Abstract
Low back pain (LBP) is largely attributed to intervertebral disc degeneration (IVDD), of which the endplate changes are an important component. However, the alterations in cell fate and properties within the endplates during degeneration remain unknown. Here, we firstly performed the single-cell RNA-sequencing analysis (scRNA-seq) of the cells focusing on degenerative human endplates. By unsupervised clustering of the 8,534 single-cell based on the gene expression, we identified nine distinct cell types. We employed Gene ontology (GO) analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways analysis, and the single-cell regulatory network inference and clustering (SCENIC) to determine the enriched pathways and transcriptional activities across seven chondrocyte subpopulations. Furthermore, two cell fates of chondrocyte differentiation were found by trajectory analysis, one was enriched in inflammation-related genes, and the other was related to extracellular matrix (ECM). Additionally, the intercellular interactions of macrophages (MA) and chondrocytes, T cells/natural killer cells (T/NK) and chondrocytes were examined by ligand-receptor pairs analysis, showing the important regulative function of FN1 from MA and CD74 from T/NK during endplate degeneration. Overall, our findings provide novel perspectives on the endplate degeneration at the single-cell level and a whole-transcriptome size.
Similar content being viewed by others
Introduction
The functional spinal units (FSUs) are the building blocks of the spine, representing segmental structures with similar properties of the entire spine1,2. Each FSU consists of two adjacent vertebrae and the connected articulation3. The intervertebral disc, which is positioned between two vertebral bodies, is an avascular cartilaginous structure composed of three components—endplates, annulus fibrosus (AF) and nucleus pulposus (NP)4. Endplates are concave structures that are 1 mm thick in humans and have two distinct layers: the cartilage endplate (CEP) made of hyaline cartilage and the bony endplate (BEP) consisting of semi-porous cancellous bone5,6. During spinal growth until skeletal maturity, the endplates have microscopic blood vessels penetrating from the vertebrae adjacent to the BEP, providing a vascular supply to the intervertebral disc5. Cartilage endplates are vital in the maintenance of disc homeostasis. Endplates are critical regulatory organs for the flow of nutrients into and out of the intervertebral disc and possess a range of physiological properties that are crucial to spinal function5,7,8.
Low back pain (LBP) is one of the most prevalent musculoskeletal system illnesses. It continues to be one of the leading causes of medical expenses and financial strain despite advances in surgery and medication. Intervertebral disc degeneration (IVDD) is thought to be the main pathogenic cause of LBP. The endplate degeneration is a significant factor leading to IVDD as an early morphological change9. Modic changes (MC) refer to endplate changes that can be identified using magnetic resonance imaging (MRI)10. Studies have shown increased outgrowth of vascular vessels and nerve fibers to endplates in degenerative spine, which is considered a possible cause of LBP5,8,11,12. Additionally, recent research has highlighted the link between chondrocytes and immunity, suggesting that immune responses may adversely affect chondrocytes either directly or indirectly13. Further understanding of the cellular composition of the endplate and its connection with cellular immunity during degeneration is necessary.
Single-cell sequencing technology has revolutionized the multidimensional analysis of various tissue cells and diseases. In particular, scRNA-seq is becoming increasingly popular in the study of tissue and cellular heterogeneity14. With the continuous improvement and innovation of high-throughput sequencing technology, scRNA-seq is being applied to investigate the molecular regulatory mechanisms involved in physiological development, pathological processes, inflammation, and immunity across a range of tissues and diseases. ScRNA-seq has been used to detect and analyze the intervertebral disc NP tissue and meniscus tissue, which has expanded our understanding of osteoporosis, IVDD and osteoarthritis (OA)15,16,17,18,19,20. For nucleus pulposus tissue, three empirically defined populations were generated: cartilage progenitor cells (CPCs), fibrochondrocyte progenitors (FCPs), and homeostatic chondrocytes (HomCs)17. In addition, unbiased chondrocyte atlases of articular cartilage have been proposed, including proliferative chondrocytes, prehypertrophic chondrocytes, hypertrophic chondrocytes, fibrocartilage chondrocytes, effector chondrocytes, regulatory chondrocytes and homeostatic chondrocytes20. These findings have significant implications for developing novel therapeutic strategies to target specific cell types or signaling pathways involved in these conditions. However, the scRNA-seq study specifically focused on endplate degeneration is lack.
In this study, we performed a cluster analysis of endplate cells during IVDD and identified nine different cell types including chondroblasts (CB), regulatory chondrocytes (RegC), homeostatic chondrocytes (HomC), prehypertrophic chondrocytes (pre-HTC), fibrocartilage chondrocytes (FC), proliferative chondrocytes (ProC), hypertrophic chondrocytes (HTC), T cells/natural killer cells (T/NK) and macrophages (MA). Macrophages and T/NK cells are both derived from hematopoietic stem cells. They collaborate in immunity, with macrophages focusing on early defense and antigen presentation, and T/NK cells managing targeted responses and cell-mediated cytotoxicity. Our findings provide a comprehensive collection of cell clusters from degenerated endplates and demonstrate the involvement of immune cells in the process of endplate degeneration. This work sheds light on the cellular composition, biological process, transcriptional activity and cell-to-cell communication of degenerative endplates.
Materials and methods
Cartilage endplate sample
Cartilage endplate tissue was obtained from two patients diagnosed with modic changes at the levels of L3-L4 or L4-L5, who were undergoing surgery for disc herniation. The patients were a 58-year-old female and a 66-year-old male, respectively. All participants included were confirmed with MRI images before surgery. This study was approved by the Life Science Ethics Review Committee of Zhengzhou University. All experiments were performed in accordance with relevant guidelines and regulations. And all the patients signed informed consent forms before participation. The cartilage endplate tissue of the intervertebral discs resected during the surgery was transported immediately to the laboratory under 4 °C cold storage for single-cell suspension preparation.
Preparation of single cell suspensions of vertebral endplate samples
The vertebral endplate tissues were washed twice with sterile PBS buffer to remove any blood and prevent contamination, left to stand, and the supernatants were aspirated. The tissues were cut quickly into small 1–2 mm pieces with pre-cooled ophthalmic scissors and placed on ice. And added pre-warmed digestive enzymes, including 1 mg/ml collagenase type I (Gibco; #17100-017), 2 mg/ml dispase II (Sigma-Aldrich; #D4693-1G) and 1 unit/ml DNase I (NEB; #M0303S), mixed gently with a pasteurizing pipette and digested in a 37 °C water bath for 20–25 min, mixed gently with a pasteurizing pipette every 5 min. At the end of digestion, added an equal volume of complete medium containing FBS to terminate the digestion. The isolated cells were filtered using 70-μm disposable filter and washed twice with PBS (Gibco; # 13151014), centrifuged for 5 min at 500×g at 4 °C to collect the cell precipitate. To remove red blood cells, incubated with 1X RBC lysis buffer (Biogems; 64010-00-100) at room temperature for 5-10 min, centrifuged for 5 min at 500×g, collected cell precipitate. Added PBS (0.04% BSA, Sigma; 9048–46-8) and washed 1–2 times, centrifuged for 5 min, discarded the supernatant and resuspend in PBS. For scRNA-seq, the cell density was subsequently changed to 106/ml.
10 × Genomics vertebral endplate cell single cell marker and library construction and sequencing
Single cell suspensions of vertebral endplate tissue were prepared and tested (cell viability > 80%). They were subjected to single cell library construction according to the Single Cell 3' Reagent Kits manual (10 × Genomics, USA). Using a microfluidic chip, cells and gel beads with cell labeling sequences were wrapped in a droplet, and the cell-wrapped droplet was collected. In the droplet, the cells were lysed so that the mRNA in the cells was connected to the cell Barcode of the bead, forming Single Cell GEMs. These were subjected to RT reactions in the droplet, followed by the disruption of emulsions and cDNA library construction. Cells and reaction reagents were placed in one lane on the microfluidic chip and gel beads in the other lane to form a GEM. Reverse transcription was performed independently in each GEM. Afterward, the labeled cDNA was mixed, then amplified, and the library was constructed.
Single-cell high-throughput sequencing
Library size was measured by a bioanalyzer (Agilent Technologies, United States) and the Qubit™ dsDNA HS Assay Kit (Thermo Fisher) was used to quantify the library products. The NovaSeq 6000 system (Illumina) was used for sequencing with the PE150 strategy.
Processing of single-cell RNA-seq raw data
Sequencing reads were aligned to the human genome reference sequences (GRCh38), and gene-level unique molecular identifier (UMI) count matrices were obtained by using Cell Ranger 3.1.0 (10X Genomics).
Clustering analysis of scRNA-seq data
Generated read counts matrices were analyzed in R (v3.5.2, https://www.R-project.org) via Seurat (v3.1.1)21 following its filtering, variable gene selection, normalization, scaling, dimensionality reduction, clustering and visualization. Cells that with gene counts greater than 200 per cell, UMI counts within outlier thresholds (Q1-1.5IQR < UMI < Q3 + 1.5IQR), and a percentage of mitochondrial genes < 20% were retained for downstream analysis. Multiple samples were integrated by CCA-based anchors method21,22. Following principal component analysis (PCA), the first 30 principal components were selected for clustering the cells. A resolution of 0.5 was used with t-SNE visualization. Differentially expressed genes (DEGs) of each cluster were identified by Seurat’s implementation function FindAllMarkers with Wilcoxon rank-sum test. According the expression of the known markers and specifically expressed genes we assigned 9 known cell types.
Sub-clustering of chondrocyte cell types
The cells of several chondrocyte cell types were performed an unsupervised sub-clustering by Seurat with the first 30 principal components. Using recently reported marker genes, we identified 9 sub cell types, including CB, RegC, HomC, pre-HTC, FC, ProC, HTC, T/NK, and MA.
Pseudotime analysis
Monocle 2 (version 2.10.1)23 was used to estimate a pseudotemporal path of chondrocyte cell differentiation. Monocle is an unsupervised algorithm that increases the temporal resolution of transcriptome dynamics using single-cell RNA-Seq data collected at multiple time points. Monocle can in principle be used to recover single-cell gene expression kinetics from a wide array of cellular processes, including differentiation, proliferation and oncogenic transformation. Cells were ordered in pseudotime along a trajectory using reduceDimension with the DDRTree method and orderCells functions.
Ligand–receptor interactions
To assess cellular crosstalk between different cell types, we used UMI count data and the Python package CellPhoneDB v2.1.224 with the human database v2.0.0 to infer cell–cell communication networks from single cell transcriptome data. Only interactions pass the p values threshold (p < 0.05) were considered.
Motif enrichment and regulatory network
The gene regulatory network of stem cells was built by SCENIC (version 1.1.2)25. With the default parameters, SCENIC generated the enrichment scores of 160 motifs. We used Wilcoxon rank-sum test to calculate the differential enriched motifs when compared between case patients and controls.
Gene functional analysis
Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) terms were applied by clusterProfiler (v3.10.1)26 with the database org.Hs.eg.db (v3.7.0).
Results
Identification of different cell types in endplate during degeneration
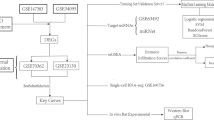
The endplate, indicated as Modic Changes in the IVDD by MRI, was isolated and digested from two patients with lumbar spine surgery. After single-cell transcriptome sequencing and quality control, a total of 8534 cells were obtained (Fig. 1A).
Identification of unique clusters of endplate cells during degeneration. (A) Schematic diagram of sample source and experimental workflow. (B) UMAP of the 8,534 cells profiled nine main cell types. (C) Percentage of each cell type in the two samples. (D) t-SNE plots showing the expression of marker genes for nine cell subclusters. (E) Heatmap of top 10 DEGs for nine clusters; each column represents a cell cluster, and each row represents a DEG for a cluster. CB, chondroblasts; RegC, regulatory chondrocytes; HomC, homeostatic chondrocytes; pre-HTC, prehypertrophic chondrocytes; FC, fibrocartilage chondrocytes; ProC, proliferative chondrocytes; HTC, hypertrophic chondrocytes; MA, macrophages; T/NK, T cells/natural killer cells.
Based on the single-cell transcriptomics data, previous literature and databases, nine distinct cell clusters were identified after unbiased analysis (Fig. 1B–D and Supplementary Fig. S1): 1) Cluster 1 (average 36.69% of all cells): CB is enriched with marker gene chondromodulin (CNMD). At the same time, the genes lumican (LUM) and decorin (DCN) representing chondrogenicity are also expressed in this cluster27; 2) Cluster 2 (average 23.34% of all cells): RegC is characteristically highly expressed marker genes complement factor H (CFH) and frizzled related protein (FRZB)20; 3) Cluster 3 (average 14.89% of all cells): HomC is specifically highly expressed marker genes ras related dexamethasone induced 1 (RASD1)20; 4) Cluster 4 (average 14.89% of all cells): pre-HTC is enriched with marker gene TNF alpha induced protein 6 (TNFAIP6)20; 5) Cluster 5 (average 7.34% of all cells): FC is particularly intensely expressed marker gene collagen type I alpha 1 chain (COL1A1)20; 6) Cluster 6 (average 1.34% of all cells): ProC is powerful expressed marker gene topoisomerase II alpha (TOP2A)28; 7) Cluster 7 (average 0.45% of all cells): HTC is enriched with marker gene collagen type X alpha 1 chain (COL10A1)20; 8) Cluster 8 (average 0.84% of all cells): T/NK) is specifically highly expressed marker gene granulysin (GNLY)29; and 9) Cluster 9 (average 0.22% of all cells): MA is most expressed marker gene lysozyme (LYZ)30. Further, the differential expression analysis showed the top 10 differentially expressed genes for each cluster (Fig. 1E). Specially, CB was newly defined in the present study, suggesting further expansion of the impression of chondrocytes.
Analyzes of biological functions across seven chondrocyte clusters
To further understand the heterogeneity and biological function of seven different chondrocyte clusters, Gene ontology (GO) analysis (Fig. 2A), cell-cycle analysis (Fig. 2B), and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enrichment analysis (Fig. 3A–F) were performed. (1) For CB, the analysis showed that its function was mainly related to cartilage development, showing a tendency towards chondrocyte proliferation and differentiation. (2) We also found that the function of FC was mainly associated with extracellular matrix (ECM) organization and ECM-receptor interaction via collagen fibers and focal adhesion, indicating the involvement of FC in degeneration. (3) As HomC, GO analysis suggested that it was mainly involved in cellular metabolism and homeostasis. In addition, it was associated with MAPK signaling pathway, by KEGG analysis. (4) It was indicated that HTC was related to the regulation of ECM, such as ECM disassembly and peptidase regulator activity. (5) pre-HTC was shown to involve in immune responses. KEGG pathways enrichment analysis demonstrated that pre-HTC was also enriched in ECM regulation. (6) In regard to ProC, it controlled cell proliferation through functions involved in the cell division process. Meanwhile, most of the cells in this cluster were in the G2/M phase of cell division, suggesting that ProC was in an active state of cell proliferation (Fig. 2B). (7) GO analysis suggested that RegC was related to protein localization and the response to reactive oxygen species and metal ion. Disturbances in transition metal ion homeostasis can lead to increased oxidative stress and reactive oxygen species production.
Gene ontology (GO) analysis of chondrocyte subsets. (A) Bar plots show the upregulated terms of seven chondrocyte clusters respectively by GO analysis. (B) The proportion of cells annotated as the different cell cycle phases in different cell clusters. NC, other chondrocytes; CB, chondroblasts; RegC, regulatory chondrocytes; HomC, homeostatic chondrocytes; pre-HTC, prehypertrophic chondrocytes; FC, fibrocartilage chondrocytes; ProC, proliferative chondrocytes; HTC, hypertrophic chondrocytes.
Bar plots show the upregulated pathways of chondrocyte clusters respectively by KEGG analysis. (A) CB, chondroblasts. (B) FC, fibrocartilage chondrocytes. (C) HomC, homeostatic chondrocytes. (D) pre-HTC, prehypertrophic chondrocytes. (E) ProC, proliferative chondrocytes. (F) RegC, regulatory chondrocytes.
Transcriptional regulatory activities in different chondrocyte subsets during endplate degeneration
The TFs could control different chondrocyte phenotypes. To further identify the transcriptional networks in different subsets, the single-cell regulatory network inference and clustering (SCENIC) method were conducted. Based on the distribution of activity across cells, the activity of each regulon in each cell was quantified, and it was then binarized as “on” or “off”. The SCENIC results indicated strong enrichment of EGR1 and JUND regulon activity in all chondrocytes; CEBPB activity in RegC; and ATF3 and BHLHE40 activity in HTC (Fig. 4A). Enrichment analysis of chondrocytes based on chondrogenic function-related TFs revealed that the highly different and specific TF networks (Fig. 4B). In detail, HomC was enriched for EGR3, RORA, MAFB and IRF1. These TFs were involved in the regulation of rhythmic genes (ERR3h and RORA)31,32 and immune response (MAFB and IRF1)33. This is consistent with the GO analysis, implying the participation of HomC in the regulation of homeostasis. In addition, the higher regulatory activity of FOXO1 and FOSL1 in HomC and FC suggested that they have the ability to regulate metabolic homeostasis (FOXO1)34,35,36 as well as cell proliferation, differentiation and transformation (FOSL1)31. The REL, one of the NF-kappaB family, exhibited increased activity in HTC, indicating the inflammatory features of HTC37. RAD21 and MYBL1 were ProC-specific regulators that were crucial for cell cycle and DNA damage repair38,39. MYC as well as CEBPG were upregulated in RegC. MYC manages the cell cycle progression, apoptosis and cellular transformation37. Additionally, EGR1 regulon regulating activities in CB were significantly increased, which is required for differentiation and mitosis. Based on the functional correlation of transcription factors and predicted target genes, we selected EGR1, FOXO1 and REL for analysis and found that EGR1-DCN, FOXO1-collagen type III alpha 1 chain (COL3A1) and REL- interferon regulatory factor 1 (IRF1) were highly related transcription factor-target gene pairs. TF and the infer target genes are co-expression show the t-SNE and related with pseudotime (Fig. 4C,D). Further suggesting that they may play important regulatory roles in CB (EGR1), FC (FOXO1) and HTC (REL), respectively. Therefore, the investigation of potential regulatory TFs will help us understand chondrocyte heterogeneity and their possible involvement in physiological processes.
Transcriptional regulation of seven chondrocyte subclusters. (A) Heatmap of regulons specific to different cell types, as inferred by the SCENIC. Regulon activity was binarized to “on” (black) or “off” (white). (B) Heatmap indicating the activity of chondrogenic function-related TFs analyzed with SCENIC in seven chondrocyte subclusters. (C) tSNE depiction of regulon activity, TF gene expression, and predicted target gene expression for EGR1 in CB, FOXO1 in FC, and REL in HTC. (D) The trend of gene expression changes with pseudotime. CB, chondroblasts; RegC, regulatory chondrocytes; HomC, homeostatic chondrocytes; pre-HTC, prehypertrophic chondrocytes; FC, fibrocartilage chondrocytes; ProC, proliferative chondrocytes; HTC, hypertrophic chondrocytes; MA, macrophages; T/NK, T cells/natural killer cells.
The trajectory branch of all chondrocyte subsets
To investigate the conversion of different chondrocyte clusters and the corresponding gene expression in endplate degeneration, we used the pseudotime trajectories to undertake a trajectory analysis of seven chondrocyte subtypes with two endpoints representing two different cell fates (Fig. 5A). By examining the dispersion patterns of several chondrocyte subtypes in the pseudotime trajectory we found: ProC was mainly located close to the root. CB, pre-HTC and FC were dispersed along the route, whereas RegC, HomC and HTC generally occupied the extremities of the two fates (Fig. 5A,B). Interestingly, ProC, pre-HTC and HTC compromised a complete evolutionary route. And ProC and HTC formed independent head ends and tail ends of the trajectory, respectively (Fig. 5C). This result confirms that these three clusters have a closer association with each other than the rest.
Trajectory branch of chondrocyte subsets. (A) Monocle pseudotime trajectory contains three branches showing the dynamic progression of chondrocyte clusters. (B) Projection of individual cell types onto the trajectory of (A). (C) ProC, pre-HTC and HTC compromise a complete evolutionary route without branches. (D) Heatmap showing the standardized kinetic curves of the genes in trajectory from root to fate1 or fate2. (E) Pseudotime kinetics of specific representative genes from the root of the trajectory to fate1 (solid line) and fate2 (dashed line). (F) Bar plots showing the top annotated GO terms in five gene sets, which are hierarchically clustered from (D). CB, chondroblasts; RegC, regulatory chondrocytes; HomC, homeostatic chondrocytes; pre-HTC, prehypertrophic chondrocytes; FC, fibrocartilage chondrocytes; ProC, proliferative chondrocytes; HTC, hypertrophic chondrocytes.
In order to determine the effect of the differential genes in cell fates, we then deconstructed the gene transformation patterns based on the findings of the trajectory analysis (Fig. 5D). Five different chondrocyte clusters were obtained based on the different gene expression. In the transition to fate1, Cluster 3-enriched genes, included CFH, chitinase 3 like 1 (CHI3L1), FRZB and ceruloplasmin (CP), that were related to regulation of complement activation, cytokine action, and immune-inflammatory response were significantly increased. However, cluster 5-enriched genes, included COL10A1 and integrin binding sialoprotein (IBSP), did not show significant enhancement during the transition to fate 1 or fate 2. COL1A1, AE binding protein 1 (AEBP1) and cartilage oligomeric matrix protein (COMP) were genes enriched in clusters 1 and 4 that were expressed in the direction of fate 2. And they were related to the ECM structure. Notably, Cluster 2-enriched genes, included Jun proto-oncogene (JUN) and FUS RNA binding protein (FUS), which were associated with cellular homeostasis and cellular metabolism, were significantly elevated during the transition to fate2 (Fig. 5D–F). Taken together, the two separate paths of differentiation provided functional diversity in chondrocytes of IVDD, one relating to cellular immunity and the other affecting the ECM. There were high correlations between the two cell fates, as well as the process of inflammatory reactions and ECM modifications. This demonstrated that the treatment of IVDD may benefit from research on the direction of chondrocytes differentiation, especially the fate 1.
Complex intercellular interaction network in degenerative endplate cells
CellPhoneDB analysis was performed to assess intercellular communication in all subpopulations, showing the higher interactions between pre-HTC and FC or CB or ProC (Fig. 6A). Then, we conducted the analysis of ligand-receptor pairs between different clusters (Fig. 6B and Supplementary Fig. S2). The results demonstrated that pre-HTC was most closely related to ProC and FC by ligand-receptor pairs (Fig. 6B). Furthermore, considering the important regulative features of macrophages and T/NK cells in local micro environment, we investigated the ligand-receptor pairs between them and other chondrocyte subsets (Fig. 6C,D). Among these chondrocyte subpopulations, the strength of MA interactions with them varied according to cell types, being the stronger with CB, pre-HTC, FC, and ProC. Notably, we found that the ligand-receptor pairs of FN1(Fibronectin)-a4b7 complex and FN1-a4b1 complex were specifically involved in the interactions of MA and all chondrocyte subsets, but not in the interactions of T/NK cells and chondrocytes (Fig. 6C). These two ligand-receptor pairs mainly participated in the cell adhesion process, indicating the important regulative function of MA. Whereas, the ligand-receptor pairs of CD74-APP, CD74-COPA, and CD74-MF were engaged in the interactions of T/NK cells and chondrocyte subsets, indicating T/NK cells may be widely involved in the regulation of endplate cell degeneration through CD74 (Fig. 6D).
Complex intercellular interaction network in degenerative endplate cells. (A) Interaction network constructed by CellPhoneDB analysis. Thicker lines indicate more interaction with other types of cells. (B) Heatmap showing the number of potential ligand-receptor pairs among the predicted cell types. (C) The detected ligand-receptor pairs of MAs and all chondrocyte subsets by CellPhoneDB. (D) The detected ligand-receptor pairs of T/NK cells and all chondrocyte subsets by CellPhoneDB. CB, chondroblasts; RegC, regulatory chondrocytes; HomC, homeostatic chondrocytes; pre-HTC, prehypertrophic chondrocytes; FC, fibrocartilage chondrocytes; ProC, proliferative chondrocytes; HTC, hypertrophic chondrocytes; MA, macrophages; T/NK, T cells/natural killer cells.
Discussion
Through scRNA-seq, we conducted a detailed analysis of endplate cells during IVDD, providing a comprehensive overview of their cellular composition and gene expression patterns. Our findings confirm previous chondrocyte research, identify key cell subsets and TFs, offer new insights into cell fates and differentiation, and demonstrate chondrocyte-macrophage communication.
We defined CB, linked to chondrogenesis and chondrocyte differentiation., and identified new genes CFH and FRZB, differing from the previous RegC definition by Quanbo Ji et al20.The role of transition metal ions in oxidative stress and inflammation is well-established40. Previously, TOP2A and CENPF were characteristic of CPC17. Based on differential genes, GO analysis, and cell trajectory analysis, we classify ProC and CPC as the same cluster.
Excess iron ions generate oxygen species (ROS) and oxidative stress, damaging cellular components and causing chronic inflammation. We found CP enrichment in RegC, involved in metal ion transport and redox processes, suggesting RegC's role in maintaining metal ion homeostasis and protecting against oxidative stress. Future studies can explore chondrocyte interactions with metal ions and oxidative stress in endplate degeneration.
SCENIC results provide insights into transcriptional activities in chondrocyte subtype. There are a few pathways for HomC to involve homeostasis and metabolism. According to previous findings, Fosl1 regulates MMPs expression41, IRF1 upregulates inflammatory factors42, and SOX4 influences cell fate in HomC43. In RegC, MYC promotes growth-associated genes and VEGFA production44,45, while CEBPG binds MYC to activate the PI2K-AKT pathway46, may link RegC to angiogenesis in IVDD. EGR1, a specific modulator of CB, plays a pivotal role in cellular processes such as survival, proliferation, and apoptosis47. DCN, one of the chondrogenic genes, is correlated with EGR1 as its predicted target gene. We speculate that CB may be involved in chondrogenesis by regulating DCN through EGR1.
Pseudo-temporal trajectory analysis shows endplate chondrocytes diverging into two fates in IVDD, one links to inflammatory and the other to homeostasis and ECM regulation. Notably, consistent with previous results, pre-HTC represents an intermediate state between ProC and HTC, indicating the continuous dynamic evolution among these chondrocyte clusters20.
CellPhoneDB analysis reveals MA’s closer relation to chondrocytes than T/NK cells, with chondrocytes-macrophages interaction relying on FN1-a4b7 complex and FN1-a4b1 complex. FN1, a major non-collagenous ECM glycoprotein, regulates cell adhesion, differentiation and growth, and is linked to the pro-fibrotic, anti-inflammatory phenotype of M2-like macrophages48,49. This suggests a unique macrophages-chondrocytes relationship may be related to cellular immunity. Interestingly, macrophages promote inflammation of NP via IL-6 and TNF in IVDD17. Here, our study provides valuable insights into the potential role of FN1 in mediating chondrocytes-macrophages crosstalk in endplate degeneration. Yamawaki et al. showed chondrocytes-macrophages contact during the early phase of cartilage regeneration50. There may be more possibilities between chondrocytes and macrophages that must be further investigated and discussed.
HomC’s lack of close associated with macrophages in cell-to-cell communication analysis is noteworthy. Relevant ligand-receptor pairs include TNFRSF1B-GRN, TNFRSF1A-GRN, HLA-C-FAM3C, TGFB1-TGFBR3 and LGALS9-CD47. TGFB1-TGFBR3 is involved in angiogenesis-related VEGF signaling pathways, with IVDD potentially triggered by reduced nutrient diffusion in vertebral endplate vasculature. HomC may also impact IVDD progression.
Single-cell sequencing of the endplate has revealed various cell types, notably immune cells. Pseudo-temporal trajectory analysis indicates chondrocytes significantly influence IVDD by maintaining homeostasis and regulating ECM, thereby altering the endplate microenvironment. The ECM ensures the intervertebral disc's mechanical integrity51, while the endplate facilitates nutrient exchange52. Changes in chondrocyte types, which modulate ECM and nutrient exchange, may drive disc degeneration. Additionally, chondrocytes are linked to immune function. Recent studies have focused on immune cell-mediated endplate changes in disc research. Our previous research demonstrated that senescent macrophages increase pSTAT3 in endothelial cells via IL10 secretion, leading to angiogenesis and sclerosis in the endplates53. CellPhoneDB analysis shows chondrocytes interact with macrophages through FN1. Further studies are needed to elucidate how chondrocytes affect macrophages. Given the crucial roles of chondrocytes and immune cells in IVDD, future treatments may involve altering chondrocyte types and regulating immune cells.
It should be noted that our study has several limitations. First, the healthy endplate tissues were absent in this study due to the ethical reasons and challenging human sample collection at a clinic. Consequently, at scRNA-seq, the data might not accurately distinguish the cell populations. Moreover, we did not use extensive clinical samples and data to validate the majority of the findings. Further large-scale experiments are needed to overcome the aforementioned difficulties and evaluate the mechanism of IVDD.
Overall, this study provides valuable reference information for future research on endplate degeneration as a potential therapeutic target. The identification of specific cell subsets and TFs may inform the development of novel therapeutic strategies that target these cells and their regulatory pathways. Additionally, the analysis of cell–cell interactions reveals a previously unrecognized link between chondrocytes and macrophages, suggesting the potential for new treatments that modulate this interaction to alleviate inflammation and promote tissue repair. Overall, our study opens up new avenues for investigation into the mechanisms underlying cartilage endplate degeneration and the development of targeted therapies for this debilitating condition.
Data availability
The datasets generated and/or analyzed during the current study are available in the Gene Expression Omnibus (GEO) repository, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE255768.
References
Faquin, W. C. Endocrine pathology Author: Lester D. R. Thompson Churchill Livingstone Elsevier, Philadelphia, PA, 2006. Diagn. Cytopathol. 36, 449–449 (2008).
Kushchayev, S. V. et al. ABCs of the degenerative spine. Insights Imaging 9, 253–274 (2018).
Udby, P. M. et al. A definition and clinical grading of Modic changes. J. Orthop. Res. 40, 301–307 (2022).
Fan, Y. et al. Serum miRNAs are potential biomarkers for the detection of disc degeneration, among which miR-26a-5p suppresses Smad1 to regulate disc homeostasis. J. Cell. Mol. Med. 23, 6679–6689 (2019).
Moore, R. J. The vertebral endplate: disc degeneration, disc regeneration. Eur. Spine J. 15, 333–337 (2006).
Pappou, I. P., Cammisa, F. P. & Girardi, F. P. Correlation of end plate shape on MRI and disc degeneration in surgically treated patients with degenerative disc disease and herniated nucleus pulposus. Spine J. 7, 32–38 (2007).
Feng, Z., Liu, Y., Yang, G., Battié, M. C. & Wang, Y. Lumbar vertebral endplate defects on magnetic resonance images: Classification, distribution patterns, and associations with modic changes and disc degeneration. Spine 43, 919–927 (2018).
Zehra, U., Cheung, J. P. Y., Bow, C., Lu, W. & Samartzis, D. Multidimensional vertebral endplate defects are associated with disc degeneration, modic changes, facet joint abnormalities, and pain. J. Orthop. Res. 37, 1080–1089 (2019).
Chen, S. et al. Grem1 accelerates nucleus pulposus cell apoptosis and intervertebral disc degeneration by inhibiting TGF-β-mediated Smad2/3 phosphorylation. Exp. Mol. Med. 54, 518–530 (2022).
Jensen, T. S., Karppinen, J., Sorensen, J. S., Niinimäki, J. & Leboeuf-Yde, C. Vertebral endplate signal changes (Modic change): a systematic literature review of prevalence and association with non-specific low back pain. Eur. Spine J. 17, 1407–1422 (2008).
Ni, S. et al. Sensory innervation in porous endplates by Netrin-1 from osteoclasts mediates PGE2-induced spinal hypersensitivity in mice. Nat. Commun. 10, 5643 (2019).
Fields, A. J., Liebenberg, E. C. & Lotz, J. C. Innervation of pathologies in the lumbar vertebral end plate and intervertebral disc. Spec. Issue Biol. 14, 513–521 (2014).
Clement, R. G. E., Hall, A. C., Wong, S. J., Howie, S. E. M. & Simpson, A. H. R. W. Septic arthritis in an in vivo murine model induced by Staphylococcus aureus : a comparison between actions of the haemolysin toxin and the effects of the host immune response. Bone Joint Res. 11, 669–678 (2022).
Potter, S. S. Single-cell RNA sequencing for the study of development, physiology and disease. Nat. Rev. Nephrol. 14, 479–492 (2018).
Li, Z. et al. Single-Cell RNA sequencing reveals the difference in human normal and degenerative nucleus pulposus tissue profiles and cellular interactions. Front. Cell Dev. Biol. 10, 910626 (2022).
Sun, H. et al. Single-cell RNA-seq analysis identifies meniscus progenitors and reveals the progression of meniscus degeneration. Ann. Rheum. Dis. 79, 408–417 (2020).
Han, S. et al. Single-Cell RNA sequencing of the nucleus pulposus reveals chondrocyte differentiation and regulation in intervertebral disc degeneration. Front. Cell Dev. Biol. 10, 824771 (2022).
Zhang, Y. et al. Single-cell RNA-seq analysis identifies unique chondrocyte subsets and reveals involvement of ferroptosis in human intervertebral disc degeneration. Osteoarthritis Cartilage 29, 1324–1334 (2021).
Ling, Z. et al. Single-Cell RNA-Seq analysis reveals macrophage involved in the progression of human intervertebral disc degeneration. Front. Cell Dev. Biol. 9, 833420 (2022).
Ji, Q. et al. Single-cell RNA-seq analysis reveals the progression of human osteoarthritis. Ann. Rheum. Dis. 78, 100–110 (2019).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888-1902.e21 (2019).
Trapnell, C. et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014).
Efremova, M., Vento-Tormo, M., Teichmann, S. A. & Vento-Tormo, R. Cell PhoneDB: Inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat. Protoc. 15, 1484–1506 (2020).
Aibar, S. et al. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods 14, 1083–1086 (2017).
Yu, G., Wang, L.-G., Han, Y. & He, Q.-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics 16, 284–287 (2012).
Chen, H. et al. Dissecting heterogeneity reveals a unique BAMBI high MFGE8 high subpopulation of human UC-MSCs. Adv. Sci. 10, 2202510. https://doi.org/10.1002/advs.202202510 (2022).
Chen, Y. et al. A high-resolution route map reveals distinct stages of chondrocyte dedifferentiation for cartilage regeneration. Bone Res. 10, 38 (2022).
Queckbörner, S. et al. Stromal heterogeneity in the human proliferative endometrium: A single-cell RNA sequencing study. J. Pers. Med. 11, 448 (2021).
Wei, B. et al. Analysis of cellular heterogeneity in immune microenvironment of primary central nervous system lymphoma by single-cell sequencing. Front. Oncol. 11, 683007 (2021).
McCleland, M. L. et al. CCAT1 is an enhancer-templated RNA that predicts BET sensitivity in colorectal cancer. J. Clin. Investig. 126, 639–652 (2016).
Ooi, W. F. et al. Epigenomic profiling of primary gastric adenocarcinoma reveals super-enhancer heterogeneity. Nat. Commun. 7, 12983 (2016).
Bernstein, B. E. et al. The NIH roadmap epigenomics mapping consortium. Nat. Biotechnol. 28, 1045–1048 (2010).
Yuan, Z. et al. Activation of FOXO1 by Cdk1 in cycling cells and postmitotic neurons. Science 319, 1665–1668 (2008).
Valis, K. et al. Hippo/Mst1 stimulates transcription of the proapoptotic mediator NOXA in a FoxO1-dependent manner. Cancer Res. 71, 946–954 (2011).
Zhao, Y. et al. Cytosolic FoxO1 is essential for the induction of autophagy and tumour suppressor activity. Nat. Cell Biol. 12, 665–675 (2010).
ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Hauf, S., Waizenegger, I. C. & Peters, J. M. Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science 293, 1320–1323 (2001).
Xie, B. et al. MYB proto-oncogene-like 1-TWIST1 axis promotes growth and metastasis of hepatocellular carcinoma cells. Mol. Ther. Oncol. 18, 58–69 (2020).
Jomova, K. & Valko, M. Advances in metal-induced oxidative stress and human disease. Toxicology 283, 65–87 (2011).
Chen, C. et al. Estrogen-induced FOS-like 1 regulates matrix metalloproteinase expression and the motility of human endometrial and decidual stromal cells. J. Biol. Chem. 295, 2248–2258 (2020).
Forrester, A. et al. A selective ER-phagy exerts procollagen quality control via a Calnexin-FAM134B complex. EMBO J. 38, e99847 (2019).
Park, J. G. et al. Loss of MAFB function in humans and mice causes Duane syndrome, aberrant extraocular muscle innervation, and inner-ear defects. Am. J. Hum. Genet. 98, 1220–1227 (2016).
Shi, Y. et al. tRNA synthetase counteracts c-Myc to develop functional vasculature. eLife 3, e02349 (2014).
Kim, J.-Y., Cho, Y.-E. & Park, J.-H. The nucleolar protein GLTSCR2 is an upstream negative regulator of the oncogenic nucleophosmin-MYC axis. Am. J. Pathol. 185, 2061–2068 (2015).
Huang, Y. et al. CEBPG promotes esophageal squamous cell carcinoma progression by enhancing PI3K-AKT signaling. Am. J. Cancer Res. 10, 3328–3344 (2020).
Lister, R. et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462, 315–322 (2009).
Libório-Ramos, S., Barbosa-Matos, C., Fernandes, R., Borges-Pereira, C. & Costa, S. Interstitial macrophages lead early stages of bleomycin-induced lung fibrosis and induce fibroblasts activation. Cells 12, 402 (2023).
Yeo, A. T. et al. Single-cell RNA sequencing reveals evolution of immune landscape during glioblastoma progression. Nat. Immunol. 23, 971–984 (2022).
Kanda, K. et al. Requirement of direct contact between chondrocytes and macrophages for the maturation of regenerative cartilage. Sci. Rep. 11, 22476 (2021).
Frost, B., Camarero-Espinosa, S. & Foster, E. J. Materials for the spine: Anatomy problems and solutions. Materials 12, 253 (2019).
Zhuang, C. et al. Osteoporosis and endplate damage correlation using a combined approach of hounsfield unit values and total endplate scores: A retrospective cross-sectional study. Clin. Interv. Aging 16, 1275–1283 (2021).
Fan, Y. et al. Senescent-like macrophages mediate angiogenesis for endplate sclerosis via IL-10 secretion in male mice. Nat. Commun. 15, 2939 (2024).
Acknowledgements
We acknowledge assistance with the access of analytic instruments from Translational Medicine Center at The First Affiliated Hospital of Zhengzhou University.
Funding
This work was supported by the National Natural Science Foundation of China (Grant Nos. 82002353, S.N.), China Postdoctoral Science Foundation (Grant No. 2020 M682360, S.N.), and Young and Middle-aged Discipline Leader Cultivation Project of He’nan Health (S.N.), National Natural Science Foundation of China (Grant Nos. 82072431, S.N.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
Chenhao Shi: conception and design, investigation, drafting of the article, analysis and interpretation of the data. Yonggang Fan: validation and visualization. Xiusheng Huang: conception and design, literature search, provision of study materials. Mingzhe Fan: collection and assembly of data. Lantian Zhao: supervision, statistical expertise. Huafeng Zhang: critically reviewing of the article. Shuangfei Ni: obtaining of funding, administrative, final approval of the article. All authors have read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Shi, C., Fan, Y., Huang, X. et al. Single-cell RNA sequencing reveals the differentiation and regulation of endplate cells in human intervertebral disc degeneration. Sci Rep 14, 21414 (2024). https://doi.org/10.1038/s41598-024-71891-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-71891-5
- Springer Nature Limited