Abstract
The time that elapsed between the initial introduction and the proliferation of an invasive species is referred to as the lag phase. The identification of the lag phase is critical for generating plans for pest management and for the prevention of biosecurity failure. However, lag phases have been identified mostly through retrospective searches of historical records. The agricultural pest fall armyworm (FAW; Spodoptera frugiperda) is native to the New World. FAW invasion was first reported from West Africa in 2016, then it spread quickly through Africa, Asia, and Oceania. Here, using population genomics approaches, we demonstrate that the FAW invasion involved an undocumented lag phase. Invasive FAW populations have negative signs of genomic Tajima’s D, and invasive population-specific genetic variations have particularly decreased Tajima’s D, supporting a substantial amount of time for the generation of new mutations in introduced FAW populations. Model-based diffusion approximations support the existence of a period with a cessation of gene flow between native and invasive FAW populations. Taken together, these results provide strong support for the presence of a lag phase during the FAW invasion. These results show the usefulness of using population genomics analyses to identify lag phases in biological invasions.
Similar content being viewed by others
Introduction
A biological invasion occurs when an introduced species from native to non-native habitat ranges establishes a stable population in a new geographic area and further proliferates and expands its distributional range1. Invasive species are one of the main causes of agricultural economic losses2 and a major threat to biodiversity3. As human trade is growing, the risk of invasion is also increasing4, especially in insects5. Recently reported cases of invasion often involve the global spread of pests6, and climate changes are predicted to further increase the likelihood of invasion7,8, placing pressure on the entire world. Importantly, a time gap is often found between the introduction and invasion. This gap is referred to as the lag phase9. Lag phases have been reported from diverse taxa including plants10, insects11, birds12, and fishes13. The cause of lag phases is unclear, but it has been speculated that a lag phase corresponds to a waiting time required for adaptive evolution by new mutation or standing genetic variation (reviewed in Bock et al.14). Since low-density populations often exhibit low growth rates, the lag phase has also been considered the time needed to surpass a specific threshold required for establishing stable populations from small founder populations15. Once an alien pest species establishes a stable population and starts spreading, eradication is typically impossible16. Therefore, taking action during the lag phase is generally regarded as the last window of opportunity to eradicate an alien pest species17.
Inferring the history of biological invasions is of utmost importance for generating prevention plans. However, the determination of the lag phase during an invasion is mainly based on the presence or absence of historical records on existing invasive species (for example, see Aikio et al.10). An obvious limitation of this approach is that the presence of alien species before the invasion does not necessarily mean that the current invasive population is their descendant (Fig. 1A, B). Moreover, historical records of initial introductions are largely incomplete for most invasive cases. Hence, we still have limited knowledge of the early phase of invasions14,18. With advances in sequencing techniques enabling whole genome resequencing from many samples, it is possible to test lag phases through population genomics approaches. A large number of SNVs (single nucleotide variations) in whole genome sequences potentially provide robust statistical power to test past demographic history, including changes in effective population size, the time of the split between populations, and the genetic closeness among populations19. Thus, genomic analysis can be used to test the lag phase through the examination of resequencing datasets encompassing a broad geographic range of both invasive and native populations. While the identification of lag phases in invasive pest species is crucial for evaluating implemented pest management and biosecurity plans, the use of genomics approaches has, indeed, been limited.
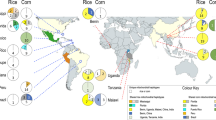
A Hypothetical evolutionary scenario of invasion with lag phase. If an introduced alien species with a historical record survived until population expansion and being invasive, a lag phase exists between introduction and invasion. B If an initially introduced alien species with a historical record experienced extinction and if a group of individuals was introduced right before population expansion and being invasive, such a lag phase does not exist. C Empirical studies demonstrated that ancestral fall armyworms were split into corn strains and rice strains with differentiated ranges of host plants24. The corn strain was further split into two distinct lineages with different mitochondrial genomes (i.e. mtA and mtB). Extant invasive populations originated from a genetically admixed population between mtA and mtB28.The lag phase corresponds to the time between the introduction of fall armyworms and the spread of the genetically admixed population. D The geographic locations from which the analyzed samples were collected by Yainna et al.28 are depicted on the map. The red and green colors indicate native and invasive areas, respectively. In the native areas, the classification of strains is also presented. The map was generated using MapChart62.
The fall armyworm, Spodoptera frugiperda (J.E. Smith) (FAW; Lepidoptera: Noctuidae: Noctuinae) is a major insect pest attacking major crops, including corn, cotton, rice, sorghum, and soybean20,21. The FAW consists of two host plant strains, the corn strain and the rice strain, named after their preferred host plants22,23. The corn strain is further divided into two sub-strains with different mitochondrial genomes, mtA and mtB, respectively24. The FAW originates from the New World (i.e., North and South America), and its invasion of West Africa was first reported in 201625. In the following years, the FAW spread rapidly in the Old World, including sub-Saharan Africa, Middle East Asian India, South East Asia, East Asia, Egypt, Oceania, and the Canary Islands (https://www.cabi.org/isc/fallarmyworm). In sub-Saharan Africa, where corn accounts for at least 30% of caloric intake26, invasive FAW larvae seriously reduce corn yield, causing losses ranging from 21% to 53% on average27.
In a recent study from our research group28, we analyzed whole genome sequences of 177 FAW individuals collected from six native areas (Brazil, French Guiana, Guadeloupe, mainland USA, Mexico, and Puerto Rico) and five invasive areas (Benin, China, India, Malawi, and Uganda). We showed that invasive populations originated from non-Mexican corn strains and that invasive populations exhibit whole genome differentiation from all native populations. We also showed that the invasive populations originated from a single population, which appears to be generated through hybridization between mtA and mtB possibly after introduction to an invasive area (Fig. 1C). Zhang et al.29 performed population genomics analysis from 280 samples collected from an increased geographic range, including five native areas (additional places in mainland USA, Brazil, Guadeloupe, Argentina, and Puerto Rico) and eight invasive areas (China, Ethiopia, Ghana, Kenya, Malawi, Rwanda, South Africa, and Zambia), and they also reported the same pattern. These studies involve samples collected within 2 years from the initial report of the FAW invasion to the sampling dates. It is improbable that the level of genomic differentiation between invasive and native populations within this timeframe could surpass that observed between all pairs of native populations (e.g., the USA and Argentina). Therefore, it is tempting to test the existence of a lag phase for the observed genomic differentiation between invasive and native populations.
Tay et al. performed genomics analyses based on 870 nuclear SNVs and mitochondrial genomes from samples collected from a wide geographic range of sampling sites, including one sample from a pre-border interception from Yunnan China to Australia in 201630. They suggested that the FAW invasion took place before 2016 and involved a complex history of multiple introductions from the New World to the Old World. The use of an intercepted FAW sample from China in 2016 alone suggests that FAWs were present in China before an official report of invasion because FAWs were not known to have arrived in China before 2016. However, it is still unclear whether the observed invasive FAWs found in Old Word are the descendants of the insects that arrived before 2016, which would indicate the presence of a lag phase.
In this study, we test the existence of a lag phase in invasive FAW populations by carrying out population genomics analyses relying on the comprehensive dataset we generated for the study of Yainna et al.28. The total number of filtered SNVs in this dataset is 27,117,672, which potentially provides sufficient statistical power to test a demographic history with lag phase. The dataset includes invasive FAW samples collected from Benin in 2017 and from India in 201831, shortly after the invasion was reported in 201625, and whose genomic sequences did not exhibit any discernible differences from those of other invasive populations28. To test for the presence of a lag phase, we applied both traditional population genetics statistics and a model-based diffusion approximation approach.
Results
The whole genome resequencing dataset28 used in this study was generated from 144 samples from Florida (24 individuals collected in 2015), Mississippi (17 individuals in 2009), Puerto Rico (15 individuals in 2009), China (two individuals in 2019), Benin (39 individuals in 2017), French Guiana (three individuals in 1992), Guadeloupe (four individuals in 2013), India (14 in 2018), and Mexico (26 in 2009) (Fig. 1D). In total, the resequencing data include 89 and 55 samples from native and invasive populations, respectively. The number of filtered SNVs was 27,117,672 in this dataset.
Increase in heterozygosity after the introduction
First, we compared heterozygosity between invasive and native populations from 412,404 SNVs for which genotypes were determined from all samples. Invasive populations had fewer heterozygous positions (15,761.40 on average) than mtA (17,571.02, p-value = 3.887 × 10−5; Wilcoxon rank-sum test) or mtB (18,833.00, p-value = 2.034 × 10−7), while the differences were only 10.30% and 16.31% for mtA and mtB, respectively (Fig. 2, see Fig. S1 for the distribution of averaged heterozygosity for each population). Intriguingly, two individuals from India showed almost the complete depletion of heterozygosity (B4 and B9). The sequencing coverage of these two samples was 13.6X and 16.7X, respectively. One individual from Puerto Rico (PR19) had particularly high heterozygosity. The numbers of homozygous positions are shown in Fig. S2.
The number of heterozygous positions for each individual was counted from positions of which genotypes are determined from all individuals. The x-axis shows each individual, and the y-axis is the number of heterozygous positions (kb). The error bars indicate 95% confidence intervals with 1000 bootstrapping replications resampled from 100 kb windows.
To test the possibility that genetic admixture between mtA and mtB contributed to heterozygosity in invasive populations, we examined the proportion of mtA-specific or mtB-specific SNVs among all SNVs for each invasive sample, in a way that these SNVs includes both fixed and polymorphic variations within mtA or mtB. mtA-specific and mtB-specific SNVs account for 8.63%–11.8% and 0.902%–1.31%, respectively, while the majority of SNVs were found in at least one sample for each of mtA or mtB samples (Fig. S3). This result implies that the genomic sequences of invasive populations are much closer to mtA than mtB and that genetic admixture could contribute at most 1% of the total heterozygosity of the invasive populations.
Logically, the slight difference in heterozygosity between native and invasive populations may result from a moderate decline in heterozygosity at an introduction or from an increase in heterozygosity after a severe bottleneck. In the former case, Tajima’s D calculated from invasive populations is expected to have a positive sign. We performed a coalescent simulation to test if an invasive population may show positive signs of Tajima’s D at 10 or 20 generations, which corresponds to around one or 2 years under laboratory conditions (1 month per generation), following a population bottleneck for the samples from Benin and India, respectively. The average heterozygosity of native populations was 0.00435. Based on the assumption that the mutation rate is mutation rate of 2.9 × 10−9 per site per generation32, the historical effective population size of native populations was estimated to be 0.00435 / (4 × 2.9 × 10−9) = 375,000. We used this calculation as a population size before the bottleneck. The simulation results showed that after 10 generations, Tajima’s D of bottlenecked populations was 0.202 and 1.066 when f was 100 and 1000, respectively (Fig. S4). After 20 generations, Tajima’s D was 0.359 and 1.513 for f values of 100 and 1000, respectively. These values of f correspond to the introduction of 3,750 individuals (375,000/100) and 375 individuals (375,000/1000), respectively. Since it is not likely that far >3750 unrelated FAW individuals could have been introduced at the same time, we assumed that bottleneck events during the introduction are sufficient to generate positive signs of Tajima’s D without a lag phase.
Tajima’s D for the populations from Benin and India was −0.512 (−0.496 to −0.531 with 95% bootstrapping confidence intervals) and −0.356 (−0.340 to −0.372 of 95% bootstrapping confidence interval), respectively, consistent with a signal of population expansion (Fig. 3). Non-Mexican native populations also displayed negative values for Tajima’s D (−1.237 on average), which were lower than those calculated for invasive populations (p-value = 0.04762). In contrast, the Mexican samples, all belonging to mtA, exhibited a positive sign for Tajima’s D. These Mexican samples had a lower number of heterozygous positions (16,005.62 on average) than the other mtA samples (18,804.36, p-value = 1.007 × 10−10). This result supports the hypothesis of the population growth of invasive FAW populations.
However, the calculated Tajima’s D from invasive populations might have a negative sign due to the effect of population expansion in native populations, which already exhibits a negative sign of Tajima’s D, rather than the expansion of invasive populations after introduction. To test this possibility, we performed coalescent simulation under an extreme scenario in which the native population experienced a 10-fold population expansion 100 generations before the introduction, followed by bottleneck events with varying f. Without bottleneck events at introduction (e.g., f = 1), Tajima’s D was lower than −0.2 (Fig. S5). When f = 100, Tajima’s D was 0.0838 and 0.274 after 10 and 20 generations following the bottleneck, respectively. When f = 1000, Tajima’s D was 0.937 and 1.429 after 10 and 20 generations following the bottleneck, respectively.This result suggests that Tajima’s D is not likely to be lowered below zero in invasive populations due to population expansion in native populations.
Then, we tested if mutations that occurred after the bottleneck contributed to the increase in heterozygosity. The total numbers of common and invasive population-specific SNVs were 9,333,445 and 1,547,174, respectively. The proportions of invasive population-specific SNVs calculated for each invasive individual were largely uniform across all invasive populations (6.40%–6.76%), with the exception of two Indian individuals, where a slight reduction was observed (5.19%–5.27%) (Fig. 4A).
A The proportion of invasive population-specific SNVs was calculated from each sample. The error bars indicate 95% confidence intervals with 1000 bootstrapping replications resampled from 100 kb windows. B Tajima’s D was calculated from SNVs common to both native and invasive populations, as well as SNVs specific to invasive populations.
If these invasive population-specific SNVs were generated after introduction, then these SNVs are expected to have lower Tajima’s D than SNVs common to both invasive and native populations because these SNVs are not expected to have sufficient time to increase the allele frequency to the common SNVs33. We observed that invasive population-specific SNVs had much lower Tajima’s D than common SNVs (Fig. 4B), which supports the presence of a lag phase providing sufficient time for the generation of new mutations.
Model-based diffusion approximation
We further tested the presence of a lag phase by estimating the time of introduction using a model-based diffusion approximation with maximum likelihood framework with the assumptions that gene flow between invasive and native populations ceased after the introduction to the Old World and that the invasive population in Benin or India was derived from one of the populations from Florida, Mississippi, or Puerto Rico, or from another genetically closely related population. We considered four models: SI (Strict Isolation), where there is a split between invasive and native populations with an immediate cessation of gene flow; IM (Isolation and Migration), involving a split without cessation of gene flow; AM (Ancient Migration), indicating a split with ongoing gene flow followed by the cessation of gene flow; and AM_bottle (Ancient Migration and Bottleneck), involving a split with gene flow followed by cessation of gene flow and a bottleneck event (Fig. S6). Please note that AM_bottle represents a realistic evolutionary scenario of invasion incorporating a lag phase through a bottleneck event34. Likelihood ratio test showed that the AM_bottle model explains best among all models in all pairs of the investigated invasive and native populations (Table S1 and Fig. S7).
The time with the cessation of gene flow (Ts) between the populations from Benin and from Florida, Mississippi, and Puerto Rico was 498.5 years (with a 95% bootstrapping confidence interval of 136.4–1353.7 years), 21.8 years (8.3–107 years), and 213 years (16.1–888 years), respectively (Fig. 5, see Fig. S8 for other parameters). When the population from India was considered as an invasive population, Ts with the population from Florida, Mississippi, and Puerto Rico was 566.9 years (156.0–1310.1 years), 15.9 years (11.7–160.3 years), and 90.6 years (17.8–583.4 years), respectively. While these confidence intervals are wide, and the native populations with the lowest Ts cannot be conclusively identified, the calculated Ts consistently exceeds the expectation without the lag phase, 1 or 2 years.
The estimated time of gene flow cessation (Ts) between pairs of invasive populations (Benin or India) and native populations (Florida, Mississippi, and Puerto Rico). The error bars indicate 95% bootstrapping confidence intervals, which were calculated using base-pair resampling with 1000 replications. The red dotted horizontal bar indicates Ts = 1, the expected value in the absence of a lag phase.
Discussion
We tested the presence of a lag phase during the FAW invasion using population genomics analyses. The slight reduction in heterozygosity and the negative sign of Tajima’s D in invasive populations support the hypothesis that FAW populations experienced a growth in population size after being introduced. The majority of the analyzed invasive FAWs had between 6.40% to 6.76% SNVs that were exclusively found from invasive populations. These SNVs had Tajima’s D values much lower than those found in both invasive and native populations, raising the possibility that these invasive population-specific SNVs were generated after the introduction of FAW in the Old World. The time of introduction (Ts), estimated using model-based diffusion approximation, indicates an introduction far predating 2016. All these results cannot be explained without the existence of a lag phase before 2016 when the invasion of FAWs was first reported.
This study emphasizes the value of population genomics approaches for determining the evolutionary history of an invasion and testing for the presence of a lag phase. The potential existence of FAWs in the Old World has been documented prior to 2016. In addition to the one pre-border intercepted FAW sample reported by Tay et al.30, the same authors presented several references stating the presence of FAWs in Vietnam in 2008 (for example35). Wiltshire also reported the presence of a likely transient population of FAW in Israel in 197736. These reports themselves do not necessarily support the existence of a lag phase for FAWs because extant invasive FAW populations may not be descendants of the reported introduced individuals30,35,36 (Fig. 1B). Furthermore, it is not practical or necessarily relevant to conduct an extensive global search for a record of the introduction of the FAW, as it is not possible to determine whether old records correspond to populations that still exist today. Only genomic analyses can accurately identify a lag phase, knowing that previous large-scale population genomics studies28,29 were able to reduce geographic sampling bias.
Lag phases may explain the ‘genetic paradox of invasion’, a dilemma invasive success cannot be easily explained because of the genetic bottleneck in introduced populations37. If a bottleneck occurs during an invasion as a result of the introduction of a small number of individuals, the invasive population may have a decreased fitness by inbreeding depression38 because small populations have a higher chance of having homozygous recessive deleterious alleles than larger populations39 before purifying selection eventually mitigates inbreeding depression. Thus, reduced fitness in invasive populations might be contradictory with ample cases of invasion. Interestingly, the majority of cases reveal that invasive populations have only a mild reduction in heterozygosity (e.g., <20%) compared with native populations, and frequently have even higher heterozygosity than native populations40. In the case of the FAW invasion, we posit the possibility that the heterozygosity of introduced populations may have increased almost to the level of native populations during the lag phase before the introduced populations were identified as invasive populations particularly because genetic admixture does not appear to be the main cause for the increased heterozygosity. If this possibility is true, a key step for successful invasions might be the increase in heterozygosity during a lag phase.
A potential criticism against our conclusion is that sampling bias might affect the pattern that was observed. Even though we analyzed samples collected from a wide geographic range of native locations, including Florida, French Guiana, Guadeloupe, Mexico, Mississippi, and Puerto Rico, it is possible that a native population that has not yet been identified shares many similarities with the invasive populations we examined but differs genetically from all of the examined native populations. In this case, it is possible that the cessation of gene flow might have occurred between multiple native populations, rather than between invasive and native populations. Then, the model selection based on diffusion approximation should not be considered as a support of a lag phase. However, if the identified invasive population-specific SNVs actually originated from native populations, it is difficult to explain why the identified invasive population-specific SNVs have much lower allele frequencies (i.e., lower Tajima’s D) than common ones. If the identified invasive population-specific SNVs exist as rare alleles in native populations and if this rarity is responsible for the very low allele frequencies in invasive populations, we need to consider an unrealistic evolutionary scenario that a substantial number of rare alleles should have been introduced and maintained in invasive populations to constitute 5.19%–6.76% of total SNVs, even though genetic drift would typically eliminate most of the rare alleles. Therefore, it is unlikely that there is a geographic population (within mtA), which is very different from all analyzed native mtA populations while being very similar to invasive populations. Indeed, previous population genomics studies showed that mtA populations (which belong to corn strain) are largely undifferentiated24,28,41,42 and that the genetic difference between corn and rice strains accounts for the majority of population structure24.
The cause of the global FAW population explosion after the lag phase is yet to be identified. Genomics studies repeatedly reported adaptive evolution associated with insecticide resistance in invasive FAWs28,43,44,45,46,47,48, raising the hypothesis that field-evolved insecticide resistance may be the primary factor in the success of FAW invasiveness. However, insecticide resistance alone might not be sufficient to explain the population explosion of introduced FAWs because native FAW populations are also under strong evolutionary pressure for insecticide resistance42,47,49,50. Future studies should focus on unveiling the direct cause of population expansion following the lag phase. Furthermore, future studies with increased sample numbers could focus on more precise estimations of the lag phase by testing parameter-rich complex demographic models.
In this study, we demonstrated that the FAW invasive process involved a lag phase, using population genomics approaches. This finding implies that introduced FAWs were present in some places in the Old World before 2016 and that their existence has been either overlooked or unreported, which contributed to the failure of preventing the FAW invasion. As mentioned before, the identification of a lag phase can be a critical step in pest management. In light of the unprecedented rise in invasion cases worldwide6, it is essential to test the presence of a lag phase from at least well-known damaging invasive species for the generation of improved prevention plans, which were chosen to be one of the global targets decided by the Convention on Biological Diversity51. We expect that the use of genomics to tackle biological invasions52 is going to be increasingly popular. We argue that genomics investigations should be conducted on invasive pest species to document cases involving lag phases in invasion. This effort will contribute to the management of pest species and the prevention of biosecurity failures. Furthermore, the detected lag phase in this study highlights the importance of monitoring programs for early detection and systematic reporting to prevent pest invasions.
Methods
Resequencing dataset
This study is based on the whole genome SNV dataset of 177 samples generated by Yainna et al.28. (Please see the Data Availability statement to find the accession numbers). Among these samples, we excluded the samples from Brazil, Uganda, and Malawi because the resequencing data were not publicly accessible when we performed the analyses, leaving a total of 144 samples analyzed. Briefly speaking about this dataset, Illumina paired-end sequencing was performed for all samples except for two Chinese samples, from which MGISEQ-2000 was used. The average read depth was approximately 20X per sample. This SNV dataset was generated by mapping against the ver7 reference genome24 and by variant calling using bowtie2 v2.3.4.153 and GATK v4.1.2.054, respectively.
Population genetics statstics
The heterozygosity was calculated from the number of nuclear biallelic heterozygous sites counted from each individual, together with bootstrapping with 1000 replications by resampling 100 kb windows. A single 100 kb window corresponds to 0.0260% of the reference genome assembly. This approach has an advantage over the use of π and θ statistics, which assume a homogeneous and panmictic population structure, because the number of heterozygous sites is not affected by population structure. Tajima’s D was calculated from 100 kb windows using VCFtools v0.1.1655. The average Tajima’s D over these windows was calculated. We determined 95% bootstrapping confidence intervals by resampling the 100 kb windows with 1000 replications. We did not calculate Tajima’s D for groups with a sample size lower than five.
Simulation
A coalescent simulation was performed using the ms software56 (Hudson 2002). Mutation and recombination rates were derived from rates estimated in two other lepidopteran species: Heliconius melpomene (L.) (mutation rate of 2.9 × 10−9 per site per generation32) and Bombyx mori (L.) (recombination rate of 1.143 × 10−8 per site per generation57). The length of the simulated DNA sequence was 1 Mb. The heterozygosity of each native sample was calculated using bcftools v1.1358, followed by calculating the average heterozygosity across all native samples. The effective population size of the native population was then determined using the classical formula of H = 4 × Ne × μ, where H is heterozygosity, Ne is the effective population size, and μ is the mutation rate. This effective population size was used as an initial population size. The population experienced varying bottleneck strengths (f = 1, 10, 100, and 1000, where f is the relative size of a population before bottleneck to the original population). Tajima’s D was calculated from 1000 individuals at 10 generations after the bottleneck using the sample_stats binary in the ms software56. Watterson’s θ per base pair was calculated from the SNV of segregating sites, which was counted by the sample_stats binary. For each f, 20 independent simulations were replicated. Watterson’s θ and Tajima’s D were averaged out across the replications for each f. Coalescent simulations were also performed in scenarios with a 10-fold increase in population size from 106, occurring 100 generations before the present time, followed by a bottleneck event 10 generations before the present time with varying f. Watterson’s θ and Tajima’s D were then averaged using the same calculation method.
The inference of demography using diffusion approximation
The presence of a lag phase was also tested by the estimation of divergence time without gene flow between invasive and native populations using ∂a∂i v1.6.359, which is based on diffusion approximations with maximum likelihood framework. We generated autosomal vcf files with 16 individuals from Benin and each of the populations from Florida (sub-sampled to n = 13), Mississippi (16), and Puerto Rico (15). The samples from Benin and India were collected in 2017 and 2018, respectively, just 1 or 2 years after the official report of the FAW invasion25. All SNV positions with missing data were discarded. The vcf files were further thinned by removing SNVs within 100 bp to reduce genetic linkages among SNVs (Fig. S9). The decay of linkage disequilibrium was estimated using PopLDdecay v3.42. Then, a Perl script (https://github.com/wk8910/bio_tools/blob/master/01.dadi_fsc/00.convertWithFSC/convert_vcf_to_dadi_input.pl) generated ∂a∂i input files for each of the vcf files. We took into consideration four models of demographic scenario: Strict Isolation (SI), Isolation-with-Migration (IM), Ancient Migration (AM), and Ancient Migration and Bottleneck (AM_bottle) (Fig. S6). In SI, an ancestral population split into two populations and these two populations did not have migrating individuals. In IM, bidirectional gene flows are allowed after the split. AM also considers that two sibling populations experienced bidirectional gene flows, which completely ceased at a certain point. AM_bottle is the same as AM, except that one population undergoes a bottleneck when gene flow ceases. In these models, the following demographic parameters were estimated. The population sizes of the two sibling populations were N1 and N2, and the rate of gene flow between these two populations were migr. The divergence time of SI and IM was Ts. In AM and AM_bottle, the divergence time was Tam + Ts, where Tam and Ts are times with gene flow and complete isolation, respectively. In AM_bottle, b is a reduction factor of N1 after the cessation of gene flow. Ts and Tam were converted to years based on the assumption that the mutation rates and the generation times are 2.9 × 10−9 per site per generation32 and 1 month, respectively.
Folded joint site frequency spectrums were projected with the sample sizes so two samples from each individual (diploid organism). Projections were set to 32 for the 16 Benin individuals, and 26, 32, and 30, for Florida, Mississippi, and Puerto Rico, respectively. The folded joint site frequency spectrum was fitted against each of the models. Then, 25 independent optimizations were performed using a hot and a cold simulated annealing procedure followed by BFGS optimization60. The grid points were set at 120, 130, and 140. The initial parameters were randomly determined. A likelihood ratio test among nest models was performed to choose the best model explaining the observed SFS. We generate 1000 bootstrapped datasets by resampling genomic scaffolds with replacement using dadiBoot.pl at 2bRAD_denovo61, followed by the estimation of the parameters and the calculation of 95% confidence intervals.
Statistics and reproducibility
All statistical analyses were performed using computer programming scripts with publicly available data. The scripts were released to the public for reproducibility.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
This study was conducted using whole genome resequencing data downloaded from NCBI (PRJNA639295, PRJNA577869, PRJNA494340, and PRJNA639296).
Code availability
Computer programming scripts used in this study are available at https://doi.org/10.5281/zenodo.12686241.
References
Prentis, P. J., Wilson, J. R. U., Dormontt, E. E., Richardson, D. M. & Lowe, A. J. Adaptive evolution in invasive species. Trends Plant Sci. 13, 288–294 (2008).
Diagne, C. et al. High and rising economic costs of biological invasions worldwide. Nature 592, 571–576 (2021).
McGeoch, M. A. et al. Global indicators of biological invasion: species numbers, biodiversity impact and policy responses. Divers. Distrib. 16, 95–108 (2010).
Seebens, H. et al. Global rise in emerging alien species results from increased accessibility of new source pools. Proc. Natl Acad. Sci. USA 115, E2264–E2273 (2018).
Roques, A. et al. Temporal and interspecific variation in rates of spread for insect species invading Europe during the last 200 years. Biol. Invasions 18, 907–920 (2016).
Tay, W. T. & Gordon, K. H. J. Going global – genomic insights into insect invasions. Curr. Opin. Insect Sci. 31, 123–130 (2019).
Ward, N. L. & Masters, G. J. Linking climate change and species invasion: an illustration using insect herbivores. Glob. Change Biol. 13, 1605–1615 (2007).
Mungi, N. A., Coops, N. C., Ramesh, K. & Rawat, G. S. How global climate change and regional disturbance can expand the invasion risk? case study of Lantana camara invasion in the Himalaya. Biol. Invasions 20, 1849–1863 (2018).
Kowarik, I. Time lags in biological invasions with regard to the success and failure of alien species. In. Plant Invasions—General Aspects and Special Problems (eds. Pyšek, P., Prach, K., Rejmánek, M. & Wade, M.) 15–38 (SPB Academic Publishing, 1995).
Aikio, S., Duncan, R. P. & Hulme, P. E. Lag-phases in alien plant invasions: separating the facts from the artefacts. Oikos 119, 370–378 (2010).
Morimoto, N., Kiritani, K., Yamamura, K. & Yamanaka, T. Finding indications of lag time, saturation and trading inflow in the emergence record of exotic agricultural insect pests in Japan. Appl. Entomol. Zool. 54, 437–450 (2019).
Aagaard, K. & Lockwood, J. Exotic birds show lags in population growth. Divers. Distrib. 20, 547–554 (2014).
Azzurro, E., Maynou, F., Belmaker, J., Golani, D. & Crooks, J. A. Lag times in Lessepsian fish invasion. Biol. Invasions 18, 2761–2772 (2016).
Bock, D. G. et al. What we still don’t know about invasion genetics. Mol. Ecol. 24, 2277–2297 (2015).
Tobin, P. C., Whitmire, S. L., Johnson, D. M., Bjørnstad, O. N. & Liebhold, A. M. Invasion speed is affected by geographical variation in the strength of allee effects. Ecol. Lett. 10, 36–43 (2007).
Mack, R. N. et al. Biotic invasions: causes, epidemiology, global consequences, and control. Ecol. Appl. 10, 689–710 (2000).
Suckling, D. M. et al. Eradication of tephritid fruit fly pest populations: outcomes and prospects. Pest Manag. Sci. 72, 456–465 (2016).
van Kleunen, M., Bossdorf, O. & Dawson, W. The ecology and evolution of alien plants. Annu. Rev. Ecol. Evol. Syst. 49, 25–47 (2018).
Beichman, A. C., Huerta-Sanchez, E. & Lohmueller, K. E. Using genomic data to infer historic population dynamics of nonmodel organisms. Annu. Rev. Ecol. Evol. Syst. 49, 433–456 (2018).
Andrews, K. L. The Whorlworm, Spodoptera frugiperda. Cent. Am. Neighb. Areas Fla. Entomol. 63, 456–467 (1980).
Sparks, A. N. A review of the biology of the fall armyworm. Fla. Entomol. 62, 82–87 (1979).
Pashley, D. P. Host-associated genetic differentiation in fall armyworm (Lepidoptera: Noctuidae): a sibling species complex? Ann. Entomol. Soc. Am. 79, 898–904 (1986).
Dumas, P. et al. Phylogenetic molecular species delimitations unravel potential new species in the pest genus Spodoptera Guenée, 1852 (Lepidoptera, Noctuidae). PLOS ONE 10, e0122407 (2015).
Fiteni, E. et al. Host-plant adaptation as a driver of incipient speciation in the fall armyworm (Spodoptera frugiperda). BMC Ecol. Evol. 22, 133 (2022).
Goergen, G., Kumar, P. L., Sankung, S. B., Togola, A. & Tamò, M. First report of outbreaks of the fall armyworm Spodoptera frugiperda (J E Smith) (Lepidoptera, Noctuidae), a new alien invasive pest in west and central Africa. PLOS ONE 11, e0165632 (2016).
Nuss, E. T. & Tanumihardjo, S. A. Maize: a paramount staple crop in the context of global nutrition. Compr. Rev. Food Sci. Food Saf. 9, 417–436 (2010).
Day, R. et al. Fall armyworm: impacts and implications for Africa. Outlooks Pest Manag. 28, 196–201 (2017).
Yainna, S. et al. The evolutionary process of invasion in the fall armyworm (Spodoptera frugiperda). Sci. Rep. 12, 21063 (2022).
Zhang, L. et al. Global genomic signature reveals the evolution of fall armyworm in the Eastern hemisphere. Mol. Ecol. 32, 5463–5478 (2023).
Tay, W. T. et al. Global population genomic signature of Spodoptera frugiperda (fall armyworm) supports complex introduction events across the Old World. Commun. Biol. 5, 1–15 (2022).
Sharanabasappa et al. First report of the Fall armyworm, Spodoptera frugiperda (J E Smith) (Lepidoptera: Noctuidae), an alien invasive pest on maize in India. Pest Manag. Hortic. Ecosyst. 24, 23–29–29 (2018).
Keightley, P. D. et al. Estimation of the spontaneous mutation rate in Heliconius melpomene. Mol. Biol. Evol. 32, 239–243 (2015).
Slatkin, M. & Rannala, B. Estimating allele age. Annu. Rev. Genom. Hum. Genet. 1, 225–249 (2000).
Ortego, J., Céspedes, V., Millán, A. & Green, A. J. Genomic data support multiple introductions and explosive demographic expansions in a highly invasive aquatic insect. Mol. Ecol. 30, 4189–4203 (2021).
Pham, V. L. In Plant Protection Magazine (Plant Protection Research Institute of Vietnam, 2019).
Wiltshire, E. P. Middle East Lepidoptera. XXXVII: notes on the Spodoptera litura (F.) group (Noctuidae Trifinae). Proc. Trans. Br. Ent. Nat. Hist. Soc. 10, 92–96 (1977).
Allendorf, F. W. & Lundquist, L. L. Introduction: population biology, evolution, and control of invasive species. Conserv. Biol. 17, 24–30 (2003).
Charlesworth, D. & Willis, J. H. The genetics of inbreeding depression. Nat. Rev. Genet. 10, 783–796 (2009).
Kimura, M., Maruyama, T. & Crow, J. F. The mutation load in small populations. Genetics 48, 1303–1312 (1963).
Estoup, A. et al. Is there a genetic paradox of biological invasion? Annu. Rev. Ecol. Evol. Syst. 47, 51–72 (2016).
Schlum, K. A. et al. Whole genome comparisons reveal panmixia among fall armyworm (Spodoptera frugiperda) from diverse locations. BMC Genom. 22, 179 (2021).
Gimenez, S. et al. Adaptation by copy number variation increases insecticide resistance in the fall armyworm. Commun. Biol. 3, 664 (2020).
Gui, F. et al. Genomic and transcriptomic analysis unveils population evolution and development of pesticide resistance in fall armyworm Spodoptera frugiperda. Protein Cell 13, 1–19 (2020).
Zhang, D. et al. Insecticide resistance monitoring for the invasive populations of fall armyworm, Spodoptera frugiperda in China. J. Integr. Agric. 20, 783–791 (2021).
Yainna, S. et al. Geographic monitoring of insecticide resistance mutations in native and invasive populations of the fall armyworm. Insects 12, 468 (2021).
Xiao, H. et al. The genetic adaptations of fall armyworm Spodoptera frugiperda facilitated its rapid global dispersal and invasion. Mol. Ecol. Resour. 20, 1050–1068 (2020).
Boaventura, D., Martin, M., Pozzebon, A., Mota-Sanchez, D. & Nauen, R. Monitoring of target-site mutations conferring insecticide resistance in Spodoptera frugiperda. Insects 11, 545 (2020).
Guan, F. et al. Whole-genome sequencing to detect mutations associated with resistance to insecticides and Bt proteins in Spodoptera frugiperda. Insect Sci. 28, 627–638 (2020).
Carvalho, R. A., Omoto, C., Field, L. M., Williamson, M. S. & Bass, C. Investigating the molecular mechanisms of organophosphate and pyrethroid resistance in the fall armyworm Spodoptera frugiperda. PLoS One 8, e62268 (2013).
Gutiérrez-Moreno, R. et al. Field-evolved resistance of the fall armyworm (Lepidoptera: Noctuidae) to synthetic insecticides in Puerto Rico and Mexico. J. Econ. Entomol. 112, 792–802 (2019).
UN Environment Programme. UN Biodiversity Conference (COP 15). http://www.unep.org/un-biodiversity-conference-cop-15 (2022).
Jaspers, C. et al. Invasion genomics uncover contrasting scenarios of genetic diversity in a widespread marine invader. Proc. Natl Acad. Sci. USA 118, e2116211118 (2021).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
McKenna, A. et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Danecek, P. et al. The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011).
Hudson, R. R. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinforma. Oxf. Engl. 18, 337–338 (2002).
Yamamoto, K. et al. A BAC-based integrated linkage map of the silkworm Bombyx mori. Genome Biol. 9, R21 (2008).
Danecek, P. et al. Twelve years of SAMtools and BCFtools. GigaScience 10, giab008 (2021).
Gutenkunst, R. N., Hernandez, R. D., Williamson, S. H. & Bustamante, C. D. inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet 5, e1000695 (2009).
Tine, M. et al. European sea bass genome and its variation provide insights into adaptation to euryhalinity and speciation. Nat. Commun. 5, 5770 (2014).
GitHub. z0on/2bRAD_denovo: Genome-wide de Novo Genotyping with 2bRAD. https://github.com/z0on/2bRAD_denovo (2022).
MapChart. Create Your Own Custom Map. https://mapchart.net/index.html (2024).
Acknowledgements
The study is supported by the department of Santé des Plantes et Environnement at Institut national de recherche pour l’agriculture, l’alimentation et l’environnement (NewHost). We are grateful to Gael J. Kergoat, Nicolas Nègre, and Emmanuelle d’Alençon for their support in this study through intensive discussions.
Author information
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Joaquín Ortego and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Hannes Schuler, Luke Grinham and Tobias Goris. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Durand, K., Yainna, S. & Nam, K. Population genomics unravels a lag phase during the global fall armyworm invasion. Commun Biol 7, 957 (2024). https://doi.org/10.1038/s42003-024-06634-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-024-06634-3
- Springer Nature Limited