Abstract
The global crisis of the COVID-19 pandemic has highlighted the need for mathematical models to inform public health strategies. The present study introduces a novel six-compartment epidemiological model that uniquely incorporates a higher isolation rate for unreported symptomatic cases of COVID-19 compared to reported cases, aiming to enhance prediction accuracy and address the challenge of initial underreporting. Additionally, we employ optimal control theory to assess the cost-effectiveness of interventions and adapt these strategies to specific epidemiological scenarios, such as varying transmission rates and the presence of asymptomatic carriers. By applying this model to COVID-19 data from India (30 January 2020 to 24 November 2020), chosen to capture the initial outbreak and subsequent waves, we calculate a basic reproduction number of 2.147, indicating the high transmissibility of the virus during this period in India. A sensitivity analysis reveals the critical impact of detection rates and isolation measures on disease progression, showing the robustness of our model in estimating the basic reproduction number. Through optimal control simulations, we demonstrate that increasing isolation rates for unreported cases and enhancing detection reduces the spread of COVID-19. Furthermore, our cost-effectiveness analysis establishes that a combined strategy of isolation and treatment is both more effective and economically viable. This research offers novel insights into the efficacy of non-pharmaceutical interventions, providing a tool for strategizing public health interventions and advancing our understanding of infectious disease dynamics.
Similar content being viewed by others
Introduction
The COVID-19 pandemic, engendered by the SARS-CoV-2 virus, has impacted the global population, accounting for more than 609 million confirmed infections and 6.5 million mortalities1. In India, the situation has been particularly severe, with information indicating over 44.5 million cases and approximately 0.52 million fatalities1. This information positions India as a preeminent focus of the pandemic, superseded only by the United States1,2.
The urgency and scale of the pandemic have necessitated the development of robust mathematical models to inform public health strategies. Traditional compartmental models, such as the susceptible-infectious-recovered (SIR) model formulated in 19273, have been instrumental in describing the dynamics of infectious diseases like dengue4, malaria5, and tuberculosis6. However, the unique epidemiological characteristics of COVID-19 have required modifications to these models7, including the integration of wavelet theory and machine learning algorithms8.
A critical feature of the COVID-19 pandemic is the important role of asymptomatic carriers in virus transmission. Asymptomatic cases, who do not exhibit symptoms, often remain undetected and have more contacts compared to symptomatic cases, facilitating extensive community spread. The challenge of identifying and testing asymptomatic individuals leads to underreporting, which exacerbates the spread of the virus and can result in superinfection incidents. Short-term predictions are crucial for supporting rapid decision-making in prevention and mitigation efforts. Effective prediction is essential for healthcare resource allocation and controlling the virus spread. Mathematical models have been central in optimizing control strategies and conducting cost-effectiveness analyses of various intervention measures, which are critical in managing infectious diseases like COVID-199,10,11,12,13,14.
Several mathematical models have been developed to address the complexities of COVID-19 transmission, particularly incorporating asymptomatic and symptomatic cases. For instance, in15, asymptomatic, undetected, and total cases were estimated during the outbreak in Wuhan, highlighting the impact of nonpharmaceutical interventions on controlling the outbreak. In11, COVID-19 epidemics in multiple countries were modeled, incorporating both reported and unreported cases and emphasizing the timing and magnitude of public policies. Additionally, in16, the impact of unreported cases in North African countries was investigated using mobility reports to assess movement restrictions’ effects. In17, the dynamics of unreported cases in Morocco was analyzed, demonstrating the importance of accurate parameter estimation. In18, both asymptomatic and symptomatic compartments were considered in a model for COVID-19 transmission dynamics in India, emphasizing the role of undetected cases in the transmission dynamics. Further models, such as9,10,14,19,20,21, explored various intervention strategies and their effectiveness in controlling the spread of COVID-19. Studies in different countries also highlighted various approaches to understanding COVID-19 impacts and outcomes22,23,24,25.
Other models have also been proposed to analyze the spread of COVID-19. For example, in26, a model incorporating contact tracing and hospitalization procedures was developed to study the dissemination of COVID-19 in India, revealing exponential growth. In12, the roles of asymptomatic, isolation, and quarantine compartments were explored, suggesting that quarantine or isolation measures were effective in reducing virus transmission. In19, a model to predict the dissemination of COVID-19 and to assess the impact of quarantine and hospitalization was presented, demonstrating the sensitivity of model parameters in preventing the spread of the virus through intervention measures. Control problems were solved in 27 to evaluate quarantine, isolation, and social distancing, along with surface disinfection and emergency provision, mitigating the dissemination of COVID-19. A model incorporating age was constructed in 28 to assess the efficacy of nonpharmaceutical interventions in mitigating COVID-19 spread and reducing the strain on healthcare infrastructure. Additionally, in.20,29,30, further insights into optimal intervention strategies and the impact of various non-pharmaceutical interventions on the dynamics of COVID-19 were provided.
The advancement in epidemiological modeling has been enriched by the development of fractional pandemic models31 and other innovations32. The exploration of approaches in differential systems33, combined with machine learning24 and metaheuristic solutions for system stability34, has opened new pathways in computational epidemiology. Moreover, the application of statistical methods in epidemic modeling32,35 highlights the importance of statistical inference for disease forecasting. Furthermore, studies like11,27,28,29,30 contributed to the understanding of COVID-19 transmission dynamics and the effectiveness of various intervention strategies.
The objectives of this study are twofold: (i) to introduce a novel six-compartment epidemiological model that integrates both reported and unreported symptomatic cases to enhance prediction precision across various epidemiological scenarios, particularly by incorporating a higher isolation rate for unreported cases once detected, addressing the challenge of initial underreporting; and (ii) to apply this model to COVID-19 data from India to evaluate the cost-effectiveness of different intervention strategies using optimal control theory. This model uniquely captures both reported and unreported symptomatic cases, integrating sensitivity analyses and cost-effectiveness evaluations to offer a comprehensive view of disease dynamics and provide valuable insights for public health policy and strategy formulation. This article is organized as follows. “Methods” delineates the approaches used in this study related to the mathematical structure of the new model as well as an analysis of its stability and inherent properties. In “Results”, our findings are provided which are associated with the model fitting to empirical data and its predictive prowess, as well as the application of optimal control methods. “Discussion and conclusions” establishes a discussion of the obtained results synthesizing our findings and providing our conclusions about the present study.
Methods
This section introduces the methods considered in our study, focusing on the formulation of the new model and the underlying principles guiding its structure. We also analyze the model, examining its mathematical properties and epidemiological implications.
Formulation of the model
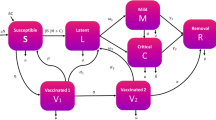
Within the range of the varied mathematical models for epidemiological transmission, the new model proposed in this study addresses nonlinear dynamics in the spread of COVID-19. We segment the human population of size \(N\) into six main compartments: \(S\), \(A\), \(I\), \(U\), \(G\), and \(R\), where \(S\) indicates the susceptible individuals, \(A\) the asymptomatic, \(I\) the infectious (reported symptomatic), \(U\) the unreported symptomatic, \(G\) the isolated, and \(R\) the recovered ones. This segmentation forms the basis of our nonlinear differential system and captures the disease transmission dynamics. The total population at any given time is expressed as \(N = S + A + I + U + G + R\), encompassing all the compartments. This model, designated as SAIUGR, allows analysis of the disease spread and effectiveness of various intervention strategies.
The SAIUGR model is formulated for the spread of COVID-19 by means of a system of nonlinear ordinary differential equations as
The SAIUGR model stated from (1) includes several parameters that describe the dynamics of disease transmission and progression. The parameter \(\Lambda\) represents the consistent influx of susceptibles due to births and migrations, while \(\mu\) denotes the natural mortality rate. Transmissibility is embodied by \(\beta\), which connects susceptible and infected cases, with transmission coefficients \(\beta \zeta _{A}\), \(\beta \zeta _{I}\), and \(\beta \zeta _{G}\) for asymptomatic, reported symptomatic, and isolated cases, respectively. These coefficients are influenced by measures such as face masks, lockdowns, and social distancing, contributing to dampening the infection rate. Transition of asymptomatic individuals to either reported symptomatic (\(I\)) or unreported symptomatic (\(U\)) categories at a rate \(\omega\), with \(\theta\) representing the proportion of symptomatic cases detected and reported, and \(1-\theta\) the fraction that remains unreported. Recovery rates are given by \(\gamma _{I}\), \(\gamma _{U}\), and \(\gamma _{G}\) for reported symptomatic, unreported symptomatic, and isolated individuals, respectively, with their corresponding mortality rates being denoted by \(\mu _{I}\), \(\mu _{U}\), and \(\mu _{G}\), respectively. These parameters account for the varying outcomes among different groups, which are crucial for understanding the disease dynamics that can be visualized in Fig. 1.
Analysis of parameters and their implications
To understand the working and mechanism of our SAIUGR model, one must examine each compartment and parameter involved, as they determine the disease spread and its eventual outcomes. Table 1 offers a systematic breakdown, describing each key parameter, along with their dvalues. Individuals, whether detected (compartment \(I\)) or undetected (compartment \(U\)), transition to the isolated compartment at rates \(\eta _{I}\) and \(\eta _{U}\) respectively.
The temporal dynamics of the SAIUGR model expressed in (1) include the mentioned six compartments: \(S\), \(A\), \(I\), \(U\), \(G\), and \(R\). Each compartment represents different stages in the disease progression. We initialize the model with non-negative values \(S(0)=S_{0}\), \(A(0)=A_{0}\), \(I(0)=I_{0}\), \(U(0)=U_{0}\), \(G(0)=G_{0}\), and \(R(0)=R_{0}\), where the numbers in parentheses denote the initial time. These initial values, representing the state of each compartment at the start of the formulation, are crucial for the integrity and realism of the model predictions.
The isolation rate for unreported symptomatic individuals (\(\eta _{U}\)) is set higher than that for reported symptomatic individuals (\(\eta _{I}\)) to represent the intensified isolation efforts that occur upon the late identification of previously unreported cases. This approach is modeled to correct for initial underreporting and to mitigate the risk of continued disease transmission from these individuals.
The SAIUGR model, grounded in empirical data and aligned with existing research, is designed as a comprehensive nonlinear dynamical system as established in (1), effectively describing the transmission and progression of COVID-19.
In the next section, we analyze the model mathematical structure and explore its predictive capabilities across different scenarios, examining its epidemiological implications.
Positivity and boundedness of the model solutions
The analysis of positivity and boundedness validates fundamental properties to maintain biological relevance and practical utility. Positivity guarantees non-negative populations across all compartments throughout an epidemic duration. Similarly, boundedness ensures that solutions do not unrealistically approach infinity. Positivity and boundedness aspects not only validate the model biological soundness but also set the stage for further stability and sensitivity explorations. Theorem 1 postulates these aspects.
Theorem 1
Let all the solutions \((S(t), A(t), I(t), U(t), G(t), R(t))\) of the structure stated in (1) have initial values \(S(0)=S_{0}\), \(A(0)=A_{0}\), \(I(0)=I_{0}\), \(U(0)=U_{0}\), \(G(0)=G_{0}\), and \(R(0)=R_{0}\). Then, these solutions are nonnegative and uniformly bounded in the region defined as
The proof of Theorem 1 is provided in Appendix A.
Derivation of the basic reproduction number
In epidemiological studies, the basic reproduction number \(R_{0}\) is essential. It represents the average number of secondary infections that an infected individual is expected to cause in a wholly susceptible population3. The value of the parameter \(R_{0}\) determines the potential spread of the disease within a population. Specifically:
-
If \(R_{0} > 1\), the disease spreads and can lead to an epidemic, but the disease may reach a higher endemic equilibrium (EE) that persists in the population at a constant level.
-
If \(R_{0} < 1\), the disease will eventually die out.
-
If \(R_{0} = 1\), that is, each infected individual, on average, produces only one secondary case and the EE persists in the population at a steady level without leading to large outbreaks.
At the disease-free equilibrium (DFE) \(E^{0} = ({\Lambda }/{\mu }, 0, 0, 0, 0, 0)\), \({\Lambda }/{\mu }\) represents \(S\) in the absence of the disease. To assess the stability of the DFE and understand the potential for disease emergence or eradication within the population, we employ the Jacobian matrices \({\mathscr {F}}\) and \({\mathscr {V}}\), which are constructed to represent the rates of new infections and transitions between compartments, respectively, for the entire system. In the context of epidemiological modeling, \({\mathscr {F}}\) captures the new infections caused by different infectious compartments, while \({\mathscr {V}}\) captures the movement between compartments and the removal of individuals from the infected classes.
To calculate \(R_0\), we use the next-generation matrix method as outlined in42,43. To do this calculation, we construct the matrices \({\mathscr {F}}\) and \({\mathscr {V}}\), which represent the rates of new infections and transitions between compartments for the entire system, and are defined as
The next-generation matrix is given by
By calculating the largest eigenvalue of \({\mathscr {F}}{\mathscr {V}}^{-1}\), we can determine the basic reproduction number as
Stability analysis of the disease-free equilibrium
The DFE of the model is determined by setting the rates of change of all compartments to zero, as per the system of equations given in (1). Specifically, this implies \({\text{d}S}/{\text{d}t}=0\), \({\text{d}A}/{\text{d}t}=0\), \({\text{d}I}/{\text{d}t}=0\), \({\text{d}U}/{\text{d}t}=0\), \({\text{d}G}/{\text{d}t}=0\), and \({\text{d}R}/{\text{d}t}=0\). In the DFE, the infected compartments are all zero, that is, \(A = 0\), \(I = 0\), \(U = 0\), \(G = 0\), and \(R = 0\). The susceptible population \(S\) at the DFE can be deduced and is typically represented as the entire population in the absence of infection, which is \(\Lambda /\mu\) under the assumption of balanced birth and death rates. Theorems 2 and 3 state the stability of this DFE of the SAIUGR model.
Theorem 2
Let \(R_{0} < 1\). Then, the DFE \(E^{0}\) of the model described in (1) is asymptotically stable in a local sense.
Theorem 3
For \(R_{0} < 1\), the DFE \(E^{0}\) of the system defined in (1) is asymptotically stable in a global sense.
The proofs of Theorems 2 and 3 are provided in Appendix A.
Stability analysis of endemic equilibrium
Let \(\Sigma ^{*}=(S^{*}, A^{*}, I^{*}, U^{*}, G^{*}, R^{*})\) be the EE point of the structure given in (1) of the SAIUGR model and
be the effective transmission rate (force of infection). In addition, suppose that \({\text{d}S}/{\text{d}t}=0\), \({\text{d}A}/{\text{d}t}=0\), \({\text{d}I}/{\text{d}t}=0\), \({\text{d}U}/{\text{d}t}=0\), \({\text{d}G}/{\text{d}t}=0\) and \({\text{d}R}/{\text{d}t}=0\). Then, we have that
where \(K_{1}=(\omega +\mu )\), \(K_{2}=(\eta _{I}+\gamma _{I}+\mu _{I}+\mu )\), \(K_{3}=(\eta _{U}+\gamma _{U}+\mu _{U}+\mu )\), and \(K_{4}=(\eta _{G}+\mu _{G}+\mu )\). Substituting all the above expressions in (2), we get the nonzero equilibrium of the system stated in (1) satisfying the linear equation in terms of \(\chi ^{*}\) as \(P_{0}\chi ^{*}+P_{1} =0\), where \(P_{0}=\mu K_{2}K_{3}K_{4}+(\gamma _{I}+\mu )\theta \,\omega K_{3}K_{4}+\gamma _{U}(1-\theta )\omega K_{2}K_{4}+(\gamma _{G}+\mu )(\eta _{I}\theta \,\omega +\eta _{U}(1-\theta )\omega K_{2})\) and \(P_{1}=\mu K_{1}K_{2}K_{3}K_{4} (1-R_{0})\), with \(P_{0}\) encapsulating various transmission and recovery dynamics, and \(P_{1}\) reflecting the impact of changes in the disease transmission rate and natural mortality rate. Clearly, \(K_{1}>0\), \(K_{2}>0\), \(K_{3}>0\), and \(K_{4}>0\). Since \((\gamma _{I}+\mu )>0\) and \((\gamma _{G}+\mu )>0\), we have \(P_{0}>0\). Similarly \(P_{1}>0\) as \(R_{0}>1\). Therefore, we can conclude that the system formulated in (1) possesses a unique positive EE point if \(R_{0}>1\), while it does not have any positive EE point if \(R_{0}<1\), which is formalized in Theorems 4 and 5. The proofs of Theorems 4 and 5 can be found in Appendix A.
Theorem 4
Let \(R_{0} > 1\). Then, the EE point \(\Sigma ^{*}\) is asymptotically stable in a local sense.
Theorem 5
For \(R_{0} > 1\), the EE point \(\Sigma ^{*}\) is asymptotically stable in a global sense.
Concluding our analysis of the SAIUGR model, we have detailed the transmission dynamics of COVID-19, providing an understanding of how the disease spreads. This analysis sets the stage for applying the model to empirical data, allowing for further validation and refinement of the results obtained.
Results
This section focuses on the empirical aspects of our study, demonstrating its application and validation through a series of analytical steps. We begin by calibrating the model to real-world COVID-19 case data from India. Then, we explore the application of optimal control theory to our epidemiological model, aiming to minimize the impact of COVID-19. We explore various control strategies and their effectiveness, both in isolation and in combination, through a cost-effectiveness analysis.
Model calibration to data
Building upon the analysis presented earlier, we now empirically calibrate the SAIUGR model using data from reported COVID-19 cases in India. This step aligns the model with real-world observations and compares theoretical forecasts against real-life epidemiological patterns.
The dataset, covering daily and cumulative confirmed cases from 30 January 2020 to 24 November 2020, a span of 300 days, was sourced from the COVID-19 API data portal (www.covid19india.org, accessed on 21 July 2024). We estimate the parameters \(\beta\), \(\eta _{I}\), \(\eta _{U}\), \(\gamma _{I}\), and \(\gamma _{U}\) during model calibration. We employ the Lsqnonlin function of MATLAB44, a non-linear least-square method, to optimize these parameters by minimizing the residual sum of squares between the observed data and model predictions.
Using the mentioned dataset, we calculated the basic reproduction number as \(R_{0} = 2.147\), using the approaches outlined in the present article. This calculation reports the severity of the COVID-19 outbreak in India at that time and is important for understanding the virus transmission dynamics as per our model. Figure 2 shows the alignment of the SAIUGR model with both the daily and cumulative confirmed COVID-19 cases in India. The empirical data are depicted using blue markers, while the simulation from the model is represented by a continuous red curve, illustrating the model fit to the real-world data.
Figure 3 extends the model predictions, projecting the likely trajectory over an impending 50-day span, concluding on 12 January 2021. In this figure, the steadfast blue trajectory symbolizes the model projections for both daily new infections and the cumulative incidence of COVID-19 cases. These predictions hint at a potential downturn in case numbers over this forecasted interval. The methodology employed in this phase, utilizing non-linear least-square fitting in MATLAB, demonstrates a rigorous approach to model calibration. The alignment of the SAIUGR model with empirical COVID-19 data substantiates the robustness of the chosen method and its effectiveness in parameter estimation.
Analysis of transcritical bifurcation
To understand the behavior of the DFE and EE in the system formulated in (1), we analyze the presence of a transcritical bifurcation at the critical point where \(R_{0} = 1\). This analysis is essential to comprehend the critical thresholds at which the system dynamics change, marking the transition from a state where the disease can be eradicated to a state where it persists within the population.
In Fig. 4, we plot the parameter \(\chi ^{*}\) against \(R_{0}\), where \(\chi ^{*}\) represents the effective transmission rate, as mentioned. For \(R_{0} < 1\), the DFE exhibits local asymptotic stability, as indicated by the solid green line. Conversely, for \(R_{0} > 1\), the DFE loses its stability, transitioning to an unstable state, depicted by the dashed red line. The ascending blue line in Fig. 4 denotes a stable EE, indicating a shift from DFE to EE, which suggests the sustained presence of the virus in the population.
From “Stability analysis of endemic equilibrium”, we know that the condition \(P_{0} \chi ^{*} + P_{1} = 0\) plays a relevant role in the stability analysis of the EE. Here, \(P_{0}\) and \(P_{1}\) are parameters derived from the model stability analysis. Specifically, \(P_{0}\) encapsulates various transmission and recovery dynamics, while \(P_{1}\) reflects the impact of changes in the disease transmission rate and natural mortality rate, as mentioned.
For \(R_{0} > 1\), the application of the Descartes rule of signs, combined with the condition that \(P_{0} > 0\) and \(P_{1} < 0\), suggests the presence of a singular EE. Conversely, the absence of an EE is noted for \(R_{0} < 1\). Figure 4 illustrates these equilibriums, showing that the system experiences a transcritical bifurcation at \(R_{0}=1\). In this figure, the green line represents the stable DFE \(E^{0}\) for \(R_{0} < 1\), while the red dashed line represents the unstable DFE point for \(R_{0} > 1\). The ascending blue line represents the stable branch of the EE point \(\Sigma ^{*}\). Therefore, \(\Sigma ^{*}\) is stable for \(R_{0} > 1\) and unstable for \(R_{0} < 1\). Similarly, \(E^{0}\) is stable for \(R_{0} < 1\) and becomes unstable for \(R_{0} > 1\). Thus, the SAIUGR model, as defined in (1), indicates that the coronavirus disease spreads in the population when \(R_{0} > 1\), as evidenced by the stable presence of the point \(\Sigma ^{*}\) in this scenario.
Sensitivity analysis
The sensitivity analysis states the importance of various parameters in the disease spread dynamics. An exposition concerning sensitivity analyses for malaria is provided in45, offering insights for enhancing the robustness of our model. This analysis grants us the capability to identify parameters that have a substantial influence on \(R_{0}\). By computing the sensitivity indexes of these control parameters, we obtain the contribution of each parameter to disease proliferation and persistence.
Adopting the methodology delineated in46, the normalized forward sensitivity index of a variable M with respect to a parameter q is represented as
Utilizing this index, one can deduce the normalized forward sensitivity index corresponding to every parameter integrated into the expression for \(R_{0}\). For example, the normalized forward sensitivity index of \(R_{0}\) concerning the parameter \(\beta\) equates to one and it is given by
Figure 5 shows the key parameters for diminishing the value of \(R_0\) beneath the threshold of one, inhibiting the viral spread.
The positive sensitivity indices, associated with \(\zeta _{A}\), \(\zeta _{I}\), \(\zeta _{G}\), \(\theta\), and \(\beta\), predominantly influence the frequency of disease manifestations, with \(\beta\) having the highest positive index, indicating its crucial role in transmission dynamics. In contrast, negative sensitivity indices linked to parameters \(\omega\), \(\eta _{I}\), \(\eta _{U}\), \(\gamma _{I}\), \(\gamma _{U}\), \(\gamma _{G}\), \(\mu _{I}\), \(\mu _{U}\), \(\mu _{G}\), and \(\mu\) attenuate the disease transmission, with \(\gamma _{U}\) showing the most negative impact. Remarkably, parameters such as the isolation rate of non-disclosed cases (\(\eta _{U}\)), recovery rates of both non-disclosed symptomatic individuals (\(\gamma _{U}\)), and isolated entities (\(\gamma _{G}\)) wield an effect on curbing the dissemination of COVID-19. This highlights the importance of improving detection and isolation measures to control the spread of the virus effectively. Furthermore, to scrutinize the impact of model parameters on \(R_{0}\), contour representations of \(R_{0}\) in relation to specific parameters are exhibited in Fig. 6. In Fig. 6a, the contour map shows that, increasing the number of individuals in isolation, through joining individuals from the reported symptomatic infected group, reduces \(R_{0}\) and hence COVID-19 spread declines. Figure 6b shows that, as the isolation of both unreported and reported symptomatic infected individuals increases, with the value of \(R_{0}\) decreasing as a result, the COVID-19 spread reduces.
To enrich our comprehension regarding the behavior of \(R_{0}\), its relation with the disease transmission rate \(\beta\) and the transition rate \(\omega\) from asymptomatic to symptomatic cases has been graphically represented in Fig. 7. The three-dimensional representation in this figure features a ’folded leaf’ structure, signifying a critical area where small variations in \(\omega\) and \(\beta\) lead to changes in \(R_0\), particularly when \(R_0\) approaches and surpasses the critical threshold of the value one, indicating increased disease spread.
Impact of parameters on prevalence of COVID 19
Next, we conduct a time series analysis to explore the effect of the parameters \(\eta _{I}\) and \(\eta _{U}\) on the total symptomatic infected and isolated populations over the period [0,400] days. Figure 8a illustrates the impact of varying isolation rates on the total symptomatic infected population (\(I+U\)). As depicted by the bell-shaped curves in different colors, each representing a unique pair of isolation rates (\(\eta _{I}\), \(\eta _{U}\)), we observe a distinct pattern. The curve with the lowest peak, in blue, corresponds to higher isolation rates (\(\eta _{I}=0.0775\), \(\eta _{U}=0.1368\)), demonstrating a reduced number of symptomatic infections. As the isolation rates decrease (moving from violet to black, and then to red), the pattern shows a peak of the curve increases, signifying a higher number of symptomatic infections. This pattern reveals a clear relation: higher isolation rates lead to a decrease in symptomatic infections, emphasizing the effectiveness of isolation measures in controlling the spread of the virus. In Fig. 8b, the focus shifts to the isolated population (\(G\)) over time, with each curve representing different combinations of isolation rates. Similar to Fig. 8a, the lowest curve in blue (\(\eta _{I}=0.002\), \(\eta _{U}=0.046\)) indicates fewer isolated individuals, relating with lower isolation rates. As the isolation rates increase (violet to black, and then to red), the number of isolated individuals rises, reaching its peak in the red curve (\(\eta _{I}=0.017\), \(\eta _{U}=0.076\)). The overlapping tails of these curves on the right side suggest that irrespective of the initial differences in isolation rates, the number of isolated individuals converges over time, reflecting the dynamic nature of the pandemic progression and the impact of varying isolation strategies.
The analysis considers a higher \(\eta _{U}\) compared to \(\eta _{I}\), reflecting the model strategy to represent a rapid response following the late identification of these cases. This higher rate aims to demonstrate the effectiveness of isolation in reducing the spread of the disease once unreported cases are detected.
Next, we explore the application of optimal control theory to our epidemiological model, aiming to minimize the impact of COVID-19. We explore various control strategies and their effectiveness, both in isolation and in combination, through a cost-effectiveness analysis.
Formulation of the optimal control problem
We implement optimal control theory to our epidemiological model to minimize the number of infected individuals. The model incorporates two control variables, \(u_1\) and \(u_2\), each targeting different aspects of disease management.
The susceptible population becomes aware of disease and their control techniques when they are provided with enough statistics, leading to a shift in population behavior. Due to the systematic dissemination of news, people have started practicing wearing face masks, adhering to social distancing measures, maintaining good cleanliness, observing quarantine protocols, and even using home isolation strategies. Therefore, we alter the rate of isolation by implementing the control factor \((1 - \varepsilon _{1}u_{1}(t))\), which measures the external efforts to prevent or control interactions between isolated \(G\) and susceptible \(S\) populations, where the parameter \(\varepsilon _1\) represents the effectiveness of these isolation measures.
The control variable \(u_1(t)\), ranging from zero to one, modifies the isolation rate and is defined by the factor \((1 - \varepsilon _{1}u_{1}(t))\). Here, \(u_1(t)\) regulates the interactions between \(G\) and \(S\) populations. A higher value of \(u_1(t)\) (closer to one) indicates more stringent isolation measures, aimed at reducing the disease transmission rate. Administering an optimal treatment strategy to persons who are isolated in the early stage of COVID-19 infection can reduce the mortality rate of the disease. The framework that is highly anticipated is when \(\varepsilon _{1}u_{1}(t)=1\), meaning that the interactions between isolated and susceptible populations are effectively avoided, resulting in a successful prevention of transmission and reducing the total transmission rate to zero. In this context, the value 0 represents a lack of response, whereas the value 1 represents a complete response as a result of the complete isolation provided.
Early in the epidemic progression, appropriate treatment strategies, inclusive of medical kit provisions and vaccination programs for isolated individuals, can mitigate the associated mortality rate. With these strategies, the epidemiological model, as set out in (1), is adjusted by integrating the treatment stated as \(-\varepsilon _{2}u_{2}(t)G\), where \(\varepsilon _{2}\) represents the treatment rate associated with the control variable \(u_{2}(t)\). Various expenses are associated with medication, such as the cost of drugs, healthcare services, isolation measures, vaccinations, and medical supplies. These costs should be considered during the medication period. Therefore, we define \(u_{2}(t)\) as a control variable that ranges from 0 to 1, with 0 indicating no reaction and 1 indicating a complete response following the therapy.
In this context, \(u_{2}\), also within the range [0,1], is introduced to optimize treatment strategies. A value of \(u_2\) near to one denotes a comprehensive implementation of treatment measures, potentially maximizing the per capita efficiency of vaccination rates. The parameter \(\varepsilon _2\) reflects the effectiveness of these treatment measures. Thus, the utilization of \(u_2\) reflects the model capacity to adjust treatment responses in line with the disease progression. The formulation for the optimal control model is presented as
subject to minimizing the objective function given by
where \(C_{1}\), \(C_{2}\), \(C_{3}\), \(C_{4}\), and \(C_{5}\) are nonnegative weight constants. Drawing upon the Pontryagin maximum principle47, the indispensable conditions for this optimal control challenge are deduced following the methodology presented in48. The integrand cumulative costs are represented by the Lagrangian function detailed as \({\mathscr {L}}(A,I,U,u_{1}(t),u_{2}(t)) = C_{1}A+C_{2}I+C_{3}U+0.5(C_{4}u^{2}_{1}+C_{5}u^{2}_{2}).\) For this problem, the Hamiltonian function \({\mathscr {H}}\) is given by
where \(\lambda _{1}\), \(\lambda _{2}\), \(\lambda _{3}\), \(\lambda _{4}\), \(\lambda _{5}\), and \(\lambda _{6}\) are the adjoint variables. The corresponding differential equations for the adjoint variables are stated as
Next, Theorem 6 establishes the existence of optimal controls. Having established the existence of optimal controls, we now aim to characterize them in Theorem 7. The proofs of Theorems 6 and 7 are provided in Appendix A.
Theorem 6
Let \(\Psi \equiv \{u_{1}(t),u_{2}(t)\text{: } 0\le u_{1}(t),u_{2}(t)\le 1,t\in [0,T]\}\). Then, under \(\Psi\), optimal controls \(u^{*}(t)=(u_{1}^{*}(t),u_{2}^{*}(t))\) exist and minimize the objective function \({\mathscr {M}}(u_{1}(t),u_{2}(t))\) stated in (4).
Theorem 7
Let optimal controls \(u_{1}^{*}(t)\) and \(u_{2}^{*}(t)\) minimize \({\mathscr {M}}(u_{1}(t),u_{2}(t))\) over the region \(\Psi\). Then, we have that \(u_{1}^{*}(t) = \min \left\{ 1, \max \left\{ 0,\bar{u_{1}}\right\} \right\}\) and \(u_{2}^{*}(t) = \min \left\{ 1, \max \left\{ 0,\bar{u_{2}}\right\} \right\}\), where \(\bar{u_{1}}\) and \(\bar{u_{2}}\) are expressed as
Optimal control model
Optimal control simulations are performed in MATLAB utilizing model parameters and fixed parameters mentioned in Table 1 over the time interval [0, 400] days. We employ the forward and backward difference approximation method for solving the extended system presented in (3), as per the approach outlined in49. We set the control weights as \(C_{1} = 1\), \(C_{2} = 1\), \(C_{3} = 1\), \(C_{4} = 50\), and \(C_{5} = 50\).
Figure 9 illustrates the effect of control interventions on the dynamics of the populations A, I, U, and G over the period [0, 400] days. Note that the application of the control measures \(u_{1}\) and \(u_{2}\) leads to a substantial reduction in the number of infections, compared to scenarios where no control interventions are implemented. Figure 10 presents the temporal profiles of the control variables \(u_{1}\) and \(u_{2}\) throughout the simulation period. The analysis indicates that maintaining these controls at their maximum value of one for a prolonged duration is essential for effectively eradicating the COVID-19 disease from the population. Figure 11 illustrates how the control profiles for \(u_1\) and \(u_2\) vary with the increasing cost associated with these control measures. Note that, as the cost of implementing the control parameters escalates, the duration for which it is necessary to maintain these controls at their maximum level of one reduces. This highlights the trade-off between the intensity and cost-effectiveness of control strategies in the management of the disease.
Our simulations underscore implications for controlling the spread of COVID-19 in the population. The control variables \(u_{1}\) and \(u_{2}\) must be maintained at a high level for extended periods to completely eradicate the disease. This insight emphasizes the importance of consistent efforts to contain the virus spread, even as the immediate threat of the pandemic recedes. When considering the rising cost, it becomes evident that the longer these controls are maintained at their maximum value, we have greater economic implications. Policymakers must weigh the economic against the public health benefits of these controls. In some contexts, the economic strain of rigorous controls might outweigh the health benefits of fewer infections, and vice-versa. Every decision must take into account the circumstances of the community and the potential impacts of these controls. Our results can also inform future research on optimal control strategies for infectious diseases. By incorporating additional parameters and constraints, models may be refined to more accurately reflect real-world complexities such as new variants, vaccination effects, and consequences of prematurely lifting controls. Therefore, our simulations offer vital insights into the control of COVID-19 spread. Understanding various control strategies and their trade-offs allows policymakers to make informed decisions that best serve their communities.
Cost-effectiveness analysis
To reduce the spread of COVID-19 effectively and economically, we focus on identifying the most cost-efficient control strategy. This identification evaluates the efficacy of two distinct and combined control strategies: isolation control (strategy \(E_{1}\)), treatment control (strategy \(E_{2}\)), and their combined application (strategy \(E_{12}\)). We employ the incremental cost-effectiveness ratio (ICER)50,51 to assess the cost-effectiveness of these strategies. ICER is calculated by dividing the cost difference between strategies X and Y by the difference in their effectiveness, measured as the number of infections averted, where \(X, Y \in \{E_{1}, E_{2}, E_{12}\}\). Thus, for two strategies X and Y, with Y being more effective than X, that is, \(\text {TA}(Y) > \text {TA}(X)\), ICER values are determined as
where TC is the total cost for each strategy calculated using the objective functional stated in (4), and TA are the total infections averted by comparing the total number of COVID-19 infections with and without the corresponding strategy.
Table 2 displays the total infections averted and the total costs for each of the control strategies \(E_{1}\) and \(E_{2}\), individually and combined, sorted by the total infections averted. We now calculate and compare the effectiveness and costs of strategies \(E_{1}\) and \(E_{2}\), as detailed in Table 3. Comparing strategies \(E_{1}\) and \(E_{2}\), ICER(\(E_{1}\)) is lower, indicating \(E_{2}\) is less cost-efficient than \(E_{1}\). Therefore, we discard \(E_{2}\) as a standalone strategy.
The comparison of \(E_{1}\) and \(E_{12}\) is presented in Table 4. The ICER for strategies \(E_{1}\) and \(E_{12}\) are calculated as
and
Since ICER(\(E_{12}\)) is less than ICER(\(E_{1}\)), this indicates that the combined strategy \(E_{12}\) is more cost-effective and efficient compared to the isolation control strategy \(E_{1}\) alone. Therefore, integrating both control strategies is identified as the most effective approach to reducing the burden of COVID-19 in the population.
Discussion and conclusions
The present study developed the SAIUGR model, a six-compartment framework for analyzing COVID-19 transmission dynamics. Unlike traditional models, this model introduces a novel compartmentalization strategy by differentiating between reported and unreported symptomatic infections. Alongside the other compartments—namely susceptible, asymptomatic infected, isolated, and recovered—the model collectively encapsulates the various stages of disease progression and response. Our study provides a more nuanced understanding of disease spread, particularly in the Indian context, where diverse social and medical factors influence disease dynamics.
The structure of the model, with its six key compartments, enabled an in-depth exploration into the complexities of COVID-19 transmission. Utilizing 11 months of empirical data from India, the basic reproduction number was calculated as 2.147. This value of 2.147, a critical indicator of the pandemic intensity and potential spread, is shaped by several key parameters within the model. Specifically, the transmission rate, transition rate from asymptomatic to symptomatic, and detection rate of symptomatic cases played significant roles in influencing the dynamics observed. These parameters provided valuable insights into the interactions between different stages of infection, underscoring the importance of effective detection and isolation strategies.
To further understand these dynamics, a sensitivity analysis was conducted using the normalized forward sensitivity index, which assessed how variations in model parameters impact infection spread. This analysis highlighted the crucial role of accurate detection and efficient isolation in managing the pandemic, revealing that enhancing these rates can significantly alter the infection trajectory. Additionally, we extended the model to include optimal control methods, exploring the impacts of isolation and treatment strategies. Simulations performed in MATLAB demonstrated the potential effectiveness of these strategies in managing the pandemic. The cost-effectiveness analysis, using the incremental cost-effectiveness ratio, compared the cost and effectiveness of various health interventions, providing insights into the additional cost required to achieve a health benefit, such as a reduction in infection rates.
Building on the insights gained from our sensitivity analysis and control strategy simulations, our findings revealed that a combined control strategy, integrating both isolation and treatment measures, is not only more effective but also economically viable compared to implementing these strategies in isolation. This highlights the advantage of a synergistic approach in managing the pandemic efficiently. Furthermore, our study investigated the phenomena of transcritical bifurcation and endemic equilibrium. Transcritical bifurcation refers to the point at which the disease-free equilibrium state transitions to an endemic equilibrium state, indicating the potential for the disease to become a persistent part of the population. Endemic equilibrium represents a state where the disease consistently exists within the population at a steady level. By analyzing these phenomena, we gained insights into the stability of disease-free equilibrium and endemic equilibrium states under varying conditions. In addition, we explored the impact of specific parameters, such as isolation rates for reported and unreported symptomatic infections, on the prevalence of COVID-19, which enhanced our understanding of the dynamics and spread of the disease.
An essential aspect of our model is the inclusion of unreported symptomatic individuals, which enhances the accuracy of our transmission dynamics predictions. These individuals, although not detected by health authorities, contribute to the transmission of the virus and increase the overall disease burden. Our simulations demonstrated that unreported cases play a critical role in the spread of COVID-19, highlighting the importance of comprehensive detection and isolation strategies. By considering both reported and unreported cases, our model provides a more accurate and holistic view of the pandemic dynamics.
While our model provides valuable insights, it is important to acknowledge its inherent limitations, particularly regarding potential underreporting and assumptions about population mixing. One limitation is the omission of spatial diffusion effects, which can play a crucial role in the spread of infectious diseases. Spatial diffusion impacts how an epidemic propagates through different regions and populations, influencing the effectiveness of intervention strategies.
Future research should incorporate spatial dynamics to provide a more comprehensive understanding of disease spread. For instance, in52, a novel approach was proposed to investigate the stability analysis and dynamics of a reaction-diffusion susceptible-vacined-infectious-recovered epidemic model, highlighting the importance of spatial components in epidemic modeling. Additionally, in53, an optimal control problem of a SIR reaction-diffusion model with inequality constraints was discussed, which can inform future studies on the spatial aspects of epidemic control measures; see also54,55,56 for other models in this line. Future research directions also involve expanding the model to account for vaccination dynamics and environmental factors, aiming to enhance the model applicability and robustness against COVID-19 and other infectious diseases.
The choice of isolation and therapy as the main controls in our study was guided by their critical roles in managing infectious diseases. Isolation reduces the contact rate between infected and susceptible individuals, directly impacting the transmission dynamics. Therapy mitigates the severity and duration of the disease, improving recovery rates and reducing the overall infection burden. These controls were selected based on their proven effectiveness in epidemiological studies and their practical applicability in real-world public health strategies.
In real-life applications, the optimal control strategies suggested by our model can be translated into actionable public health measures. For instance, the optimal isolation strategy can be implemented through timely and widespread testing, contact tracing, and mandatory quarantine for infected individuals. The optimal therapy strategy can be applied by ensuring adequate healthcare resources, timely medical interventions, and efficient vaccination programs. This strategys, if implemented effectively, can reduce the spread of COVID-19 and other infectious diseases, thereby protecting public health and saving lives.
In conclusion, our research contributes to epidemiological modeling by demonstrating the importance of a multifaceted approach in understanding and combating COVID-19, as well as in preparing for future public health crises.
Data availibility
The data used in this study are available upon request from the corresponding author.
Code availability
The codes employed in this investigation are available upon request to the authors.
References
World Health Organization. World Health Statistics 2022. https://www.who.int/data/gho/publications/world-health-statistics (2022).
Kuppalli, K. et al. India’s COVID-19 crisis: A call for international action. Lancet 397, 2132–2135 (2021).
Kermack, W. O. & McKendrick, A. G. A contribution to the mathematical theory of epidemics. Proc. R. Soc. Lond. 115, 700–721 (1927).
Ortiz, S. et al. Identification of hazard and socio-demographic patterns of dengue infections in a Colombian subtropical region from 2015 to 2020: Cox regression models and statistical analysis. Trop. Med. Infect. Dis. 8, 30 (2023).
Mohammed-Awel, J. & Gumel, A. B. Mathematics of an epidemiology-genetics model for assessing the role of insecticide resistance on malaria transmission dynamics. Math. Biosci. 312, 33–49 (2019).
Kim, S., de Los Reyes, V. A. A. & Jung, E. Mathematical model and intervention strategies for mitigating tuberculosis in the Philippines. J. Theor. Biol. 443, 100–112 (2018).
Pan, J. et al. Why controlling the asymptomatic infection is important: A modelling study with stability and sensitivity analysis. Fractal Fract. 6, 197 (2023).
Tat Dat, T. et al. Epidemic dynamics via wavelet theory and machine learning with applications to COVID-19. Biology 9, 477 (2023).
Mandal, M. et al. A model based study on the dynamics of COVID-19: Prediction and control. Chaos Solitons Fractals 136, 109889 (2020).
Khajanchi, S., Sarkar, K., Mondal, J., Nisar, K. S. & Abdelwahab, S. F. Mathematical modeling of the COVID-19 pandemic with intervention strategies. Results Phys. 25, 104285 (2021).
Liu, Z., Magal, P. & Webb, G. Predicting the number of reported and unreported cases for the COVID-19 epidemics in China, South Korea, Italy, France, Germany and United Kingdom. J. Theor. Biol. 509, 110501 (2021).
Bajiya, V. P., Bugalia, S. & Tripathi, J. P. Mathematical modeling of COVID-19: Impact of non-pharmaceutical interventions in India. Chaos 30, 113143 (2020).
Salman, A. M. et al. An optimal control of SIRS model with limited medical resources and reinfection problems. Malays. J. Fundam. Appl. Sci. 18, 332–342 (2022).
Ali, M., Shah, S. T., Imran, M. & Khan, A. The role of asymptomatic class, quarantine, and isolation in the transmission of COVID-19. J. Biol. Dyn. 14, 389–408 (2020).
Huo, X., Chen, J. & Ruan, S. Estimating asymptomatic, undetected and total cases for the COVID-19 outbreak in Wuhan: A mathematical modeling study. BMC Infect. Dis. 21, 476 (2021).
Djilali, S., Benahmadi, L., Tridane, A. & Niri, K. Modeling the impact of unreported cases of the COVID-19 in the North African countries. Biology 9, 373 (2020).
Hamou, A. A., Rasul, R. R., Hammouch, Z. & Özdemir, N. Analysis and dynamics of a mathematical model to predict unreported cases of COVID-19 epidemic in Morocco. Comput. Appl. Math. 41, 289 (2022).
Samui, P., Mondal, J. & Khajanchi, S. A mathematical model for COVID-19 transmission dynamics with a case study of India. Chaos Solitons Fractals 140, 110173 (2020).
Gao, S. et al. A mathematical model to assess the impact of testing and isolation compliance on the transmission of COVID-19. Infect. Dis. Model. 8, 427–44 (2023).
Ullah, S. & Khan, M. A. Modeling the impact of non-pharmaceutical interventions on the dynamics of novel coronavirus with optimal control analysis with a case study. Chaos Solitons Fractals 139, 110075 (2020).
Prathumwan, D., Trachoo, K. & Chaiya, I. Mathematical modeling for prediction dynamics of the coronavirus disease 2019 (COVID-19) pandemic, quarantine control measures. Symmetry 12, 1404 (2020).
Perez-Lillo, N., Lagos-Alvarez, B., Muñoz-Gutierrez, J., Figueroa-Zúñiga, J. & Leiva, V. A statistical analysis for the epidemiological surveillance of COVID-19 in Chile. Signa Vitae 18, 19–30 (2022).
Ospina, R., Leite, A., Ferraz, C., Magalhaes, A. & Leiva, V. Data-driven tools for assessing and combating COVID-19 outbreaks based on analytics and statistical methods in Brazil. Signa Vitae 18, 18–32 (2022).
Sardar, I., Akbar, M. A., Leiva, V., Alsanad, A. & Mishra, P. Machine learning and automatic ARIMA/Prophet models-based forecasting of COVID-19: Methodology, evaluation, and case study in SAARC countries. Stoch. Environ. Res. Risk Assess. 37, 345–359 (2023).
Martin-Barreiro, C., Cabezas, X., Leiva, V., Ramos de Santis, P., Ramirez-Figueroa, J.A., & Delgado, E. Statistical characterization of vaccinated cases and deaths due to COVID-19 Methodology and case study in South America. AIMS Math. 8, 22693–22713 (2023).
Kanchanarat, S., Chinviriyasit, S. & Chinviriyasit, W. Mathematical assessment of the impact of the imperfect vaccination on diphtheria transmission dynamics. Symmetry 2022, 14 (2000).
Lemecha Obsu, L. & Feyissa, Balcha S. Optimal control strategies for the transmission risk of COVID-19. J. Biol. Dyn. 14, 590–607 (2020).
Tuite, A. R., Fisman, D. N. & Greer, A. L. Mathematical modelling of COVID-19 transmission and mitigation strategies in the population of Ontario, Canada. Can. Med. Assoc. J. 192, E497-505 (2020).
Mondal, J. & Khajanchi, S. Mathematical modeling and optimal intervention strategies of the COVID-19 outbreak. Nonlinear Dyn. 109, 177–202 (2022).
Ankamma Rao, M. & Venkatesh, A. SEAIQHRDP mathematical model analysis for the transmission dynamics of COVID-19 in India. J. Comput. Anal. Appl. 31, 96–116 (2023).
Umapathy, K., Palanivelu, B., Leiva, V., Dhandapani, P. B. & Castro, C. On fuzzy and crisp solutions of a novel fractional pandemic model. Fractal Fract. 7, 528 (2023).
Fierro, R., Leiva, V. & Balakrishnan, N. Statistical inference on a stochastic epidemic model. Commun. Stat. Simul. Comput. 44, 2297–2314 (2015).
Dhandapani, P. B., Thippan, J., Martin-Barreiro, C., Leiva, V. & Chesneau, C. Numerical solutions of a differential system considering a pure hybrid fuzzy neutral delay theory. Electronics 11, 1478 (2022).
Hamidi, F. et al. Metaheuristic solution for stability analysis of nonlinear systems using an intelligent algorithm with potential applications. Fractal Fract. 7, 78 (2023).
Ospina, R., Gondim, J. A. M., Leiva, V. & Castro, C. An overview of forecast analysis with ARIMA models during the COVID-19 pandemic: Methodology and case study in Brazil. Mathematics 11, 3069 (2023).
Ferguson, N. et al. Impact of non-pharmaceutical interventions (NPIs) to reduce COVID-19 mortality and healthcare demand. Report 9, Imperial College 2020. https://doi.org/10.25561/77482.
Li, R. et al. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV-2). Science 368, 489 (2020).
Gumel, A.B., Ruan, S., Day, T., Watmough, J., Brauer, F., van den Driessche, P., Gabrielson, D., Bowman, C., Alexander, M.E., Ardal, S., Wu, J., & Sahai. Modeling strategies for controlling SARS outbreaks. Proc. Royal Soc. of Lond. B 271, 2223–2232 (2004).
Nadim, S. S., Ghosh, I. & Chattopadhyay, J. Short-term predictions and prevention strategies for COVID-19: A model-based study. Appl. Math. Comput. 404, 126251 (2021).
Biswas, S. K. et al. COVID-19 pandemic in India: A mathematical model study. Nonlinear Dyn. 102, 537–553 (2020).
Khajanchi, S., Sarkar, K., Mondal, J., & Perc, M. Dynamics of the COVID-19 pandemic in India. arXiv:2005.06286 (2020).
Diekmann, O., Heesterbeek, J. A. & Roberts, M. G. The construction of next-generation matrices for compartmental epidemic models. J. R. Soc. Interface 7, 873–885 (2010).
Van den Driessche, P. & Watmough, J. Reproduction numbers and sub-threshold endemic equilibria for compartmental models of disease transmission. Math. Biosci. 180, 29–48 (2002).
Xue, D. MATLAB Programming: Mathematical Problem Solutions (De Gruyter, 2020).
Chitnis, N., Hyman, J. M. & Cushing, J. M. Determining important parameters in the spread of malaria through the sensitivity analysis of a mathematical model. Bull. Math. Biol. 70, 1272–1296 (2008).
Rodrigues, H.S., & Monteiro, M.T.T. Torres DFM. Sensitivity analysis in a dengue epidemiological model. In Conference Papers in Mathematics, 721406 (2013).
Pontryagin, L. S. et al. The Mathematical Theory of Optimal Processes (Wiley, 1962).
Bajiya, V. P., Bugalia, S., Tripathi, J. P. & Martcheva, M. Deciphering the transmission dynamics of COVID-19 in India: Optimal control and cost-effective analysis. J. Biol. Dyn. 16, 665–712 (2022).
Workman, J. T. & Lenhart, S. Optimal Control Applied to Biological Models (CRC Press, 2007).
Venkatesh, A. & Ankamma, Rao M. Mathematical model for COVID-19 pandemic with implementation of intervention strategies and cost-effectiveness analysis. Results Control Optim. 4, 100345 (2024).
Venkatesh, A., Ankamma Rao, M., & Vamsi, D.K.K. A comprehensive study of optimal control model simulation for COVID-19 infection with respect to multiple variants. Commun. Math. Biol. Neurosci. 75 (2023).
Salman, A. M., Mohd, M. H. & Muhammad, A. A novel approach to investigate the stability analysis and the dynamics of reaction-diffusion SVIR epidemic model. Commun. Nonlinear Sci. Numer. Simul. 126, 107517 (2023).
Jang, J., Kwon, H. D. & Lee, J. Optimal control problem of an SIR reaction-diffusion model with inequality constraints. Math. Comput. Simul. 171, 136–151 (2020).
Dhandapani, P. B., Leiva, V., Martin-Barreiro, C. & Rangasamy, M. On a novel dynamics of a SIVR model using a Laplace-Adomian decomposition based on a vaccination strategy. Fractal Fract. 7, 407 (2023).
Rangasamy, M., Chesneau, C., Martin-Barreiro, C. & Leiva, V. On a novel dynamics of SEIR epidemic models with a potential application to COVID-19. Symmetry 14, 1436 (2022).
Rangasamy, M., Alessa, N., Dhandapani, P. B. & Loganathan, K. Dynamics of a novel IVRD pandemic model of a large population with efficient numerical methods. Symmetry 2022, 14 (1919).
Hurwitz, A. Ueber die Bedingungen, unter welchen eine Gleichung nur Wurzeln mit negativen reellen Theilen besitzt. Math. Ann. 46, 273–284 (1895).
Castillo-Chavez, C. & Song, B. Dynamical models of tuberculosis and their applications. Math. Biosci. Eng. 1, 361–404 (2004).
Khajanchi, S., Das, D. K. & Kar, T. K. The dynamics of tuberculosis transmission with exogenous reinfections and endogenous reactivation. Phys. A 497, 52–71 (2018).
Rana, P. S. & Nitin, S. The modeling and analysis of the COVID-19 pandemic with vaccination and treatment control: A case study of Maharashtra, Delhi, Uttarakhand, Sikkim, and Russia in the light of pharmaceutical and non-pharmaceutical approaches. Eur. Phys. J. Special Top. 231, 3629–3648 (2022).
LaSalle, J. P. Stability theory for ordinary differential equations. J. Differ. Equ. 4, 57–65 (1968).
Schechter, M. Principles of Functional Analysis (American Mathematical Society, 2001).
Acknowledgements
The authors would like to thank the editors and anonymous reviewers for their valuable comments and suggestions, which helped us to improve the quality of this article. This research was partially supported by the Vice-rectorate for Research, Creation, and Innovation (VINCI) of the Pontificia Universidad Católica de Valparaíso (PUCV), Chile, grants VINCI 039.470/2024 (regular research), VINCI 039.493/2024 (interdisciplinary associative research), VINCI 039.309/2024 (PUCV centenary), and FONDECYT 1200525 (Víctor Leiva), from the National Agency for Research and Development (ANID) of the Chilean government under the Ministry of Science, Technology, Knowledge, and Innovation; and by Portuguese funds through the CMAT—Research Centre of Mathematics of University of Minho, Portugal, within projects UIDB/00013/2020 (https://doi.org/10.54499/UIDB/00013/2020) and UIDP/00013/2020 (https://doi.org/10.54499/UIDP/00013/2020) (Cecilia Castro).
Author information
Authors and Affiliations
Contributions
Conceptualization: V.A., A.R.M., V.S., P.B.D., V.L., C.M.-B., C.C.; data curation: V.A., A.R.M., V.S., P.B.D., C.M.-B., C.C.; formal analysis: V.A., A.R.M., V.S., P.B.D., V.L., C.M.-B., C.C.; investigation: V.A., A.R.M., V.S., P.B.D., V.L., C.M.-B., C.C.; methodology: V.A., A.R.M., V.S., P.B.D., V.L., C.M.-B., C.C.; writing-original draft: V.A., A.R.M., V.S., P.B.D., C.M.-B.; writing-review and editing: V.L., C.C. All authors have read and agreed to the published version of the article.
Corresponding authors
Ethics declarations
Competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this article.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Proofs of Theorems from “Methods”
Proofs of Theorems from “Methods”
Proof of Theorem 1
Proof
Let (S(t), A(t), I(t), U(t), G(t), R(t)) be any solution of the expression formulated in (1), where \(t \in [0, t_{0}]\). We show that this solution lies in \({\mathbb {R}}_{+}^{6}\) for the specified interval. From the formulas presented in (1), we have that \({\text{d}S}/{\text{d}t}=\Lambda -(\zeta _{A}\,A+\zeta _{I}\,I+\zeta _{G}\,G)\frac{S}{N}-\mu S\) and subsequently, \({\text{d}S}/{\text{d}t}=\Lambda -\left( (\zeta _{A}\,A+\zeta _{I}\,I+\zeta _{G}\,G)\frac{1}{N}+\mu \right) S=\Lambda -\psi (t)S,\) where \(\psi (t) = \beta (\zeta _{A}\,A+\zeta _{I}\,I+\zeta _{G}\,G)({1}/{N})+\mu\). Integrating, we obtain that \(S(t) = S_{0} \exp (-\int _{0}^{t} \psi (s)\,\text{d}s) \int _{0}^{t}\exp (\int _{0}^{s}\psi (u)\,\text{d}u) \text{d}s\ge 0,\) confirming \(S(t)\ge 0\). Additionally, from the expressions established in (1), we find \({\text{d}A}/{\text{d}t} = (\zeta _{A}\,A+\zeta _{I}\,I+\zeta _{G}\,G)({S}/{N}) - (\omega + \mu )A \ge -(\omega + \mu )A\), which infers \({\text{d}A}/{\text{d}t}\ge -(\omega + \mu )A\), and thereby \(A(t) \ge A_{0} \exp \left( -\int _{0}^{t} (\omega + \mu ) \, \text{d}s\right) \ge 0\), validating \(A(t)\ge 0\). Also from the formulas given in (1), we get \({\text{d}I}/{\text{d}t} = \theta \omega \,A - (\eta _{I} + \gamma _{I}+\mu _{I}+\mu ) I \ge -(\eta _{I} + \gamma _{I}+\mu _{I}+\mu ) I\), and then \(I(t) \ge I_{0} \exp \left( -\int _{0}^{t} (\eta _{I} + \gamma _{I}+\mu _{I}+\mu ) \, \text{d}s\right) \ge 0\), certifying \(I(t)\ge 0\). Similarly, from expressions presented in (1) for the compartment \(U(t)\), we attain \({\text{d}U}/{\text{d}t} = (1-\theta )\omega \,A - (\eta _{U}+\gamma _{U}+\mu _{U}+\mu )U \ge -(\eta _{U}+\gamma _{U}+\mu _{U}+\mu )U\), leading to \(U(t) \ge U_{0} \exp \left( -\int _{0}^{t} (\eta _{U} + \gamma _{U} + \mu _{U} + \mu ) \, \text{d}s\right) \ge 0\), verifying \(U(t)\ge 0\). For the compartment \(G(t)\), again from (1), we deduce \({\text{d}G}/{\text{d}t} = \eta _{I}\,I+\eta _{U}\,U-(\gamma _{G}+\mu _{G}+\mu ) G \ge -(\gamma _{G} + \mu _{G} + \mu ) G\), and thereby \(G(t) \ge G_{0} \exp \left( -\int _{0}^{t} (\gamma _{G} + \mu _{G} + \mu ) \, \text{d}s\right) \ge 0\), confirming \(G(t)\ge 0\). Regarding the compartment \(R(t)\), we have \({\text{d}R}/{\text{d}t} = \gamma _{I}\,I + \gamma _{U} U + \gamma _{G}\,G - \mu R \ge -\mu R\), which gives \(R(t) \ge R_{0} \exp \left( -\int _{0}^{t} \mu \, \text{d}s\right) \ge 0\), validating \(R(t)\ge 0\).
Now, we prove the boundedness of the solutions (S, A, I, U, G, R) of the expression stated in (1). From the positivity of the solutions, we get \({\text{d}S}/{\text{d}t}\le \Lambda -\mu S\), which implies that \(\lim _{t\rightarrow \infty }\sup S\le {\Lambda }/{\mu }\) and consequently \(S\le {\Lambda }/{\mu }\). Consider the total population \(N(t) = S(t) +A(t)+ I(t)+ U(t) + G(t)+ R(t)\). After differentiating the expressions described in (1), we get \({\text{d}N}/{\text{d}t}\le \Lambda -\mu N\), which gives \(\lim _{t\rightarrow \infty }\sup N\le {\Lambda }/{\mu }\). Thus, we get \(N\le {\Lambda }/{\mu }\) and consequently \(S + A + I + U + G + R \le {\Lambda }/{\mu }\). Hence, all the solution trajectories (S, A, I, U, G, R) satisfying the primary values of (1) are uniformly bounded in \({\mathbb {R}}_{+}^{6}\) and in the region \(\rho\). As a result, in a finite amount of time, all the solutions of (1) initiating in \({\mathbb {R}}_{+}^{6}\) enter into the region \(\rho\). \(\square\)
Proof of Theorem 2
Proof
To begin, we determine the eigenvalues of the Jacobian matrix \(J_{E^{0}}\) at \(E^{0}\) for the system depicted in (1). This matrix is given by
The characteristic equation derived from the matrix \(J_{E^{0}}\) is given by \(\det (J_{E^{0}} - \lambda I) = 0\), which further reduces to\((\lambda +\mu )(\lambda +\mu )(\lambda +(\eta _{U}+\gamma _{U}+\mu _{U}+\mu ))(\lambda ^{3} + b_{1}\lambda ^{2} + b_{2}\lambda + b_{3}) = 0\), where
Note that \((\eta _{I}+\gamma _{I}+\mu _{I}+\mu )(\eta _{U}+\gamma _{U}+\mu _{U}+\mu )(\gamma _{G}+\mu _{G}+\mu )>(\beta \zeta _{A}-(\omega + \mu )),\) which leads to \((\eta _{I}+\gamma _{I}+\mu _{I}+\mu )(\eta _{U}+\gamma _{U}+\mu _{U}+\mu )(\gamma _{G}+\mu _{G}+\mu )-(\beta \zeta _{A}-(\omega + \mu ))>0\), that is, \(b_{1}>0\). Since we have that \((\eta _{I}+\gamma _{I}+\mu _{I}+\mu )(\eta _{U}+\gamma _{U}+\mu _{U}+\mu )(\gamma _{G}+\mu _{G}+\mu )+(\omega + \mu )((\eta _{I}+\gamma _{I}+\mu _{I}+\mu )+(\eta _{U}+\gamma _{U}+\mu _{U}+\mu ))(\gamma _{G}+\mu _{G}+\mu )+\beta \zeta _{G}(1-\theta )\omega \eta _{U}>(\beta \zeta _{I}\theta \,\omega +\beta \zeta _{A}((\eta _{I}+\gamma _{I}+\mu _{I}+\mu )+(\eta _{U}+\gamma _{U}+\mu _{U}+\mu ))(\gamma _{G}+\mu _{G}+\mu )),\) this implies that \((\eta _{I}+\gamma _{I}+\mu _{I}+\mu )(\eta _{U}+\gamma _{U}+\mu _{U}+\mu )(\gamma _{G}+\mu _{G}+\mu )+(\omega + \mu )((\eta _{I}+\gamma _{I}+\mu _{I}+\mu )+(\eta _{U}+\gamma _{U}+\mu _{U}+\mu ))(\gamma _{G}+\mu _{G}+\mu )+\beta \zeta _{G}(1-\theta )\omega \eta _{U}-\beta \zeta _{I}\theta \,\omega -\beta \zeta _{A}((\eta _{I}+\gamma _{I}+\mu _{I}+\mu )+(\eta _{U}+\gamma _{U}+\mu _{U}+\mu ))(\gamma _{G}+\mu _{G}+\mu )>0,\) that is, \(b_{2}>0\). If \(R_{0} <1\), then \(1- R_{0} >0\). Thus, we have \((\eta _{I}+\gamma _{I}+\mu _{I}+\mu )(\eta _{U}+\gamma _{U}+\mu _{U}+\mu )(\gamma _{G}+\mu _{G}+\mu )(\omega +\mu )(1-R_{0})>0\), that is, \(b_{3}>0\) for \(R_{0} <1\). Similarly, we can prove that \(b_{1} b_{2} > b_{3}\) for \(R_{0} <1\). Hence, we get \(b_{1}, b_{2},b_{3} > 0\) and \(b_{1} b_{2} > b_{3}\), for \(R_{0} <1\).
Observe that the Routh–Hurwitz criterion57 states that, if \(b_{1}, b_{2},b_{3} > 0\) and \(b_{1} b_{2} > b_{3}\), then the DFE is locally asymptotically stable. Therefore, by this criterion, the DFE of the structure stated in (1) is locally asymptotically stable if \(R_{0} <1\). \(\square\)
Proof of Theorem 3
Proof
From the model defined in (1), observe that \(S\) and \(R\) are disease-free compartments, while \(A\), \(I\), \(U\), and \(G\) are diseased compartments. Thus, the model can be written as \({\text{d}X}{\text{d}t} = P(X,Y)\), \({\text{d}Y}{\text{d}t} = Q(X,Y)\), and \(Q(X, 0) = 0\), where \(X = (S, R) \in {\mathbb {R}}_{+}^{2}\), \(Y = (A, U, I, G) \in {\mathbb {R}}_{+}^{4}\). The global stability of the DFE \(E^{0}\) can be derived through the method presented in58. To prove the global asymptotic stability of \(E^{0}\), the following two conditions must be satisfied: (i) \({\text{d}X}/{\text{d}t} = P(X^{*}, 0)\), where \(X^{*}\) is globally asymptotically stable; and (ii) \(Q(X,Y) = M Y - \bar{Q}(X,Y)\), where \(\bar{Q}(X, Y) \ge 0\) and \(M = D_{Y} G(X^{*}, 0)\) is the Metzler matrix. If the structure defined in (1) satisfies the assumed conditions in (i) and (ii) above, then the equilibrium point \(E^{0}\) is globally asymptotically stable for \(R_{0} < 1\). Hence, for the model formulated in (1), \(P(X, Y)\) and \(Q(X, Y)\) can be rewritten as
From the expression \(Q(X, Y)\), it is easily shown that \(Q(X, 0) = 0\). The model stated in (1) in its part (i) can be written as
which implies that \(S(t) = {\Lambda }/{\mu } + (S(0) - {\Lambda }/{\mu })\exp (-\mu t)\) and \(R(t) = R(0)\exp (-\mu t)\). As \(t \rightarrow \infty\), \(S(t) = {\Lambda }/{\mu }\) and \(R(0) = 0\). Thus, \(X^{*}\) is asymptotically stable globally for \({\text{d}X}/{\text{d}t} = P(X, 0)\). Therefore, the first condition (i) is satisfied for the system formulated in (1). Now, the matrices \(M\) and \(\bar{Q}(X,Y)\), which are derived from the model presented in (1), can be written as
Note that all non-diagonal elements of the matrix \(M\) are non-negative as the terms represent transition rates between compartments, which cannot be negative. Since all non-diagonal elements of matrix \(M\) are non-negative, \(M\) is a Metzler matrix. As \(S(t) \le N(t)\), \(\frac{S}{N} \ge 0\). Therefore, \(\bar{Q}(X,Y) \ge 0\). Given that the two conditions (i) and (ii) above are satisfied, the disease-free steady state \(E^{0}\) of the system given in (1) is globally asymptotically stable for \(R_{0} < 1\). \(\square\)
Proof of Theorem 4
Proof
Now, we employ the center manifold theory to analyze the local asymptotic stability of the EE point \(\Sigma ^{*}\). \(\beta\) is considered as a bifurcation parameter that, given \(R_0 = 1\), yields \(\beta = \beta ^{*}\) given by
where \(K_{1} = \omega + \mu\), \(K_{2} = \eta _{I} + \gamma _{I} + \mu _{I} + \mu\), \(K_{3} = \eta _{U} + \gamma _{U} + \mu _{U} + \mu\), and \(K_{4} = \eta _{G} + \mu _{G} + \mu\). The Jacobian of the system (1) around the DFE \(E^{0}\) is given by
which implies that
One eigenvalue of the Jacobian matrix \(J^{*}_{E^{0}}\) is zero, having algebraic multiplicity one, meaning it does not repeat and is associated with a single linearly independent eigenvector. The other eigenvalues of the Jacobian matrix are either negative or have negative real parts. This suggests that the equilibrium point \(E^{0}\) is stable in the directions corresponding to these eigenvalues, as negative real parts indicate stability in those directions. To analyze the system dynamics near the critical value \(\beta ^{*}\), where \(R_{0} = 1\), we apply the center manifold theorem. This reduces the dimensionality of the system and focuses on the dynamics on the center manifold, influenced by the zero eigenvalue. This approach helps us study the stability and bifurcation behavior of the system around \(\beta = \beta ^{*}\). We use both right and left eigenvectors corresponding to the zero eigenvalue to fully understand the system dynamics. The right eigenvector \(\varvec{p}\) satisfies \(J^{*}_{E^{0}} \varvec{p} = 0\), representing the direction of the transformation. The left eigenvector \(\varvec{q}\) satisfies \(\varvec{q}^{\top } J^{*}_{E^{0}} = 0\), crucial for analyzing the system’s response and sensitivity.
The right eigenvector \(\varvec{p} = [p_{1}, p_{2}, p_{3}, p_{4}, p_{5}, p_{6}]^{\top }\) and the left eigenvector \(\varvec{q} = [q_{1}, q_{2}, q_{3}, q_{4}, q_{5}, q_{6}]^{\top }\) have components
To facilitate our analysis, we introduce the following notations for the SAIUGR model system (1): \(S = y_{1}\), \(A = y_{2}\), \(I = y_{3}\), \(U = y_{4}\), \(G = y_{5}\), \(R = y_{6}\), and let \({\text{d}y_{j}}/{\text{d}t} = g_{j}\) for \(j=1,2,3,4,5,6\). These notations will simplify the representation of the system’s equations and the subsequent calculations.
Next, we need to calculate the second-order partial derivatives of \(g_{j}\) at \(E^{0}\) to understand the nonlinear interactions in the system. The non-zero second-order partial derivatives at \(E^{0}\) are given by
with all the remaining partial derivatives at \(E^{0}\) being zero, we will determine the coefficients \(a\) and \(b\) using the second-order partial derivatives calculated above and the center manifold theory, as described in Theorem 4.1 from58. These coefficients are given by
Using the left and right eigenvectors and the values of all the nonzero second-order partial derivatives at \(\beta =\beta ^{*}\), we obtain
Hence, \(a < 0\) and \(b > 0\) at \(\beta = \beta ^{*}\). Therefore, by58,59, a transcritical bifurcation occurs at \(R_{0} = 1\), and the EE point \(\Sigma ^{*}\) is locally asymptotically stable for \(R_{0} > 1\). \(\square\)
Proof of Theorem 5
Proof
To prove this, we construct a Lyapunov function \(L\) defined over the set \(\Phi ^{*}\) and mapping to the real numbers \({\mathbb {R}}\) as \(L\text{: } \{(S^{*}, A^{*}, I^{*}, U^{*}, G^{*}) \in \Phi ^{*}\} \rightarrow {\mathbb {R}}\), given by
where \(\Phi ^{*} = \{(S^{*}, A^{*}, I^{*}, U^{*}, G^{*}) \in {\mathbb {R}}^{5}\text{: } S(t), A(t), I(t), U(t), G(t) > 0\}\).
To establish global asymptotic stability, it is necessary to demonstrate that \({\text{d}L}/{\text{d}t}\) is negative definite. This is achieved by differentiating \(L\) along the solution trajectories of the system previously described obtaining
At EE point \(\Sigma ^{*}\), we have
Substituting the above expressions in (5), we get
Utilizing the method outlined in60, the function \(\widehat{g}(S, A, I, U, G)\) is non-positive. This implies that \(\widehat{g} \le 0\) for all values of \(S, A, I, U, G,R > 0\).Since the arithmetic mean is greater or equal to the geometric mean), we see that
Consequently, it is established that \({\text{d}L}/{\text{d}t} < 0\) and \({\text{d}L}/{\text{d}t} = 0\) if and only if \(S = S^{*}\), \(A = A^{*}\), \(I = I^{*}\), \(U = U^{*}\), and \(G = G^{*}\). Therefore, the EE point \(\Sigma ^{*}\) is the unique single set where \({\text{d}L}/{\text{d}t} = 0\). Now, applying the invariance principle as detailed in61, we can demonstrate that \(E^{*}\) is global asymptotic stability within \(\Phi ^{*}\) when \(R_{0} > 1\). \(\square\)
Proof of Theorem 6
Proof
Through Theorem 1, it is stated that all solutions of the structure delineated in (1) remain nonnegative and uniformly bounded within the region \(\rho\). Consequently, the nonnegativity of the objective function \({\mathscr {M}}\) is apparent, and it also remains bounded. Hence, there exists a minimization of the system of optimal controls \(v^{\tau }(t) = (u_{1}^{\tau }(t), u_{2}^{\tau }(t))\) within \(\Psi\), such that \(\lim _{\tau \rightarrow \infty }{\mathscr {M}}(u^{\tau }(t)) = \inf _{u\in {\mathscr {M}}}{\mathscr {M}}(u(t))\). Thus, the optimal controls in \(M\) are uniformly bounded over \(L^{\infty }\) and uniformly bounded over \(L^{2}\). Since \(L^{2}([0,T])\) is a reflexive space62, \(u^{*}(t) = (u_{1}^{*}(t), u_{2}^{*}(t))\) exists in \({\mathscr {M}}\) such that \(u_{1}^{\tau }(t) \rightarrow u_{1}^{*}(t)\) and \(u_{2}^{\tau }(t) \rightarrow u_{2}^{*}(t)\) weakly as \(\tau \rightarrow \infty\) in \(L^{2}([0,T])\). Then, the state system \({\mathscr {X}}^{\tau } = (S^{\tau }, A^{\tau }, I^{\tau }, U^{\tau }, G^{\tau }, R^{\tau })\) is uniformly bounded corresponding to \(v^{\tau }(t)\). Note that the uniform boundedness implies the uniform boundedness of the differentiation of \({\mathscr {X}}^{\tau }\) and equi-continuity of the corresponding state system \({\mathscr {X}}^{\tau }\). By the Arzelà-Ascoli theorem, there exists a subsequence \({\mathscr {X}}^{\tau } = (S^{\tau }, A^{\tau }, I^{\tau }, U^{\tau }, G^{\tau }, R^{\tau })\) such that \({\mathscr {X}}^{\tau } \rightarrow {\mathscr {X}}^{*}\) uniformly on [0, T]. Now, consider a proper subsequence that converges and corresponds to optimal controls \(u_{1}^{*}(t)\) and \(u_{2}^{*}(t)\) such that the state solution \({\mathscr {X}}^{*}\) is achieved. The lower semi-continuity of the \(L^{2}\) norm and weak convergence indicates that
Consequently, \(u^{*}(t)\) is an optimal control. \(\square\)
Proof of Theorem 7
Proof
To prove this theorem, we use the optimal conditions \({\partial {\mathscr {H}}}/{\partial u_{1}}=0\) and \({\partial {\mathscr {H}}}/{\partial u_{2}}=0\). Then, we get
Since \(0 \le u_{1},u_{2} \le 1\), we have \(u_{1} = u_{2} = 0\) for \(\bar{u_{1}} < 0\) and \(\bar{u_{2}} < 0\) and \(u_{1} = u_{2} = 1\) if \(\bar{u_{1}} > 1\) and \(\bar{u_{2}} > 1\). Otherwise, \(u_{1} = \bar{u_{1}}\) and \(u_{2} = \bar{u_{2}}\). Thus, we get optimum values of \({\mathscr {M}}\) for these optimal controls. \(\square\)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ambalarajan, V., Mallela, A.R., Sivakumar, V. et al. A six-compartment model for COVID-19 with transmission dynamics and public health strategies. Sci Rep 14, 22226 (2024). https://doi.org/10.1038/s41598-024-72487-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-72487-9
- Springer Nature Limited