculated by Eq. (1)26,27,28.
Encapsulation efficiency of nanoparticles
Determining the intracellular uptake of aprepitant-loaded drug delivery systems
The cellular uptake of aprepitant-loaded drug carrier systems was investigated using PLGA (PLGA-Apr) and Eudragit RS 100 (ERS-Apr) nanoparticles labeled with coumarin 6 (C6). Coumarin-labeled nanoparticles were prepared by mixing PLGA-Apr or ERS-Apr solution with 0.2 mg/ml C6 before adding aprepitant solution. HUVEC cells were seeded into 6-well plates at a density of 5 × 104 cells per well, and after 24 h, the existing medium was replaced with the medium containing the free C6 and 0.5 µM C6-labeled nanoparticles. Cells were incubated with NPs for different periods of time (0.5, 2, and 24 h). Cellular uptake was determined by immunofluorescence imaging and fluorescence intensity measurement on the Cytation 3 Multi-Mode Reader29.
Cell culture
Human monocyte THP-1 (American Type Culture Collection (ATCC), TIB-202) and human umbilical vein vascular endothelial HUVEC (ATCC, CRL-1730) cell lines were cultured in an incubator at 37 °C (5% CO2, 95% humidity) with 10% fetal bovine serum (FBS), 1% penicillin/streptomycin containing RPMI-1640, and F12-K media, respectively. HUVEC growth medium was supplemented with 0.03 mg ml−1 endothelial growth factor.
The culture flask was divided into subcultures after two to three days, or when the cell density reached 70%. The viability and number of cells were determined with a cell counting device (Cedex, Roche) by staining with trypan blue solution before the experiment. Passages with a cell viability of 90% or less were excluded. Cells between passages 5 and 10 were used.
Determination of cytotoxicity in HUVEC cells with the WST-1 method
HUVEC cells were cultured in a medium containing 10% FBS and 1% penicillin–streptomycin, and cell counting was performed. 1 × 104 cells per well were planted in 96-well plates. Following a 24-h incubation period, concentration groups of 1, 10, 100, and 400 µg mL−1 aprepitant, lyophilized nanoparticle formulations of aprepitant (ERS-Apr, PLGA-Apr), solvent control (ERS, PLGA), and doxorubicin (positive control) were applied. The cytotoxicity of SP was screened at 0.001, 0.01, 0.1, 1, 10, and 100 nM concentrations. Following the incubation periods (24 and 48 h), 10 µL of WST-1 reagent was added to the cells in each 96-well plate according to the WST-1 kit (catalog no. 11644807001, Roche) procedure instructions, and the cells were incubated for 3 h. At the end of the test, absorbance was measured at 420 nm on the Cytation 3 Multi-Mode Reader device (Biotek). The results were calculated using the viability formula and expressed as a percentage of cell viability30,31.
Determination of IC50 concentrations in HUVEC cells
A xCELLigence real-time cell analyzer device (RTCA DP) (Roche Applied Science, Germany) was used to determine the IC50 concentrations of the samples. This system generates real-time data by measuring electrical impedance from the e-plate and converting it to a unitless 'cell index (CI)' value to assess cell viability. The CI value increases along with the electrical response as the cells proliferate and cover the bottom of the E-plate2.
In order to calculate the IC50 values of doxorubicin, aprepitant, and lyophilized nanoparticle formulations of aprepitant (ERS-Apr, PLGA-Apr), 100 µL of SP-containing medium was first added to the E-plates, and the background of the wells was read. After the device's first reading, the E-plates were removed, and 100 µL of medium containing the cells was subsequently added. Cells were seeded in each well at a density of 10,000/well. After 24 h, the device was stopped, and 10, 100, and 200 µg mL−1 of the samples were applied to the E-plate wells in 100 µL medium. CI values were collected for 48 h. The RTCA DP Software 1.2.1 program was used to determine IC50 values based on the CI values30.
Determination of anti-angiogenic effects
In order to determine the anti-angiogenic activity, invasion and migration studies were conducted on HUVEC cells. For invasion studies, CIM plates coated with matrigel were used to examine the impact of different concentrations of aprepitant and its lyophilized nanoparticle formulations (ERS-Apr, PLGA-Apr) on HUVEC cells. These plates have a two-chamber system with a microporous membrane that allows cells to pass through.
In the RTCA DP system, HUVEC cells were placed in the upper chamber, which was coated with matrigel. These cells were exposed to a serum-free medium containing 100 nM SP. Subsequently, they migrated through the matrigel membrane and reached the lower chamber, where the medium contained IC50 values of 109 µg/mL for aprepitant, 11.2 µg/mL for ERS-Apr, and 30.9 µg/mL for PLGA-Apr. The changes in their movement and behavior were continuously monitored and recorded as CI data. This allowed for the assessment of how these compounds affected the cells' migration capabilities. Matrigel prevented non-invasive cells from crossing the membrane, allowing only invasive cells to pass through, making it possible to distinguish between them.
For cell migration assessment, CIM plates without a matrigel coating were used to measure migration ability. The results were used to determine the anti-invasion and anti-migration effects of different concentration groups, and graphical representations were created using the RTCA DP Software 1.2.1 program32,33,34,35.
Determination of anti-migration morphologically
This study utilized a cell migration assay kit, which included a special plate with stoppers for seeding cells. These stoppers ensured that the central point in all 96-wells remained empty during cell seeding. After seeding, the stoppers were removed from each well using a special tool, and this allowed for an equal area for cell migration in each well.
In the 96-well plate, HUVEC cells were plated at a density of 5 × 104 cells/mL, with each well containing 100 µL of medium and 100 nM SP. Following a 24-h incubation period to facilitate cell adhesion to the plate, the medium was substituted with 100 µL of a solution containing IC50 concentrations, which were 109 µg/mL for aprepitant, 11.2 µg/mL for ERS-Apr, and 30.9 µg/mL for PLGA-Apr. At 0, 24, and 48 h, the stoppers were removed from the wells using the special tool. These wells were then stained with Hoechst 33258 fluorescent dye, and images were captured at 0, 24, and 48 h. Fluorescence intensity measurements in the range of 350–461 nm were also conducted and graphed using a Cytation 3 Multi-Mode Reader (Biotek)36.
Determination of VEGF mRNA expression levels associated with angiogenesis using real-time polymerase chain reaction (RT-PCR)
The RT-PCR method was employed to investigate the mRNA expression levels of VEGF, a gene associated with angiogenesis. Initially, HUVEC cells were seeded in 6-well plates at a density of 1 × 106 cells in a medium containing 100 nM SP. Following a 24-h incubation period, the original medium was substituted with a medium containing IC50 concentrations of 109 µg/mL for aprepitant, 11.2 µg/mL for ERS-Apr, and 30.9 µg/mL for PLGA-Apr. The cells were then incubated for an additional 48 h. At the end of the incubation period, total RNA was isolated from HUVEC cells using an isolation robot (MagNA Pure Compact, Roche).
The amount of RNA obtained was assessed through spectrophotometric measurements at optical densities of 260 nm and 280 nm using a NanoDrop 2000 (Thermo Scientific). For cDNA synthesis, the Transcriptor High Fidelity cDNA Synthesis Kit (catalog no. 05091284001, Roche) procedure was followed. The cDNAs obtained were utilized for amplification in the LightCycler 480 RT-PCR instrument, using the LightCycler 480 Probes Master Kit (catalog no. 04 707 494 001, Roche). Both PCR mixes and cDNAs were individually loaded into 96-well plates. Temperature settings suitable for the monochromatic hydrolysis probe method were applied. After loading, the plate was placed in the LightCycler 480 RT-PCR device for quantitative measurements. The housekeeping gene GAPDH was used, and results were analyzed (SP4 version 1.5.0.39), with VEGF gene expression normalized to GAPDH gene expression levels34,37. Primer designs for the genes used can be found in Table 2.
Differentiation of THP-1 cells into macrophages and polarization into M2c macrophage subtype
THP-1 cells were cultured in RPMI-1640 medium supplemented with 1% penicillin–streptomycin and 10% fetal bovine serum. Following cell proliferation, cell counts were conducted, and the cells were then placed into inserts with 0.4 µm pores, specially designed for co-culture studies. These inserts contained a culture medium enriched with 100 ng/mL PMA and 1% FBS. After a 24-h incubation period, the cells were washed with PBS to eliminate PMA and subsequently rested in fresh medium for 48 h38. Subsequently, to initiate the transformation into the M2c polarized macrophage subtype, the existing medium was substituted with a medium containing 1 µM hydrocortisone, and the cells were cultured in this medium for an additional 72 h39.
Determination of VEGF protein levels
To assess the impact of concentration groups on VEGF protein levels in HUVEC cells, immunofluorescence labeling technique was utilized. Cells were seeded in 24-well plates at a density of 25,000 cells per well and incubated for 48 h with the respective IC50 concentrations of aprepitant and aprepitant's lyophilized nanoparticle formulations. After incubation, immunofluorescence labeling was conducted following the Immunofluorescence Application Solutions Kit protocol (Cell Signaling Technology, catalog no. 12727).
Initially, cells were washed with a 1xPBS solution and fixed with 4% formaldehyde at room temperature for 15 min. Subsequently, cells were permeabilized with cold 100% methanol at − 20 °C for 10 min. To prevent non-specific binding, the blocking solution was used, and cells were incubated for 1 h. Afterward, VEGF Receptor 2 (D5B1) Rabbit mAb (PE Conjugate) (Cell Signaling Technology, catalog no. 12634) was diluted 1:500 with an antibody dilution solution for labeling. Following incubation, protein levels were determined by immunofluorescence imaging and fluorescence intensity measurements using the Cytation 3 Multi-Mode Reader device 30.
Statistical analysis
The statistical analysis of the results was performed using one-way ANOVA and Tukey’s post hoc test with GraphPad Prism 8.0 software. The results are the means of three independent experiments expressed as the mean ± standard deviation. p values represent the significance of the results compared to the control group (p > 0.05 n.s., *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001) or M2c group (p > 0.05 n.s., #p < 0.05, ##p < 0.01, ###p < 0.001, and ####p < 0.0001).
Results and discussion
Evaluation of formulation and characterization studies
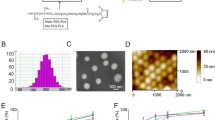
Particle size, particle size distribution (PDI), and zeta potential are used to evaluate nanoparticle characteristics. Table 3 summarizes the pertinent information. The particle size achieved in the blank formulation was 188.70–201.43 nm, whereas the aprepitant-loaded nanoparticles had a particle size of 208.50–238.67 nm. In particle size analysis, it is important to note that particle size increases with API loading. The literature backs up this conclusion18.
It has been discovered that as the particle size increases, the PDI values increase in parallel. For monophasic systems, the PDI value indicating the particle size distribution is between 0.01 and 0.7; A value greater than 0.7 indicates a very wide distribution of particle size, while a value close to zero indicates a limited PS distribution 23. When the PDI values given in Table 3 are examined, it is understood that highly monodisperse nanoparticle systems are prepared.
Table 3 shows the zeta potential (ZP) of several formulations. In colloidal systems, the particles repel each other if the ZP has a high positive or negative value. If distributed as a colloidal dispersion in a liquid, a colloidal system with a ZP value of less than 30 mV is considered a stable formulation 40. The limited flocculation zone is defined by ZPs between ± 5.0 and ± 15.0 mV, while the maximal flocculation zone is defined by ZPs between ± 5.0 and ± 3.0 mV 41.
As a result, when all particle size, ZP and PDI values were evaluated, appropriate results were obtained in terms of drug use.
Encapsulation efficiency
Following validation of the UV–visible spectrophotometric method developed, linearity was determined to be at a concentration range of 2.0–10.0 μg mL−1 with a linearity of y = 0.1043 x + 0.0463 (r2 = 0.9989). The method was decided to be precise due to relative standard deviation (RSD) values of < 2% for repeatability and intermediate precision. Recovery and accuracy of the method were satisfactory owing to the < 2% RSD value. Accuracy values (mean ± SD) of 99.5045 ± 0.6034%, 99.0409 ± 0.1918% and 99.9041 ± 0.4023% for concentrations of 4.0, 5.0, and 6.0 μg/mL, respectively, were determined for the UV–visible spectrophotometric method. The limit of detection (LOD) was determined to be 0.3200 μg/mL, while the limit of quantitation (LOQ) was 0.9699 μg/mL. Consequently, the easy and inexpensive procedure proposed in this study can be used for routine and simultaneous aprepitant determination28,42,43.
Because higher loading results in a lesser quantity of NPs for a given treatment dose, the drug encapsulation efficiency of nanoparticles is a crucial element in formulations44. The high values of encapsulation for PLGA and Eudragit RS (ERS) 100-based nanoparticles achieved are probably due to the lipophilic nature of the aprepitant, which presents low affinity to water phases and thus tends to migrate to the organic phase (Table 3)45. As a result, EE results were obtained in terms of drug administration in all nanoparticles.
Quantification of aprepitant-loaded drug delivery systems' cellular uptake
For evaluating the efficiency of drug delivery systems and comprehending their mechanisms of action, cellular uptake studies are helpful. The extent and kinetics of cellular uptake are revealed by methods like fluorescence microscopy, flow cytometry, and quantitative assays, which enable researchers to develop more effective drug delivery systems and improve therapeutic outcomes46.
ERS-Apr and PLGA-Apr were conjugated with coumarin to examine the increased cellular uptake of aprepitant. Free coumarin and conjugated ERS-Apr-C and PLGA-Apr-C nanoparticles were both incubated with HUVEC cells for varying amounts of time. Fluorescence microscopy analysis revealed that cells treated with ERS-Apr-C and PLGA-Apr-C uptake nanoparticles more effectively than cells treated with free coumarin (Fig. 1).
Uptake of nanoparticles labeled with coumarin into HUVEC cells depending on time (A) and fluorescence intensity-time graph of nanoparticles labeled with coumarin into HUVEC cells (B). For cellular uptake experiments, a two-way ANOVA was used to test the change of fluorescence intensity versus time (0.5, 2, 24 h). The data are presented as mean ± SD (n = 3), normalized to cell numbers. A Bonferroni post-hoc test was used to calculate significant differences between groups (**p < 0.01, ***p < 0.001, ****p < 0.0001).
Due to its hydrophobic and fluorescent properties, coumarin-6 is commonly employed as a tracking molecule to investigate the pharmacokinetics and tissue uptake of drug-loaded nanoparticles29,47. According to the experimental results, HUVEC cells exhibited higher sensitivity to ERS-Apr-C compared to both ERS-Apr-C and PLGA-Apr-C, as opposed to free coumarin. Cellular uptake was observed to be the fastest and most prominent with ERS-Apr-C.
Evaluation of cytotoxicity in HUVEC cells
The WST-1 assay measures mitochondrial dehydrogenase activity in live cells by producing formazan from a tetrazolium salt. The amount of formazan generated is directly linked to the number of metabolically active live cells. This formazan dye can be quantified with a spectrophotometer, providing a direct correlation with the count of metabolically active cells in the culture. Compared to the widely used MTT assay, the WST-1 test is easier to use, more sensitive, and less harmful to cells during testing48,49,50.
The WST-1 assay was employed both to assess the cytotoxic effects of nanoparticles, solvents, the positive control doxorubicin, and the NK1R agonist SP and to narrow down the concentration ranges (400, 100, 10, and 1 µg/mL) before determining IC50 values. The cytotoxicity intervals with observed effects were evaluated, and concentration ranges of 10, 100, and 200 µg/ml were selected for determining the IC50 (Fig. 2).
Cytotoxic effects of aprepitant, doxorubicin, ERS-Apr, PLGA-Apr, ERS, and PLGA on HUVEC cells. Cell viability results obtained from cells treated with various concentrations over 24 and 48 h were used to draw cytotoxicity graphs. The data is presented as mean ± SD (n = 8) and statistically evaluated using one-way ANOVA and Tukey post hoc test (significant difference: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001; no difference: p > 0.05 ns).
SP is a neuropeptide predominantly released from sensory nerve terminations and exhibits a selective affinity for the NK1R. Its functional roles encompass diverse physiological processes, including neuroinflammation, emesis, mood disorders, and nociceptive signal transmission. SP is widely distributed throughout the nervous system51. In vitro investigations have established the upregulation of both SP and the NK1R in multiple types of tumor cell lines, including malignant gliomas, breast carcinomas, and metastatic melanomas52.
To determine the optimal concentration of SP required to induce NK1R expression in HUVEC cells, a range of concentrations (100, 10, 1, 0.1, 0.01, and 0.001 nM) were applied to the cells for 24 and 48 h. The resulting cell viability data, depicted in Fig. 3, revealed that SP did not induce significant cytotoxicity. As a result, 100 nM was identified as suitable for supplementation in the culture medium to induce NK1R.
Cytotoxic effects of SP concentrations on HUVEC cells. Cell viability data obtained from cells treated with various concentrations for both 24 and 48 h were utilized to draw the cytotoxicity graph. The data is presented as mean ± SD (n = 8) and underwent statistical evaluation using one-way ANOVA and Tukey's post hoc test (no significant difference: p > 0.05 ns).
Determination of the IC50 concentrations
In order to determine the IC50 values for subsequent experiments in HUVEC cells, the cell index (CI) data of cells treated with aprepitant and aprepitant-loaded nanoparticles at concentrations of 200, 100, and 10 µg/mL were recorded using the xCELLigence RTCA-DP system over a 48-h period. In this system, cells adhered to the plate surface obstruct the contact between the electrode and the culture medium, thereby impeding the flow of electrons. This impedance level, referred to as CI, is influenced by factors such as cell number, shape, size, and the strength of cell adhesion to the underlying layer of the plate53.
The IC50 values at 24 h, as determined by the real-time cell analysis system, were as follows: doxorubicin: 77.6 nM; ERS-Apr: 11.9 µg/mL; PLGA-Apr: 94.3 µg/mL; ERS: 37.5 mg/mL; PLGA: 28.8 mg/mL; and aprepitant: 134 µg/mL and at 48 h were as follows: doxorubicin: 109.4 nM; ERS-Apr: 11.2 µg/mL; PLGA-Apr: 30.9 µg/mL; ERS: 76.7 mg/mL; PLGA: 64.5 mg/ml; and aprepitant: 109 µg/ml (Fig. 4).
Cell proliferation curves of HUVEC cells treated with different aprepitant, ERS-Apr, PLGA-Apr, ERS, PLGA, and doxorubicin concentrations (200, 100, and 10 μg/mL or nM) using the RTCA DP analysis system for 48 h (A). IC50 values of the compounds at 24 h and 48 h were calculated based on cell proliferation data (B).
The absence of similar studies investigating the anti-proliferative effects of aprepitant on HUVEC cells within the existing literature underscores the novelty of our research. In similar studies conducted with cervical cancer cells54 and osteosarcoma cells55, the IC50 values of aprepitant at 24 h were determined to be 31.6 µM and 31.55 µM, respectively. The comparison between the IC50 values implies a heightened susceptibility of cancer cells to aprepitant in contrast to HUVEC cells.
It is known that the solubility of aprepitant in water is very low, which limits its oral bioavailability due to its low solubility and dissolution process13,56,57. When comparing IC50 values, it becomes evident that ERS-Apr displays greater cytotoxicity against HUVEC cells than the original aprepitant formulation. Similarly, aprepitant-loaded PLGA nanoparticles also exhibit increased cytotoxicity compared to the original formulation. In the comparison of IC50 values for aprepitant-loaded nanoparticles, ERS-Apr proves to be more cytotoxic to HUVEC cells than PLGA-Apr. Furthermore, at 24 h, the cytotoxic effect of ERS-Apr (11.9 µg/ml) is observed to occur more rapidly than that of PLGA-Apr (94.3 µg/ml). These findings are consistent with the experimental data presented in Fig. 1, highlighting the concurrence between the cellular uptake levels of aprepitant-loaded drug delivery systems and the observed cytotoxic effects of ERS-Apr and PLGA-Apr at 24 h.
It is important to note that one of the primary limitations of chemotherapeutic drugs is their lack of specificity, potentially affecting healthy tissues58. Cell index data obtained from the cytotoxicity study with doxorubicin indicates that when compared to aprepitant-loaded nanoparticles, aprepitant-loaded ERS and PLGA nanoparticles provide a more controlled cytotoxicity response.
ERS polymers offer various formulations with pH-dependent solubilities, aiding developers in designing precisely controlled release profiles59. PLGA, an FDA-approved polymer, is renowned for its biocompatibility, non-toxicity, targeting capabilities, and sustained drug release capacity, making it a cornerstone in the creation of drug-loaded nanoparticles for targeted cancer therapy60. To ensure the safety of the carrier systems on HUVEC cells, experiments were also conducted using ERS and PLGA without any loaded substances. Remarkably, the IC50 values for both ERS and PLGA at 24 and 48 h exceeded 400 µg/ml, indicating no significant cytotoxicity in cell culture. Consequently, it can be deduced that ERS and PLGA nanoparticles did not exhibit cytotoxicity in HUVEC cells.
Evaluation of antiangiogenic effects
Effects of aprepitant, ERS-Apr and PLGA-Apr on HUVEC cell migration
Angiogenesis, a physiological process characterized by the formation of new blood vessels from pre-existing ones, plays a pivotal role in tumor progression and development. Investigating the migration of endothelial cells in response to VEGF, a well-established contributor to neovascularization, holds significant importance in enhancing our comprehension of the underlying biological and molecular mechanisms61.
Real-time cell analysis devices serve as invaluable tools for scrutinizing cell migration dynamics without necessitating cell labeling. In this analysis, the observed increase in impedance data acquired from the device is directly proportional to the number of cells migrating towards the lower substrate surface62.
In the migration studies, the impact of M2c supernatant, aprepitant, and aprepitant-loaded nanoparticles (ERS-Apr and PLGA-Apr) on HUVEC cell migration were investigated. The changes in cell migration relative to the control were as follows: 185.57, 75.02, 87.90, and 80.56%, respectively (Fig. 5A). Notably, in the presence of M2c, cell migration significantly increased compared to the control (*p < 0.05), but this increase was effectively mitigated by aprepitant, ERS-Apr, and PLGA-Apr, demonstrating significant inhibition compared to M2c (##p < 0.01).
The impact of different concentrations of aprepitant, ERS-Apr + M2c, and PLGA-Apr + M2c on HUVEC cell migration, either alone (A) or in M2c co-culture (B). The migration slope graphs illustrate the relationship between aprepitant, ERS-Apr, and PLGA-Apr concentrations applied in the presence or absence of M2c, plotted against 24-h cell index values in HUVEC cells. The graphs represent the mean ± standard deviation of CI data (n = 4). Statistical analysis was conducted using one-way ANOVA, followed by multiple comparisons with post hoc Tukey tests. Results were categorized as follows: no difference (p > 0.05), significant difference compared to the control (*p < 0.05), and significant difference compared to the M2c group (#p < 0.05, ##p < 0.01).
The composition of the tumor microenvironment varies depending on tissue origin and stages of progression. Typically, the tumor microenvironment consists of tumor vascular systems, extracellular matrix, cancer-associated fibroblasts, and activated immune cells. These components interact with cancer cells not only through paracrine signaling but also through cell-to-cell contact mechanisms63. Therefore, in cancer research, it is considered essential to allow interaction with microenvironment components to create a more realistic tumor model. Consequently, co-culture models involving different cell types are frequently used64. Hence, the migration study was also conducted in an M2c co-culture setting. Changes in migration were observed in the following groups: M2c supernatant, aprepitant + M2c, ERS-Apr + M2c, and PLGA-Apr + M2c, with values of 180.96, 163.39, 153, and 104.2%, respectively (Fig. 5B). In this context, the presence of M2c led to a significant increase in cell migration compared to the control (*p < 0.05). Interestingly, this enhanced migration was notably inhibited only by PLGA-Apr in the presence of M2c (#p < 0.05).
The migration experiment was supplemented with a morphological assessment. When examining the immunofluorescence intensity data representing HUVEC cell migration at 24 h, the changes compared to the control in the M2c, aprepitant, ERS-Apr, PLGA-Apr, aprepitant + M2c, ERS-Apr + M2c, and PLGA-Apr + M2c groups were determined to be 197.64, 90.99, 98.52, 92.26, 144.75, 135.19, and 124.75%, respectively (Fig. 6). Similarly, at 48 h, the changes in the same groups were 203.99, 86.02, 98.90, 94.98, 183.56, 160.26, and 155.98%, respectively. The increase in cell migration in the M2c group was significantly inhibited in all experimental groups at both 24 and 48 h (####p < 0.0001). While the experimental results align with those obtained through real-time cell analysis, morphological analysis exhibits lower sensitivity.
Morphological migration analysis results. The 24- and 48-h impact of aprepitant, ERS-Apr, and PLGA-Apr on HUVEC cell migration, both alone or in M2c co-culture, is visually presented (A) (objective: 10×). The slope graph is based on the immunofluorescence intensity measurement results obtained from morphological imaging (B). The graphs represent the mean ± standard deviation of immunofluorescence intensity data (n = 3). Statistical analysis for the 24- and 48-h data involved an independent one-way ANOVA followed by post hoc Tukey tests for multiple comparisons (no significant difference: p > 0.05; significant difference compared to control: **p < 0.01 and ****p < 0.0001, significant difference compared to M2c group: ####p < 0.0001).
Effects of aprepitant, ERS-Apr and PLGA-Apr on HUVEC cell invasion
Angiogenesis, the formation of new blood vessels from preexisting ones, is a crucial process in both health and disease. It relies on endothelial cells' ability to migrate and invade tissues65. Many drugs developed for cancer treatment target metastatic properties of cancer cells, including migration, chemotaxis, and invasion, which are part of invasive processes like angiogenesis. Inhibiting the invasion of HUVEC endothelial cells that interact with cancer cells in such processes is an interesting target in cancer therapy. The effects of aprepitant and aprepitant-loaded nanoparticles on HUVEC cell invasion were studied using a real-time cell analysis system with matrix-coated CIM plates.
Based on the results of the HUVEC cell invasion analysis, the presence of M2c led to notable alterations in HUVEC cell invasion in various groups: the M2c supernatant, aprepitant + M2c, ERS-Apr + M2c, and PLGA-Apr + M2c, resulting in percentage changes of 123.16%, 82.95%, 73.55%, and 56.49%, respectively, relative to the control (Fig. 7). It was evident that the presence of M2c significantly increased cell invasion when compared to the control (*p < 0.05). Furthermore, it is noteworthy that this observed increase was significantly suppressed in all groups when compared to M2c-alone conditions.
The effects of aprepitant, ERS-Apr, and PLGA-Apr concentrations on HUVEC cell invasion in M2c polarize macrophage co-culture (A). An invasion slope graph (B) was plotted for aprepitant, ERS-Apr, and PLGA-Apr concentrations applied in the presence of M2c, based on 24-h cell index values in HUVEC cells. The graphs represent mean ± standard deviation of cell index data (n = 4). Statistical analysis was conducted using a one-way ANOVA followed by a post hoc Tukey test. No difference: p > 0.05; significant difference compared to the control: *p < 0.05, **p < 0.01, and ****p < 0.0001, significant difference compared to the M2c group: ####p < 0.0001).
Evaluation of the VEGF levels
VEGF, the most potent endogenous pro-angiogenic factor in humans66,67,68,69, plays a pivotal role in triggering angiogenesis by binding to type III receptor tyrosine kinases on cell surfaces, leading to receptor dimerization and activation through phosphorylation pathways70. Ligands such as VEGFA and VEGFB, upon binding to the VEGF receptor, initiate downstream signaling pathways, enhancing endothelial cell proliferation, migration, invasion, and vascular permeability through signaling molecules like ERK1/2 and AKT6,7,8,970,71,72,73. Consequently, therapeutic angiogenesis studies have predominantly focused on disrupting the VEGF-VEGFR pathways74.
In the presence of M2c, the effects of aprepitant and aprepitant-loaded nanoparticles on VEGFA and VEGFB gene expression were examined using the RT-PCR method. According to the results, VEGFA gene expression was notably changed in the presence of M2c by aprepitant, ERS-Apr, and PLGA-Apr, with reductions of 0.43, 0.05, and 0.03-fold, respectively. Similarly, VEGFB gene expression in the same groups showed reductions of 0.33, 0.36, and 0.03-fold, respectively (Fig. 8). These results were statistically significant across all groups (****p < 0.0001).
The effects of aprepitant, ERS-Apr, and PLGA-Apr concentrations in M2c co-culture on the HUVEC VEGFA and VEGFB gene expressions. The graph represents the mean ± standard deviation of normalized RT-PCR fold change data (n = 3). Statistical analysis was conducted independently for each gene using a one-way ANOVA followed by a post hoc Tukey test (significant difference compared to the control: ****p < 0.0001).
VEGFR-2 serves as the primary mediator of VEGF's effects on proliferation, migration, survival, and permeability69,75,76. It is known that signal transduction is modulated through VEGFR-2 signaling activation by VEGF in HUVECs77. Therefore, the effects of aprepitant and aprepitant-loaded nanoparticles on VEGFR-2 were investigated at the protein level. For this purpose, HUVEC cells were treated with aprepitant, ERS-Apr, and PLGA-Apr, followed by labeling with VEGFR-2 antibody and immunofluorescence intensity measurement.
Analysis of immunofluorescence intensity data, reflecting the impact of aprepitant and nanoparticles on VEGFR-2 protein in the presence of M2c, revealed reductions of 61.96%, 59.65%, and 50.45% in the aprepitant, ERS-Apr, and PLGA-Apr groups, respectively, in comparison to the control (Fig. 9). Importantly, VEGFR-2 exhibited a significant decrease across all concentration groups (****p < 0.0001).
The effects of aprepitant, ERS-Apr, and PLGA-Apr concentrations in M2c co-culture on HUVEC VEGFR-2 protein levels (objective: 10X) (A). The slope graph drawn is based on immunofluorescence intensity measurement results obtained from morphological imaging (B). The graph represents the mean ± standard deviation of immunofluorescence intensity data (n = 3), normalized to cell numbers. Statistical analysis was conducted using a one-way ANOVA followed by a post hoc Tukey test for multiple comparisons (significant difference compared to control: ****p < 0.0001).
Conclusions
This study investigated aprepitant-loaded PLGA and Eudragit RS 100 nanoparticles as potential therapies targeting cancer angiogenesis. Our findings demonstrate efficient encapsulation of aprepitant, ensuring stable and uniform particles suitable for drug delivery. Characterization studies showed optimal particle sizes for cellular uptake and stable zeta potentials crucial for maintaining colloidal stability. Fluorescence microscopy revealed enhanced uptake of aprepitant-loaded nanoparticles by HUVEC cells compared to free aprepitant, indicating improved bioavailability. Cytotoxicity tests showed dose-dependent effects on HUVEC cell viability, with low IC50 values suggesting potent anti-proliferative activity. In vitro assays demonstrated that aprepitant-loaded nanoparticles effectively inhibited HUVEC cell migration and invasion, especially in environments mimicking tumor conditions with M2c supernatant. This was associated with downregulation of VEGFA and VEGFB gene expressions and reduced VEGFR-2 protein levels, critical for tumor angiogenesis.
Overall, aprepitant-loaded PLGA and Eudragit RS 100 nanoparticles show promise as targeted therapies for disrupting cancer angiogenesis. Utilizing nanoparticle technology to enhance drug delivery efficacy presents a strategic approach to reducing tumor vascularization and potentially improving cancer treatment outcomes. Future research should validate these findings in animal models and clinical trials to confirm their therapeutic potential and safety.
Data availability
The data that support the findings of this study are available in the methods of this article.
References
Anttila, J. V. et al. Contrasting the impact of cytotoxic and cytostatic drug therapies on tumour progression. PLoS Comput. Biol. 15, e1007493 (2019).
Serkova, N. J. & Eckhardt, S. G. Metabolic imaging to assess treatment response to cytotoxic and cytostatic agents. Front. Oncol. 6, 152 (2016).
Rixe, O. & Fojo, T. Is cell death a critical end point for anticancer therapies or is cytostasis sufficient?. Clin. Cancer Res. 13, 7280–7287 (2007).
Muñoz, M. & Coveñas, R. The neurokinin-1 receptor antagonist aprepitant: An intelligent bullet against cancer?. Cancers (Basel) 12, 2682 (2020).
Rizwanullah, M., Perwez, A., Mir, S. R., Rizvi, M. M. A. & Amin, S. Exemestane encapsulated polymer-lipid hybrid nanoparticles for improved efficacy against breast cancer: Optimization, in vitro characterization and cell culture studies. Nanotechnology 32, 415101 (2021).
Rizwanullah, M. et al. Polymer-lipid hybrid nanoparticles of exemestane for improved oral bioavailability and anti-tumor efficacy: An extensive preclinical investigation. Int. J. Pharm. 642, 123136 (2023).
Muñoz, M., Coveñas, R., Esteban, F. & Redondo, M. The substance P/NK-1 receptor system: NK-1 receptor antagonists as anti-cancer drugs. J. Biosci. 40, 441–463 (2015).
Munoz, M., Rosso, M. & Covenas, R. The NK-1 receptor: A new target in cancer therapy. Curr. Drug Targets 12, 909–921 (2011).
Ramjiawan, R. R., Griffioen, A. W. & Duda, D. G. Anti-angiogenesis for cancer revisited: Is there a role for combinations with immunotherapy?. Angiogenesis 20, 185–204 (2017).
Yao, Y., Xu, X.-H. & Jin, L. Macrophage polarization in physiological and pathological pregnancy. Front. Immunol. 10, 792 (2019).
Atri, C., Guerfali, F. Z. & Laouini, D. Role of human macrophage polarization in inflammation during infectious diseases. Int. J. Mol. Sci. 19, 1801 (2018).
Lin, L. et al. CCL18 from tumor-associated macrophages promotes angiogenesis in breast cancer. Oncotarget 6, 34758 (2015).
Ren, L., Zhou, Y., Wei, P., Li, M. & Chen, G. Preparation and pharmacokinetic study of aprepitant–sulfobutyl ether-β-cyclodextrin complex. AAPS PharmSciTech 15, 121–130 (2014).
Mudshinge, S. R., Deore, A. B., Patil, S. & Bhalgat, C. M. Nanoparticles: Emerging carriers for drug delivery. Saudi Pharm. J. 19, 129–141 (2011).
Nagarwal, R. C., Kant, S., Singh, P. N., Maiti, P. & Pandit, J. K. Polymeric nanoparticulate system: A potential approach for ocular drug delivery. J. Controlled Release 136, 2–13 (2009).
Öztürk, A. A., Yenilmez, E. & Özarda, M. G. Clarithromycin-loaded poly (lactic-co-glycolic acid)(PLGA) nanoparticles for oral administration: effect of polymer molecular weight and surface modification with chitosan on formulation, nanoparticle characterization and antibacterial effects. Polymers (Basel) 11, 1632 (2019).
Öztürk, A. A., Martin-Banderas, L., Cayero-Otero, M. D., Yenilmez, E. & Yazan, Y. New approach to hypertension treatment: Carvediol-loaded PLGA nanoparticles, preparation, in vitro characterization and gastrointestinal stability. Lat. Am. J. Pharm. 37, 1730–1741 (2018).
Şenel, B. & Öztürk, A. A. New approaches to tumor therapy with siRNA-decorated and chitosan-modified PLGA nanoparticles. Drug Dev. Ind. Pharm. 45, 1835–1848 (2019).
Öztürk, A. A., Yenilmez, E., Şenel, B., Kıyan, H. T. & Güven, U. M. Effect of different molecular weight PLGA on flurbiprofen nanoparticles: Formulation, characterization, cytotoxicity, and in vivo anti-inflammatory effect by using HET-CAM assay. Drug Dev. Ind. Pharm. 46, 682–695 (2020).
Martín-Banderas, L. et al. Engineering of Δ9-tetrahydrocannabinol delivery systems based on surface modified-PLGA nanoplatforms. Colloids Surf. B Biointerfaces 123, 114–122 (2014).
Alper Öztürk, A., Namlı, İ & Aygül, A. Cefaclor monohydrate-loaded colon-targeted nanoparticles for use in COVID-19 dependent coinfections and intestinal symptoms: Formulation, characterization, release kinetics, and antimicrobial activity. Assay Drug Dev. Technol. 19, 156–175 (2021).
Ekinci, M., Öztürk, A. A., Santos-Oliveira, R. & İlem-Özdemir, D. The use of Lamivudine-loaded PLGA nanoparticles in the diagnosis of lung cancer: Preparation, characterization, radiolabeling with 99mTc and cell binding. J. Drug Deliv. Sci. Technol. 69, 103139 (2022).
Öztürk, A. A., Aygül, A. & Şenel, B. Influence of glyceryl behenate, tripalmitin and stearic acid on the properties of clarithromycin incorporated solid lipid nanoparticles (SLNs): Formulation, characterization, antibacterial activity and cytotoxicity. J. Drug Deliv. Sci. Technol. 54, 101240 (2019).
Öztürk, A., Güven, U. & Yenilmez, E. Flurbiprofen loaded gel based topical delivery system: Formulation and in vitro characterization with new developed UPLC method. ACTA Pharmaceutica Sciencia 56, 81–105 (2018).
Ozturk, A. & Güven, U. Cefaclor monohydrate loaded microemulsion formulation for topical application: Characterization with new developed UPLC method and stability study. J. Res. Pharm. 23, 426–440 (2019).
Öztürk, A. A. et al. Dexketoprofen trometamol-loaded poly-lactic-co-glycolic acid (PLGA) nanoparticles: Preparation, in vitro characterization and cyctotoxity. Trop. J. Pharm. Res. 18, 1–11 (2019).
Öztürk, A. & Aygül, A. Design of cefaclor monohydrate containing nanoparticles with extended antibacterial effect by nano-spray dryer: A nanoenglobing study. J. Res. Pharm. 24, 100–111 (2020).
Öztürk, A. A., Namlı, İ, Güleç, K. & Görgülü, Ş. Design of lamivudine loaded nanoparticles for oral application by nano spray drying method: A new approach to use an antiretroviral drug for lung cancer treatment. Comb. Chem. High Throughput Screen 23, 1064–1079 (2020).
Jiménez-López, J. et al. A novel nanoformulation of PLGA with high non-ionic surfactant content improves in vitro and in vivo PTX activity against lung cancer. Pharmacol. Res. 141, 451–465 (2019).
Kaya-Tilki, E. & Dikmen, M. Neuroprotective effects of some epigenetic modifying drugs’ on Chlamydia pneumoniaeinduced neuroinflammation: A novel model. PLoS One 16, e0260633 (2021).
Kaya Tilki, E., Engür Öztürk, S., Özarda, M. G., Cantürk, Z. & Dikmen, M. Investigation of the neuroprotective and neuritogenic effects of halotolerant Penicillium flavigenum-derived sorbicillin-like compounds on PC-12 Adh cells. Cytotechnology 73, 801–813 (2021).
Dikmen, M., Canturk, Z., Tilki, E. K. & Engur, S. Evaluation of antiangiogenic and antimetastatic Effects of Penicillium chrysogenum Secondary Metabolites. Indian J. Pharm. Sci. 79, 49–57 (2017).
Canturk, Z., Artagan, Ö. & Dıkmen, M. Anticancer effects of secondary metabolites of Penicillium chrysogenum var. chrysogenum on colon adenocarcinoma cells. Fresenius Environ. Bull. 25, 6190–6197 (2016).
Engür, S. & Dikmen, M. The evaluation of the anti-cancer activity of ixazomib on Caco2 colon solid tumor cells, comparison with bortezomib. Acta Clin. Belg. 72, 391–398 (2017).
Roshan Moniri, M. et al. Dynamic assessment of cell viability, proliferation and migration using real time cell analyzer system (RTCA). Cytotechnology 67, 379–386 (2015).
Dikmen, M., Cantruk, Z., Engur, S. & Tilki, K. Inhibitory effects of secondary metabolites of halotolerant Aspergillus terreus on angiogenesis. Bıomed. Res.-Indıa. 28, 3613–3618 (2017).
Engür, S., Dikmen, M. & Öztürk, Y. Comparison of antiproliferative and apoptotic effects of a novel proteasome inhibitor MLN2238 with bortezomib on K562 chronic myeloid leukemia cells. Immunopharmacol. Immunotoxicol. 38, 87–97 (2016).
Yuan, A. et al. Opposite effects of M1 and M2 macrophage subtypes on lung cancer progression. Sci. Rep. 5, 14273 (2015).
Engür-Öztürk, S. & Dikmen, M. Proteasome inhibitor immunotherapy for the epithelial to mesenchymal transition: assessing the A549 lung cancer cell microenvironment and the role of M1, M2a and M2c ‘hydrocortisone-polarised’macrophages. Mol. Biol. Rep. 49, 4777–4793 (2022).
Emami, J., Mohiti, H., Hamishehkar, H. & Varshosaz, J. Formulation and optimization of solid lipid nanoparticle formulation for pulmonary delivery of budesonide using Taguchi and Box-Behnken design. Res. Pharm. Sci. 10, 17 (2015).
Müller, R. H., Mäder, K. & Gohla, S. Solid lipid nanoparticles (SLN) for controlled drug delivery—A review of the state of the art. Eur. J. Pharm. Biopharm. 50, 161–177 (2000).
Yurtdaş Kırımlıoğlu, G. & Öztürk, A. A. Levocetirizine dihydrochloride-loaded chitosan nanoparticles: Formulation and ın vitro evaluation. Turk. J. Pharm. Sci. 17, 27–35 (2020).
Öztürk, A. A. & Kirimlioğlu, G. Y. Preparation and in vitro characterization of lamivudine loaded nanoparticles prepared by acid and/or ester terminated PLGA for effective oral anti-retroviral therapy. J. Res. Pharm. 23, 897–913 (2019).
Shen, S., Wu, Y., Liu, Y. & Wu, D. High drug-loading nanomedicines: Progress, current status, and prospects. Int. J. Nanomed. 12, 4085 (2017).
Durán-Lobato, M., Martín-Banderas, L., Gonçalves, L. M. D., Fernández-Arévalo, M. & Almeida, A. J. Comparative study of chitosan-and PEG-coated lipid and PLGA nanoparticles as oral delivery systems for cannabinoids. J. Nanoparticle Res. 17, 1–17 (2015).
Rennick, J. J., Johnston, A. P. R. & Parton, R. G. Key principles and methods for studying the endocytosis of biological and nanoparticle therapeutics. Nat. Nanotechnol. 16, 266–276 (2021).
Lin, T.-T. et al. Development and characterization of sorafenib-loaded PLGA nanoparticles for the systemic treatment of liver fibrosis. J. Controlled Release 221, 62–70 (2016).
Akkouch, A., Zhang, Z. & Rouabhia, M. Engineering bone tissue using human dental pulp stem cells and an osteogenic collagen-hydroxyapatite-poly (l-lactide-co-ɛ-caprolactone) scaffold. J. Biomater. Appl. 28, 922–936 (2014).
Ngamwongsatit, P., Banada, P. P., Panbangred, W. & Bhunia, A. K. WST-1-based cell cytotoxicity assay as a substitute for MTT-based assay for rapid detection of toxigenic Bacillus species using CHO cell line. J. Microbiol. Methods 73, 211–215 (2008).
Schneider, R. et al. White mineral trioxide aggregate induces migration and proliferation of stem cells from the apical papilla. J. Endod. 40, 931–936 (2014).
Bardelli, C. et al. Recurrent major depressive disorder: imbalance of neurokinin (NK)-1 and NK-2 receptor expression in monocytes. Pharmacol. Res. 68, 24–30 (2013).
Harford-Wright, E., Lewis, K. M., Vink, R. & Ghabriel, M. N. Evaluating the role of substance P in the growth of brain tumors. Neuroscience 261, 85–94 (2014).
Hamidi, H., Lilja, J. & Ivaska, J. Using xCELLigence RTCA instrument to measure cell adhesion. Bio. Protoc. 7, e2646 (2017).
Mozafari, M., Ebrahimi, S., Darban, R. A. & Hashemy, S. I. Potential in vitro therapeutic effects of targeting SP/NK1R system in cervical cancer. Mol. Biol. Rep. 49, 1067–1076 (2022).
Alsaeed, M. A., Ebrahimi, S., Alalikhan, A., Hashemi, S. F. & Hashemy, S. I. The potential in vitro inhibitory effects of Neurokinin-1 Receptor (NK-1R) antagonist, aprepitant, in osteosarcoma cell migration and metastasis. Biomed. Res. Int. 2022, 8082608 (2022).
Nanaki, S. et al. Aprepitant drug in ternary pharmaceutical solid dispersions with Soluplus® and poloxamer 188 prepared by melt mixing. Sci. 1, 48 (2019).
Ridhurkar, D. N. et al. Inclusion complex of aprepitant with cyclodextrin: Evaluation of physico-chemical and pharmacokinetic properties. Drug Dev. Ind. Pharm. 39, 1783–1792 (2013).
Anirudhan, T. S. & Sandeep, S. Synthesis, characterization, cellular uptake and cytotoxicity of a multi-functional magnetic nanocomposite for the targeted delivery and controlled release of doxorubicin to cancer cells. J. Mater. Chem. 22, 12888–12899 (2012).
Kriplani, P. & Guarve, K. Eudragit, a nifty polymer for anticancer preparations: A patent review. Recent Pat. Anticancer Drug Discov. 17, 92–101 (2022).
Saraf, A., Dubey, N., Dubey, N. & Sharma, M. Enhancement of cytotoxicty of diallyl disulfide toward colon cancer by Eudragit S100/PLGA nanoparticles. J. Drug Deliv. Sci. Technol. 64, 102580 (2021).
Cerezo, A. B., Hornedo-Ortega, R., Álvarez-Fernández, M. A., Troncoso, A. M. & García-Parrilla, M. C. Inhibition of VEGF-induced VEGFR-2 activation and HUVEC migration by melatonin and other bioactive indolic compounds. Nutrients 9, 249 (2017).
Bird, C. & Kirstein, S. Real-time, label-free monitoring of cellular invasion and migration with the xCELLigence system. Nat. Methods 6, v–vi (2009).
Horman, S. R., To, J. & Orth, A. P. An HTS-compatible 3D colony formation assay to identify tumor-specific chemotherapeutics. J. Biomol. Screen 18, 1298–1308 (2013).
Liu, T., Lin, B. & Qin, J. Carcinoma-associated fibroblasts promoted tumor spheroid invasion on a microfluidic 3D co-culture device. Lab. Chip 10, 1671–1677 (2010).
Figueiredo, A. M. et al. Endothelial cell invasion is controlled by dactylopodia. Proc. Natl. Acad. Sci. 118, e2023829118 (2021).
Cebe-Suarez, S., Zehnder-Fjällman, A. & Ballmer-Hofer, K. The role of VEGF receptors in angiogenesis; complex partnerships. Cell. Mol. Life Sci. 63, 601–615 (2006).
Cook, K. M. & Figg, W. D. Angiogenesis inhibitors: Current strategies and future prospects. CA Cancer J. Clin. 60, 222–243 (2010).
Dulak, J. Nutraceuticals as anti-angiogenic agents: Hopes and reality. J. Physiol. Pharmacol. Suppl. 56, 51–69 (2005).
Giles, F. J. The vascular endothelial growth factor (VEGF) signaling pathway: A therapeutic target in patients with hematologic malignancies. Oncologist 6, 32–39 (2001).
Koch, S., Tugues, S., Li, X., Gualandi, L. & Claesson-Welsh, L. Signal transduction by vascular endothelial growth factor receptors. Biochem. J. 437, 169–183 (2011).
Bernatchez, P. N., Soker, S. & Sirois, M. G. Vascular endothelial growth factor effect on endothelial cell proliferation, migration, and platelet-activating factor synthesis is Flk-1-dependent. J. Biol. Chem. 274, 31047–31054 (1999).
Cines, D. B. et al. Endothelial cells in physiology and in the pathophysiology of vascular disorders. Blood J. Am. Soc. Hematol. 91, 3527–3561 (1998).
Pettersson, A. et al. Heterogeneity of the angiogenic response induced in different normal adult tissues by vascular permeability factor/vascular endothelial growth factor. Lab. Investig. 80, 99–115 (2000).
Farzaneh Behelgardi, M., Zahri, S., Mashayekhi, F., Mansouri, K. & Asghari, S. M. A peptide mimicking the binding sites of VEGF-A and VEGF-B inhibits VEGFR-1/-2 driven angiogenesis, tumor growth and metastasis. Sci. Rep. 8, 17924 (2018).
Ferrara, N. & Kerbel, R. S. Angiogenesis as a therapeutic target. Nature 438, 967–974 (2005).
Holmes, K., Roberts, O. L., Thomas, A. M. & Cross, M. J. Vascular endothelial growth factor receptor-2: Structure, function, intracellular signalling and therapeutic inhibition. Cell Signal 19, 2003–2012 (2007).
Cerezo, A. B., Winterbone, M. S., Moyle, C. W. A., Needs, P. W. & Kroon, P. A. Molecular structure-function relationship of dietary polyphenols for inhibiting VEGF-induced VEGFR-2 activity. Mol. Nutr. Food Res. 59, 2119–2131 (2015).
Acknowledgements
This research was supported by Anadolu University Scientific Research Project Commission (Project number: 2103S115). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
E. Kaya-Tilki and A. A. Ozturk conceived and designed the research; E. Kaya-Tilki, A. A. Ozturk and S. Engur-Ozturk performed the research and acquired the data, E. Kaya-Tilki, and M. Dikmen analyzed and interpreted the data. All authors were involved in drafting and revising the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Kaya-Tilki, E., Öztürk, A.A., Engür-Öztürk, S. et al. Enhanced anti-angiogenic effects of aprepitant-loaded nanoparticles in human umbilical vein endothelial cells. Sci Rep 14, 19837 (2024). https://doi.org/10.1038/s41598-024-70791-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-70791-y
- Springer Nature Limited