Abstract
Alzheimer’s disease (AD), characterized by cognitive decline, is increasingly recognized as a disorder marked by synaptic loss and dysfunction. Despite this understanding, the underlying pathophysiological mechanisms contributing to synaptic impairment remain largely unknown. In this study, we elucidate a previously undiscovered signaling pathway wherein the S-nitrosylation of the Cdk5 activator p39, a post-translational modification involving the addition of nitric oxide to protein cysteine residues, plays a crucial role in synaptic dysfunction associated with AD. Our investigation reveals heightened p39 S-nitrosylation in the brain of an amyloid precursor protein (APP)/presenilin 1 (PS1) transgenic mouse model of AD. Additionally, soluble amyloid-β oligomers (Aβ), implicated in synaptic loss in AD, induce p39 S-nitrosylation in cultured neurons. Notably, we uncover that p39 protein level is regulated by S-nitrosylation, with nitric oxide S-nitrosylating p39 at Cys265 and subsequently promoting its degradation. Furthermore, our study demonstrates that S-nitrosylation of p39 at Cys265 significantly contributes to amyloid-β (Aβ) peptide-induced dendrite retraction and spine loss. Collectively, our findings highlight S-nitrosylation of p39 as a novel aberrant redox protein modification involved in the pathogenesis of AD, suggesting its potential as a therapeutic target for the disease.
Similar content being viewed by others
Introduction
Alzheimer’s disease (AD) stands as a formidable challenge in contemporary healthcare, characterized by progressive cognitive decline and profound synaptic dysfunction1,2,3. As our understanding of AD deepens, it becomes increasingly evident that synaptic loss plays a pivotal role in the pathophysiology of this neurodegenerative disorder. While the overarching impact of synaptic loss in AD is well-established, the specific molecular mechanisms orchestrating this phenomenon remain elusive.
Cyclin-dependent kinase 5 (Cdk5) is a proline-directed serine/threonine kinase, playing pivotal roles in various brain functions. Its contributions span neurogenesis, neuronal migration, dendrite and axon development, as well as synapse formation and synaptic plasticity4,5,6. However, deregulation of Cdk5 has been implicated in the pathogenesis of several neurological diseases, including Alzheimer’s disease (AD), Parkinson’s disease (PD), and certain psychiatric disorders4,5,6,7. Hence, maintaining precise regulation of Cdk5 kinase activity is crucial for ensuring normal development and optimal functioning of the nervous system.
Cyclin-dependent kinase 5 (Cdk5), unlike other members of the CDK family, relies on neural-specific regulatory proteins—p35 and p39, along with their proteolytic derivatives p25 and p29, for activation8,9. While p35 and p39 share over 50% sequence homology, they exhibit distinct spatial and temporal expression patterns. Notably, p35 is prominently expressed from embryonic to postnatal stages, whereas p39 shows minimal detection until the postnatal stage, increasing during postnatal development10. In addition, the distribution of p39 is highest in the brain stem, cerebellum, and spinal cord, whereas p35 predominates in the cerebral cortex10. Despite the pivotal role of Cdk5 in neuronal functions within the central nervous system, research has predominantly focused on p35 and its associated Cdk5 activity. However, during synaptogenesis and synaptic maintenance, p35 expression is down-regulated, making p39 as the primary activator during postnatal development in mouse and rat brains. This suggests a crucial role for p39 expression in postnatal neuronal differentiation and the formation of neuronal networks. Studies in p39−/− mice have revealed aberrant dendritic morphology in pyramidal neurons, characterized by shortened length and reduced arborization11. Additionally, histone acetylation-mediated transcription selectively upregulates p39, enhancing Cdk5 activity during neuronal differentiation in the postnatal brain. Loss of p39 results in disrupted axonal growth and impaired dendritic spine formation12. Despite these insights, the specific involvement of deregulated p39 in the pathogenesis of neurodegenerative diseases like Alzheimer’s (AD) and Parkinson’s (PD) remains largely unknown.
Here in our current study, we discovered that p39 can undergo S-nitrosylation, a post-translational modification in which nitric oxide (NO) is attached to free cysteine residues, both in vitro and in vivo. This S-nitrosylation prompted p39 degradation in a proteasome-dependent manner. Our identification of cysteine 265 (Cys265) as the specific site for S-nitrosylation prompted us to generate a p39-C265A mutant, where Cys265 is substituted with alanine (Ala), resulting in resistance to GSNO-induced S-nitrosylation and subsequent degradation. Importantly, our study revealed an increase in p39 S-nitrosylation levels within Aβ-treated cultured neurons as well as in the brains of APP/PS1 mice, suggesting a connection with Aβ-induced dendrite and spine loss. Furthermore, the expression of p39-C265A in hippocampal neurons successfully restored dendritic spine and synaptic dysfunction observed in APP/PS1 mice. These results reveal a previously unknown crosstalk between NO signaling and p39, implying that S-nitrosylation-mediated p39 degradation played a contributing role in synaptic dysfunction associated with Alzheimer’s disease pathogenesis.
Results
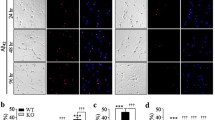
p39 is S-nitrosylated in vitro and in vivo
Previous reports highlighted NO’s role in restricting Cdk5 activity by inducing p35 S-nitrosylation, leading to its degradation and decreased protein level13. Given the sequence similarity between p39 and p35, we hypothesized that NO might similarly regulate Cdk5 activity via p39 S-nitrosylation. To investigate this, we initially assessed whether p39 could undergo S-nitrosylation upon exposure to physiological NO donors. Our experiments involved HEK293T cells overexpressing p39 incubated with the NO donors S-Nitroso-L-glutathione (GSNO) or S-nitrosocysteine (SNOC), followed by the biotin switch assay. Notably, both GSNO and SNOC led to the formation of S-nitrosylated p39 (SNO-p39) (Fig. 1A). Remarkably, a 5-min incubation with GSNO led to a robust increase of SNO-p39 in HEK293T cell lysates overexpressing p39 (Fig. 1B), indicating rapid and spontaneous p39 S-nitrosylation. Further, treating cultured cortical neurons with GSNO for various durations also revealed endogenous p39 S-nitrosylation (Fig. 1C). In particular, ascorbate-dependent basal S-nitrosylation of endogenous p39 was detected in WT but not nNOS-KO adult mouse brains, suggesting that p39 is S-nitrosylated in vivo in a nNOS-dependent manner (Fig. 1D). These findings collectively provide compelling evidence that p39 can be S-nitrosylated by NO, both in vitro and in vivo.
A NO S-nitrosylated p39 in vitro. HEK293T cells overexpressing p39 were incubated with NO donor GSNO or SNOC for 30 min and subsequently lysed and subjected to the biotin switch assay. The biotinylated proteins were pulled down with neutravidin-agarose, followed by western blot analysis. All data are normalized to control group and represent mean ± SEM from 4 independent experiments. **p = 0.0012, ***p = 0.0006; unpaired Student’s t test. B Lysates of HEK293T cells overexpressing p39 were incubated with NO donor GSNO for different time points and subsequently subjected to the biotin switch assay. All data are normalized to control group and represent mean ± SEM from 4 independent experiments. *p = 0.019 (5 min), *p = 0.028 (15 min), *p = 0.031 (30 min), *p = 0.041 (45 min), **p = 0.0066 (60 min); one-way ANOVA with the Student–Newman–Keuls test. C NO S-nitrosylated endogenous p39 in cultured neurons in vitro. Cultured mouse cortical neurons at DIV7-9 was incubated with NO donor GSNO for the indicated time points, and then subjected to biotin switch assay. All data are normalized to control group and represent mean ± SEM from 4 independent experiments. *p = 0.010 (5 min), **p = 0.0011 (15 min), ***p = 0.00079 (30 min), *p = 0.021 (60 min), **p = 0.0076 (120 min); one-way ANOVA with the Student–Newman–Keuls test. D S-nitrosylation of p39 was detected in mouse brain in vivo. Brain lysates of adult WT and nNOS-knockout (nNOS-KO) mice were subjected to the biotin switch assay in the presence or absence of ascorbate as indicated. All data are normalized to control group and represent mean ± SEM from 4 independent experiments. **p = 0.0066; one-way ANOVA with the Student–Newman–Keuls test.
NO negatively regulates p39 protein level in vitro and in vivo
S-Nitrosylation serves as a regulatory mechanism primarily altering protein function through modulating interactions with binding partners, enzymatic activities, or controlling protein abundance13,14,15,16,17,18,19. Our prior work established that S-nitrosylation of p35 enhances its ubiquitination, leading to proteasome-dependent degradation13. The observed decrease in p39 protein levels upon prolonged exposure of cultured neurons to the NO donor GSNO (Fig. 1C) prompted us to explore whether NO regulates p39 protein stability. Treatment of mouse cortical neurons with GSNO resulted in a concentration- and time-dependent decrease in p39 levels, without causing proteolytic cleavage of p39 to p29 (Fig. 2A, B). This effect was corroborated by consistent reductions in p39 protein levels over time upon incubation with another physiological NO donor, SNOC (Fig. 2C). To determine if NO directly influences p39 degradation, we treated cortical neurons with GSNO in the presence of the proteasome inhibitor, MG132. Remarkably, MG132 pre-treatment effectively blocked the GSNO-induced reduction in p39 level (Fig. 2D), suggesting that NO negatively regulates p39 level by promoting its degradation. To investigate whether inhibition of endogenous NOS protects p39 from degradation, we examined p39 protein levels in the hippocampus of nNOS-KO mice. Interestingly, p39 protein levels were elevated in the hippocampus of 3-month-old nNOS-KO mice (Fig. 2E). Additionally, we treated cultured cortical neurons with the NOS inhibitor, L-NAME; this blockade of NO production resulted in a significant increase in p39 levels (Fig. 2F). These findings suggest that NO decreases p39 abundance through proteasome-mediated degradation, both in vitro and in vivo.
A GSNO treatment decreased p39 protein level in a concentration-dependent manner. Cultured cortical neurons at DIV7-9 were treated with GSNO at different concentrations for 2 h and then subjected to western blot analysis. All data are normalized to control group and represent mean ± SEM from 4 independent experiments. **p = 0.0020 (50 µM), **p = 0.0013 (100 µM), **p = 0.0016 (200 µM), **p = 0.0018 (500 µM); one-way ANOVA with the Student–Newman–Keuls test. B GSNO treatment decreased p39 protein level in a time-dependent manner. Cultured cortical neurons at DIV7-9 were treated with 100 µM GSNO for the indicated time points and then subjected to western blot analysis. All data are normalized to control group and represent mean ± SEM from 7 independent experiments. *p = 0.029 (5 min), *p = 0.019 (15 min), ***p = 0.00053 (30 min), ***p = 0.00021 (60 min), ***p = 0.00026 (120 min); one-way ANOVA with the Student–Newman–Keuls test. C SNOC treatment decreased p39 protein level in a time-dependent manner. Cultured cortical neurons at DIV7-9 were treated with 100 µM SNOC for the indicated time points and then subjected to western blot analysis. All data are normalized to control group and represent mean ± SEM from 4 independent experiments. **p = 0.0018 (60 min), ***p = 2.62E-6 (120 min); one-way ANOVA with the Student–Newman–Keuls test. D GSNO decreased p39 level protein in a proteasome-dependent manner. Cultured cortical neurons were treated with GSNO for the indicated times in the presence or absence of the proteasome inhibitor, MG132 and then subjected to western blot analysis for p39 protein. All data are normalized to control group and represent mean ± SEM from 5-7 independent experiments. **p = 0.008 (5 min), **p = 0.0010 (15 min), **p = 0.0082 (30 min), ***p = 0.00050 (60 min), ***p = 0.00070 (120 min); one-way ANOVA with the Student–Newman–Keuls test. E Hippocampal lysates of 3-month-old WT or nNOS-KO mice were subjected to western blot analysis using the indicated antibodies. Quantification of p39 protein levels (ratio of specific protein to actin) in WT and nNOS-KO mice. Data are normalized to WT controls and represent the mean ± SEM of four independent experiments, n = 5 mice per condition. *p = 0.033; unpaired Student’s t test. F L-NAME treatment increased p39 level. Cultured cortical neurons at DIV7-10 were treated with L-NAME for 4 h and subjected to Western blot analysis with the indicated antibodies. Quantification of p39 levels. Data represent the mean ± SEM of 5 independent experiments. *p = 0.018; unpaired Student’s t test.
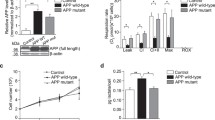
S-Nitrosylation of p39 at the Cys265 residue promotes its proteasome-dependent degradation
To explore whether NO triggers proteasome-dependent degradation of p39 directly through protein S-nitrosylation, we initially aimed to identify the specific site(s) of S-nitrosylation on p39. Given that S-nitrosylation typically occurs at cysteine residues20, we scrutinized the p39 amino acid sequence, revealing the presence of eight cysteine residues (Supplementary Fig. 1A), which are conserved cross mouse, rat, and human species. Subsequently, we generated mutants where each of these eight cysteine residues was replaced with alanine (p39-CxxxA mutants). Upon transfection of HEK293T cells with human wild-type p39 (p39-WT) or its cysteine mutant constructs, we treated the cells with GSNO and performed biotin switch assay. Strikingly, while GSNO treatment robustly S-nitrosylated p39-WT, mutation of Cys265 alone significantly abolished S-nitrosylation, indicating Cys265 as the primary site of S-nitrosylation (Fig. 3A, B, Supplementary Fig. 1B, C). Notably, the other cysteine mutants showed S-nitrosylation level akin to p39-WT, confirming the presence of a single major S-nitrosylation site at Cys265 on p39.
A Cys265 is the major target site for p39 S-nitrosylation. HEK293T cells were transfected with expression constructs encoding the WT or specific cysteine mutants of p39 as indicated. Cells were then incubated with GSNO for 30 min, lysed and subjected to the biotin switch assay. B Quantification of SNO-p39 level. Data are normalized to WT (GSNO treatment) and represent mean ± SEM from four independent experiments. ***p = 1.95E-5 (-WT), ***p = 5.80E-6 (C265A); one-way ANOVA with the Student–Newman–Keuls test. C HEK293T cells were transfected with p39-WT or p39-C265A mutant; 24 h after transfection, cells were treated with GSH/GSNO in the presence of CHX for 4 h and subjected to western blot analysis. D Quantification of p39 level (relative to the actin level). Data are normalized to WT (GSH treatment) and represent mean ± SEM from four independent experiments. ***p = 0.0007, ###p = 0.0005; one-way ANOVA with the Student–Newman–Keuls test. E GSNO treatment increased p39 polyubiquitination level. HEK293T cells were transfected with p39-WT or p39-C265A mutant; 24 h after transfection, cells were treated with GSH/GSNO in the presence of MG132. The protein lysate was subjected to immunoprecipitation using p39 antibody followed by western blot analysis for polyubiquitin. F GSNO treatment increased p39 level in the proteasome. Cultured cortical neurons were treated with GSNO in the presence of MG132 and proteasome was isolated and then subjected to western blot analysis. Data are normalized to control treatment and represent mean ± SEM from 4 independent experiments. *p = 0.020; unpaired student’s t test.
To directly evaluate whether the reduction in p39 level induced by NO (as observed in Fig. 2) resulted from SNO-p39 formation, we overexpressed p39-WT or its S-nitrosylation-deficient mutant, p39-C265A in HEK293T cells. Subsequently, we treated cells with GSH/GSNO in the presence of the protein synthesis inhibitor cycloheximide (CHX). Remarkably, GSNO treatment significantly accelerated the turnover of p39-WT, whereas p39-C265A protein turnover remained unaffected (Fig. 3C, D). We also observed increased polyubiquitination of p39-WT, but not p39-C265A following GSNO treatment (Fig. 3E), indicating that S-nitrosyaltion of p39 at Cys265 promotes its degradation through a ubiquitin/proteasome-dependent mechanism. Additionally, we isolated proteasomes through differential centrifugation and found that p39 was enriched in proteasomes after GSNO treatment (Fig. 3F). Given the recent identification of Praja-2 (PJA2) as the E3 ligase of p3513, we explored whether NO-induced degradation of p39 in the nervous system is mediated by PJA2. However, silencing PJA2 expression in cultured neurons did not alter p39 protein levels, suggesting the involvement of another E3 ligase in p39 degradation. These results compellingly indicate that S-nitrosylation specifically at Cys265 promotes the proteasome-dependent degradation of p39.
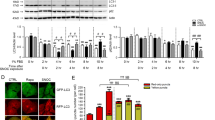
Hypernitrosylation of p39 in the brain of APP/PS1 mice
We sought to explore the potential functional significance of p39 S-nitrosylation in neuronal functions, considering the association between neuronal nitric oxide synthase (nNOS/NOS1) activation, excessive S-nitrosylation, and various neurodegenerative diseases, including Alzheimer’s disease (AD)18,21,22,23. To examine SNO-p39 level in an AD mouse model, we analyzed whole-brain samples from APP/PS1 mice and their control littermates at different postnatal stages (6, 9, 12, & 24 M). Notably, we observed a significant increase in SNO-p39 level in the AD mouse brain compared to control mice (Fig. 4A, B, Supplementary Fig. 2A, B). As a positive control, we also detected elevated SNO-Cdk5 level in the APP/PS1 mouse brain, consistent with previous reports18. Intriguingly, we noted a mild decrease in p39 protein level in the whole-brain lysates of APP/PS1 mice (Fig. 4A, B), suggesting a reduction in p39 abundance due to SNO-p39 formation. To reinforce this hypothesis, we conducted further assessments of p39 protein levels specifically in the mouse hippocampus, known for its high expression of nNOS. Our findings revealed a significant decrease in p39 levels in APP/PS1 mice (Fig. 4C, D). These findings collectively indicate that increased S-nitrosylation of p39 correlates with reduced p39 protein level in the AD mouse model, suggesting a potential role for SNO-p39 in the pathogenesis of Alzheimer’s disease.
A SNO-p39 level was increased in APP/PS1 mouse brain. Adult mouse brain (6 M & 24 M) lysate from APP/PS1 mice or their control littermates was subjected to the biotin switch assay. The biotinylated proteins were pulled down with neutravidin-agarose, followed by western blot analysis for the indicated antibodies. B Quantification of SNO-p39 level. Data are normalized to control group and represent mean ± SEM from 5 animals of three independent experiments. *p = 0.026 (6 M), *p = 0.048 (24 M); Student’s t test. C p39 protein level was decreased in APP/PS1 mouse hippocampus at different ages. APP/PS1 and control mouse hippocampal lysate (6 M, 9 M &12 M) was subjected to the western blot analysis using indicated antibodies. D Quantification of p39 protein level from APP/PS1 mouse hippocampus and their littermate control WT mice at different ages. Data are normalized to control group and represent mean ± SEM from 5 animals of three independent experiments. *p = 0.010 (6 M), ***p = 0.00016 (9 M), ***p = 0.00023 (24 M); one-way ANOVA with the Student–Newman–Keuls test.
S-Nitrosylation of p39 contributes to Aβ-induced dendrite retraction and dendritic spine loss
We aimed to elucidate whether p39 S-nitrosylation serves as a molecular mechanism underlying Alzheimer’s disease (AD) pathogenesis. Observing a substantial increase in p39 protein level during development (from P10 to P20), which persisted into adulthood in the mouse cortex and hippocampus (Supplementary Fig. 3A–D), indicated the pivotal role of p39 in maintaining neuronal network functions. Previous reports have underscored the significance of upregulated p39, but not p35, in regulating dendritic morphology, spine morphogenesis, and neuronal network assembly in the adult mouse brain11,12. Moreover, soluble amyloid-β peptide oligomers (Aβ), which are generated by the proteolytic cleavage of amyloid precursor protein (APP), are believed to be a major causative agent of synaptic impairment during AD progression24,25. It was also reported that AD brains manifest a significant increase in Aβ and a subsequent dramatic increase in NO production as well as dendrite and spine loss in mouse brains18. We hypothesized that p39 S-nitrosylation might mediate Aβ-induced synaptic dysfunction. Indeed, exposure to Aβ substantially elevated SNO-p39 level in cultured cortical neurons, concurrently reducing p39 protein level (Supplementary Fig. 3E–H). Additionally, NMDA-induced overstimulation of NMDA-type glutamate receptors leads to excessive NO production. Consistently, exposure to NMDA resulted in increased p39 S-nitrosylation and subsequent degradation in cultured neurons (Supplementary Fig. 3I, J), highlighting the dynamic nature of this post-translational modification. This supports the notion that excessive NO production induced by Aβ production or neuronal hyper-activation, mediated by S-nitrosylation of p39, promotes its degradation.
In both animal models and human AD brains, synaptic damage including dendrite and spine retraction represents an early neuropathological hallmark26,27. In line with this notion, our findings demonstrated that 24-h exposure to Aβ oligomers induced dendritic defects in hippocampal neurons. Cultured hippocampal neurons expressing p39-WT exhibited reduced dendritic complexity characterized by shortened and fewer dendrites upon Aβ exposure. Intriguingly, expression of the S-nitrosylation-deficient p39-C265 A mutant prevented this effect (Fig. 5A–D). Additionally, p39-WT-transfected neurons experienced approximately 27% dendritic spine loss after Aβ exposure, whereas neurons expressing p39-C265A remained resistant to Aβ-induced spine loss (Fig. 5E, F). To ascertain that NO production and its subsequent S-nitrosylation directly mediate these effects, we treated cultured cortical neurons with the NOS inhibitor, L-NG-nitroarginine methyl ester (L-NAME). Remarkably, we observed that blockade of NO production abolished Aβ exposure-induced p39 S-nitrosylation (Supplementary Fig. 4A, B). Moreover, L-NAME treatment effectively reversed Aβ-induced spine loss (Supplementary Fig. 4C, D). To corroborate these findings at the functional level, we conducted electrophysiological studies. Quantitative analysis revealed that expression of the S-nitrosylation-deficient p39-C265A mutant rescued the deficits in miniature excitatory postsynaptic current (mEPSC) frequency and amplitude observed in Aβ-exposed neurons (Fig. 5G–I). To further validate these findings in vivo, we introduced the S-nitrosylation-deficient p39 mutant (p39-C265A) into hippocampal neurons from 9-month-old WT and APP/PS1 mice using our Sindbis viral system28. After 24 h, we evaluated dendritic spine morphology and conducted electrophysiological studies (Fig. 6A). Remarkably, we consistently observed reduced spine density in neurons from APP/PS1 mice, and the expression of the p39-C265A mutant effectively rescued the deficits in dendritic spine morphology (Fig. 6B, C). Furthermore, functional quantitative analysis revealed that the expression of p39-C265A restored both the frequency and amplitude deficits in mEPSCs observed in hippocampal neurons from APP/PS1 mice (Fig. 6D–F). Taken together, our findings provide compelling evidence that S-nitrosylation of p39 plays a role in Aβ-induced dendritic retraction and dendritic spine loss, offering insight into a potential mechanism underlying synaptic dysfunction in AD.
A Representative images of hippocampal pyramidal neurons overexpressing p39-WT or its S-nitrosylation-deficient mutant (p39-C265A) treated with vehicle or Aβ. Scale bar, 20 µm. B–D SNO-p39 contributed to Aβ-induced dendrite retraction. Sholl analysis (B) and quantification of total dendrite length (C), dendrite number (D); n = 20 neurons from 4 independent experiments; ***p = 2.80E–5 (C), ###p = 1.27E–5 (C), ***p = 0.0003 (D), ###p = 3.88E-6 (D); one-way ANOVA with the Student–Newman–Keuls test. E, F SNO-p39 contributed to Aβ-induced spine loss. Cultured hippocampal neurons transfected with p39-WT-T2A-GFP or p39-C265A-T2A-GFP construct at DIV7 and fixed at DIV15. Representative images (E) are shown. Scale bar, 20 µm. Quantification of spine density (F); n = 30-40 dendrites from 10 to 15 neurons of 4 independent experiments; ***p = 2.43E–8, ###p = 3.64E–11; one-way ANOVA with the Student–Newman–Keuls test. G–I SNO-p39 contributed to Aβ-induced synaptic dysfunction. Cultured hippocampal neurons transfected with p39-WT-T2A-GFP or p39-C265A-T2A-GFP construct at DIV7 and mEPSCs were recorded at DIV15. Quantification of frequency (H) and amplitude (I) of mEPSC; Data represent mean ± SEM from 20 to 30 neurons of 4 independent experiments; ***p = 2.93E–5 (H), ###p = 0.0009(H), ***p = 0.0002 (I), ##p = 0.001(I); one-way ANOVA with the Student–Newman–Keuls test.
A Schematic drawing of dendritic spine quantification and electrophysiological recording experiments in APP/PS1 mice. B, C Overexpression of p39 in the hippocampus of 9-month-old APP/PS1 mice rescued their dendritic spine loss; n = 12 neurons with 36 dendritic spines from 3 to 4 independent experiments; scale bar, 10 µm; All data represent mean ± SEM from 20 to 30 neurons of 4 independent experiments; ***p < 0.0001, #p = 0.03, ###p < 0.0001; one-way ANOVA with the Student–Newman–Keuls test. D–F Expression of p39-C265A corrected the synaptic transmission failure of APP/PS1 mice. Hippocampal neurons were infected with GFP or p39-C265A-T2A-GFP or p39-C265A-T2A-GFP with Sindbis virus system and mEPSCs were recorded 24 h after infection. Quantification of frequency (E) and amplitude (F) of mEPSCs. All data represent mean ± SEM from 20 to 30 neurons of 4 independent experiments; **p = 0.0032 (E), #p = 0.022 (E), ###p < 0.0001 (E), ***p < 0.0001 (F), #p = 0.019 (F), ###p < 0.0001 (F); one-way ANOVA with the Student–Newman–Keuls test.
Discussion
Previous studies shed light on the crucial involvement of p35 and p39, the neuron-specific activators of Cdk5, in neuronal dendrite development and synaptic growth, with its dysregulation implicated in neurodegenerative diseases11,12. It has been demonstrated that under pathological conditions, p35/p39 undergo calpain-dependent cleavage, generating a proteolytic product termed p25/p299. However, it is reported that compared to p35, p39 is much more resistant to calpain-dependent cleavage6. Moreover, the mechanisms underlying the role of deregulated p39 in disease processes remained elusive. Notably, excitotoxic stressors, such as soluble amyloid-β peptide oligomers (Aβ) or overstimulation of NMDA-type glutamate receptors (NMDARs), elevate intracellular Ca2+ level, subsequently activating neuronal nitric oxide synthase (nNOS/NOS1)-the primary source of nitric oxide (NO) in neurons. NO and its derivatives have been implicated in various neurodegenerative diseases21,29,30,31,32,33. NO exerts its effects in neurons by stimulating soluble guanylate cyclase to produce cGMP or by S-nitrosylating critical cysteine residues in multiple target proteins, thus modulating their activity20. Our previous findings have revealed that NO regulates Cdk5 activity through S-nitrosylation of p35, leading to its degradation and reduced protein level13. Given the high sequence homology and similar properties shared between p35 and p39, it is plausible to speculate that p39 might represent a novel target of NO. Moreover, both nNOS and p39 are localized at the cell surface34,35, suggesting a close spatial proximity that could enable endogenous NO to modify p39, forming S-nitrosylated p39 (SNO-p39). Here, our investigation unveils aberrant redox regulation of p39 protein level via S-nitrosylation in APP/PS1 mice and Aβ-treated neuron cultures. We demonstrate the susceptibility of p39 to S-nitrosylation both in vitro and in vivo, and S-nitrosylation promotes its proteasome-dependent degradation. Notably, we identified Cys265 as the pivotal cysteine residue susceptible to pathological and physiological regulation by nitric oxide (NO). Furthermore, we provide compelling evidence that S-nitrosylation of p39 mediates Aβ-induced dendritic retraction and spine loss. More importantly, the deficits in dendritic spine density and neurotransmission observed in APP/PS1 mice were effectively alleviated by the expression of the S-nitrosylation-deficient mutant p39-C265A. Together, these findings strongly implicate p39 S-nitrosylation as a contributing factor in the pathogenesis of Alzheimer’s disease.
Cdk5 activity is known to regulate the dendrite and spine morphogenesis, the major sites of excitatory synaptic transmission in the brain36. It is indeed multifaceted and can produce seemingly contradictory effects, influenced by various factors including its activators, substrate preferences, and their specific roles in different brain stages. Cdk5’s actions on spine density can vary, demonstrating both positive and negative regulatory effects. For instance, Cdk5’s phosphorylation of TrkB is necessary for spine morphogenesis induced by BDNF and glutamate, highlighting its positive role in these processes37. In addition, Cdk5 activity is involved in the regulation of spine morphogenesis and inhibition of Cdk5 activity reduces spine density in nucleus accumbens38. Conversely, phosphorylation of ephexin1 or WAVE1 by Cdk5 leads to a reduction in dendritic spines, indicating a negative influence on spine formation39,40. Moreover, the pseudo kinase CaMKv and the scaffold protein liprinα1 are required for dendritic spine morphogenesis and maturation, whereas Cdk5-dependent phosphorylation of CaMKv at Thr345 and liprinα1 at Thr701 inhibit their activity in these processes41,42. In addition, while it is well accepted that increased Cdk5 activity has been implicated in AD pathogenesis, proteomic and transcriptomic analyses have suggested a downregulation of Cdk5 levels in AD patients43,44. Additionally, other studies have identified the dual roles of Cdk5 in the regulation of ALDH1A1, revealing an initial neuroprotective role of Cdk5 in combating oxidative stress under excitotoxic conditions45. Furthermore, it has been reported that negative modulation of cellular Cdk5 activity by its inhibitor, roscovitine, or Cdk5 antisense stimulates Aβ production in APP processing46. A possible explanation of the sophisticated and sometimes contradictory functions of Cdk5 in modulating synaptic functions under physiological and pathological conditions may be the different expression profile and substrate preference of the two activators, p35 and p39. Indeed, it is reported that p39 associated Cdk5 activity preferentially affects phosphorylation of some specific Cdk5 substrates. Besides, p39 and p35 play non-overlapping and even opposing roles in the postnatal brain12. Similarly, in our study, we found that NO orchestrates similar regulatory mechanisms for p39 as it does for p35, yet their roles in dendritic and spine morphology regulation differ. Our work highlights that NO-induced S-nitrosylation of p39 in APP/PS1 mouse brains promotes its degradation, contributing to Aβ-triggered dendrite retraction and spine loss. Contrastingly, our previous findings revealed that p35 S-nitrosylation and subsequent degradation increased dendritic spine density13. This suggests a dichotomy in the functions of SNO-p35 and SNO-p39 in spine density regulation. The complexity of NO signaling’s impact on dendritic spine density may stem from the intricate spatiotemporal coordination required for precise Cdk5 activity in ensuring proper synaptic function. In our previous study, we explored the functional role of p35 S-nitrosylation in WT and nNOS-KO mice at postnatal 3 months (~P90), at which stage p35 protein level is quite low and found that p35 level is abnormally upregulated upon NO depletion. This underscores the critical role of NO signaling in maintaining structural and functional synaptic strength under physiological conditions. Here, we mainly focused our study under pathological conditions and detected p39 S-nitrosylation in old WT and APP/PS1 mice (6M-24M), at which stage p39 is the major activator of Cdk5 activity. Indeed, we detected enhanced SNO-p39 and SNO-Cdk5, but not SNO-p35 in APP/PS1 mouse brain. Previously, many studies focused on the hyper-activation of p35 associated Cdk5 in several psychiatric disorders and neurodegenerative diseases and proposed the inhibition of Cdk5 activity can alleviate the synaptic losses and cognitive impairments in certain pathological conditions47,48,49. Here our findings unveil a novel molecular mechanism underlying AD pathogenesis, hinting at the potential of targeting SNO-p39 as a unique therapeutic approach for mitigating spine damage in AD and other neurodegenerative conditions. Notably, treatments such as the CC chemokine-CCL5, known to upregulate p3950, contribute to hippocampal spine and synapse formation, enhancing learning and memory51. Additionally, selective upregulation of p39 through histone acetylation-mediated transcription presents a promising avenue for drug development aimed at rectifying synaptic failures in AD51.
Our findings unveil three distinct mechanisms through which NO signaling may regulate Cdk5 activity. Firstly, S-nitrosylation of Cdk5 at Cys83 inhibits its kinase activity, likely by disrupting its binding to ATP. Secondly, S-nitrosylation of p35 and p39 leads to their degradation and reduced protein abundance, representing the second and third mechanisms, respectively. Interestingly, previous studies by the Lipton group have demonstrated that NO-related species activate Cdk5 activity via S-nitrosylation, leading to synaptic loss in AD. These seemingly contradictory results may be reconciled by the concept of transnitrosylation, wherein S-nitrosylated proteins transfer the NO group to other targets, thus providing a deeper understanding of NO’s actions52. Lipton’s group demonstrated that NO-related species activate Cdk5 activity via S-nitrosylation, leading to transnitrosylation and subsequent activation of Drp1. This process results in excessive mitochondrial fragmentation and damage, leading to compromised energy production and ultimately contributing to synaptic loss in AD18,19. Furthermore, the cascade of transnitrosylation reactions involves SNO-Uch-L1 to Cdk5 to Drp1, and notably does not involve the canonical enzymatic effects of Uch-L1 or Cdk552. These findings suggest that the kinase activity of Cdk5 may not be directly involved in the synaptic damage previously observed. Instead, S-nitrosylation rapidly modifies Cdk5 to transnitrosylate other target proteins within minutes, indicating a short half-life of SNO-Cdk5. Given the physiological interaction between Cdk5 and p39, it is of great interest to investigate whether SNO-Cdk5 S-nitrosylates p39 via transnitrosylation. Considering the fact that Cdk5/p35/p39 have many different substrates, it is possible that Cdk5 may also transnitrosylate these substrates to influence multiple biological progresses. Our findings also indicate that p39 S-nitrosylation occurs at Cys265 and promotes its degradation. Notably, previous research demonstrated that NO S-nitrosylates p35 and promotes its ubiquitin-dependent degradation in a PJA2-dependent manner. This raises the intriguing possibility that S-nitrosylation at Cys265 on p39 may induce conformational changes in the protein structure, rendering p39 more susceptible to the E3 ligase PJA2 and targeting it for degradation via the proteasome system. However, silencing of PJA2 expression in cultured neurons did not alter p39 protein levels, suggesting the involvement of an alternative, unidentified E3 ligase for p39. It is also noteworthy that while Cys265 is conserved between p35 and p39, we only identified it as the major S-nitrosylation site on p39. One possible explanation could be the differential expression pattern and subcellular localization between p35 and p39. Notably, p35 is prominently expressed from embryonic to postnatal stages, whereas p39 shows minimal detection until the postnatal stage, increasing during postnatal development. This suggests that p39 may serve as the primary Cdk5 activator in the adult mouse hippocampus (Supplementary Fig. 3C, D). Additionally, p39 is more frequently targeted to the cell surface owing to its myristoylation motif53, rendering it more accessible to NO produced by nNOS. In this context, investigating the distinct regulatory mechanisms of p35 and p39 S-nitrosylation, both under physiological and pathological conditions, presents an intriguing avenue for further exploration. It will also be interesting to explore the mechanism of p39 S-nitrosylation and determine whether it is enzymatic or non-enzymatic. In conclusion, our study highlights the involvement of SNO-p39 in the synaptic dysfunction observed in AD mouse models. This discovery emphasizes the significance of further investigating the molecular and cellular mechanisms both upstream and downstream of SNO-p39. Such comprehensive exploration holds the potential to reveal novel therapeutic strategies for the treatment and prevention of AD, as well as other neurological diseases.
Methods
Animals
The study utilized male/female C57/BL6 wild-type mice and male APPswe/PS1dE9 double-transgenic mice (APP/PS1). The male APP/PS1 mice and their wild-type (WT) counterparts were purchased from Changzhou Cavens Laboratory Animal Co., Ltd. (Changzhou, China). The nNOS transgenic mice were gift from Prof. Dongya Zhu (Nanjing Medical University). All mice were housed in a temperature-controlled room with a 12/12 h light/dark cycle, with humidity controlled as 55%, provided with food and water ad libitum. All animal-related procedures, including surgery and care, adhered to approved protocols by the Animal Care and Use Committees at Shanghai Jiao Tong University School of Medicine (SYXK(Hu)2018-0027).
Reagents and plasmids
All chemicals were purchased from Sigma unless stated otherwise. Antibodies against p39 (1:1,000, 3275), p35 (1:1,000, 2680), Cdk5 (1:2,000, 2506) and actin (1:1,000, 4967) as well as horseradish peroxidase-conjugated goat anti-mouse and anti-rabbit antibodies were from Cell Signaling Technology. Additional primary antibodies included those against polyubiquitin (1:1,000, SC-8017, Santa Cruz) and Rpt1 (1:1000, Ab22678, Abcam). The human p39 plasmid was purchased from Miaoling Biology and Aβ monomer was from rPeptide.
Site-directed mutagenesis
Site-directed mutagenesis was performed with oligonucleotide primers designed to substitute the corresponding cysteine residue(s) with alanine residue(s) using overlap PCR. All mutations were verified by plasmid DNA sequencing.
Cell culture and plasmid transfection
HEK293T cells were cultured in DMEM (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum plus antibiotics. Cells were transfected with expression constructs using Lipofectamine3000 reagent (Invitrogen). Primary cortical/hippocampal neurons were prepared from embryonic day (E) 18 mouse embryos, seeded on cultured plates coated with poly-L-lysine (50 µg/ml) and maintained in Neurobasal medium supplemented with 2% B27 and 0.5 mM glutamine (Invitrogen). Cultured mouse hippocampal neurons were used for morphological observations and electrophysiological recordings. The morphometric analysis of dendrite complexity was performed using the ImageJ software (National Institutes of Health). For dendritic spine density quantification, three dendrite segments of secondary apical dendrites from each neuron were analyzed with Metamorph software. Meanwhile, cultured mouse cortical neurons were used for biochemical analyses. For drug treatment, 100 µM GSH/GSNO, 100 µM SNOC, 10 µM MG132, 10 µg/ml CHX, 25 µM NMDA and 250 nM Aβ oligomers, was added to the cultured neurons unless stated otherwise.
Biotin switch assay
The biotin switch assay was performed to detect protein S-nitrosylation13,16. Briefly, embryonic or postnatal mouse brains or cultured cortical neurons or HEK293T cells overexpressing p39 or its cysteine mutants were lysed in HENT buffer (250 mM HEPES, 1 mM EDTA, 0.1 mM Neocuproine, and 1% Triton X-100, pH = 7.7). The cell lysates were then incubated with 10 mM S-methyl methanethiosulfonate (MMTS) for 20 min at 50 °C to block free cysteine, and excess MMTS was removed by three passes through a G25 Sephadex spin column. The samples were then incubated with 5 mM ascorbate to specifically reduce S-nitrosylated cysteine to free cysteine and 0.4 mM N-[6-(biotinamido)hexyl]-30-(20-pyridyldithio) propionamide (biotin-HPDP) at room temperature with rotation for 1 h to label the reduced cysteines. Unreacted biotin-HPDP was removed by acetone precipitation, and the biotinylated samples were then re-suspended and incubated with 40 µl NeutrAvidin agarose for 1 h. The pellets were washed five times with neutralization buffer with 0.6 M NaCl, eluted by 2x SDS sample buffer and subjected to western blot analysis.
Isolation of proteasomes by differential centrifugation
Cultured neurons were washed with DPBS and then collected in ice-cold DPBS. Cell pellets were collected by centrifugation at 350 × g for 5 min and then suspended in lysis buffer and lysed by sonication. The supernatant was first centrifuged at 10,000 × g for 10 min, then at 100,000 × g for 30 min and finally at 150,000 × g for 1 h. The final pellet was proteasome fraction. Then rinse the proteasome pellet with wash buffer, and resuspend the proteasome pellet by sonication. All procedures were conducted on ice or at 4 °C.
Aβ oligomer preparation
Aβ oligomer was prepared as described before24,54. Briefly, 0.5 mg monomeric Aβ1–42 was dissolved in 22.2 μL DMSO and then diluted in ice-cold phenol-red free F-12 medium to yield a final Aβ concentration of 100 μM. The Aβ solution was incubated at 4 °C for 24 h and then centrifuged at 14,000 × g for 10 min. The supernatant was frozen in liquid nitrogen and stored at −20 °C for up to 1 month.
Stereotaxic surgery and electrophysiology
GFP, p39-WT-T2A-GFP and p39-C265A-T2A-GFP were sub-cloned into Sindbis viral vector pSinREP5 and viral particles were produced following previous studies28. Sindbis viral particles were delivered into the mouse hippocampal CA1 region (AP: −2.00 mm, ML: ±1.70 mm, DV: −1.4 mm, relative to the bregma) for 24 h using a Quintessential Stereotaxic Injector (Stoelting Company) with an injection volume of 0.5 μl at 0.1 μl min−1 through a customized ∼100-μm borosilicate glass microcapillary tip. After surgery, the animals were placed under a heating lamp until reawakening. Acute hippocampal slices were prepared from male APP/PS1 mice and their WT littermates. Animals were deeply anesthetized by isoflurane and decapitated. The brain containing the hippocampus was quickly removed and placed into cold (0–4 °C) oxygenated physiological solution containing (in mM): 82.75 NaCl, 2.4 KCl, 6.8 MgCl2, 0.5 CaCl2, 1.4 NaH2PO4, 23.8 NaHCO3, 23.7 dextrose, and 65 sucrose. Coronal slices were cut at 300 μm thickness using a Vibratome (VT1200S). These slices were kept at 31.0 ± 0.5 °C in oxygenated ACSF containing (in mM): 125 NaCl, 2.5 KCl, 1.25 NaH2PO4, 25 NaHCO3, 1 MgCl2, 12.5 dextrose, and 2 CaCl2 for 1–2 h before recordings. Whole-cell recordings were obtained from CA1 pyramidal neurons under visual guidance using transmitted light illumination. The patch recording pipettes (4 − 7 MΩ) were filled with intracellular solution containing 115 mM cesium methanesulfonate, 20 mM CsCl, 10 mM HEPES, 2.5 mM MgCl2, 4 mM Na2ATP, 0.4 mM Na3GTP, 10 mM sodium phosphocreatine, 0.6 mM EGTA, and 0.1 mM spermine (pH 7.25) for voltage-clamp recordings. To record mEPSCs, 50 μM pictrotoxin and 0.5 μM TTX were added to the ACSF solution gassed with 5% CO2/95% O2. To ensure the detection of the smallest synaptic events, mEPSCs were analyzed using only the high-quality patch-clamp recordings with the low baseline noise (≤2 pA) obtained by gently breaking-in high-resistance seal (>10 GΩ) patches.
Statistics and reproducibility
All data are presented as mean ± SEM from at least three independent experiments. The significance of differences was determined by unpaired Student’s t test or one-way analysis of variance (ANOVA) with the Student– Newman–Keuls test. The level of significance was set at p < 0.05. No samples or animals were excluded. All samples from each group were analyzed to confirm a normal distribution and equal variance.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data supporting the findings of this study are available within the paper, its Supplementary Information and Supplementary Data 1.
References
Tzioras, M., McGeachan, R. I., Durrant, C. S. & Spires-Jones, T. L. Synaptic degeneration in Alzheimer disease. Nat. Rev. Neurol. 19, 19–38 (2023).
Yang, X. et al. Synaptic mechanism in Alzheimer’s disease: A selective degeneration of an excitatory synaptic pathway in the CA1 hippocampus that controls spatial learning and memory in Alzheimer’s disease. Mol. Psychiatr. 23, 167–167 (2018).
Lista, S. & Hampel, H. Synaptic degeneration and neurogranin in the pathophysiology of Alzheimer’s disease. Expert Rev. Neurother. 17, 47–57 (2017).
Dhavan, R. & Tsai, L. H. A decade of CDK5. Nat. Rev. Mol. Cell Bio 2, 749–759 (2001).
Su, S. C. & Tsai, L. H. Cyclin-dependent kinases in brain development and disease. Annu Rev. Cell Dev. Bi 27, 465–491 (2011).
Pao, P. C. & Tsai, L. H. Three decades of Cdk5. J. Biomed. Sci. 28, 79 (2021).
Cheung, Z. H. & Ip, N. Y. Cdk5: a multifaceted kinase in neurodegenerative diseases. Trends Cell Biol. 22, 169–175 (2012).
Lee, M. S. et al. Neurotoxicity induces cleavage of p35 to p25 by calpain. Nature 405, 360–364 (2000).
Patzke, H. & Tsai, L. H. Calpain-mediated cleavage of the cyclin-dependent kinase-5 activator p39 to p29. J. Biol. Chem. 277, 8054–8060 (2002).
Wu, D. C. et al. The expression of Cdk5, p35, p39, and Cdk5 kinase activity in developing, adult, and aged rat brains. Neurochem Res 25, 923–929 (2000).
Ouyang, L. et al. p39-associated Cdk5 activity regulates dendritic morphogenesis. Sci Rep-Uk 10, 18746 (2020).
Li, W. Q. et al. p39 is responsible for increasing Cdk5 activity during postnatal neuron differentiation and governs neuronal network formation and epileptic responses. J. Neurosci. 36, 11283–11294 (2016).
Zhang, P., Fu, W. Y., Fu, A. K. Y. & Ip, N. Y. S-nitrosylation-dependent proteasomal degradation restrains Cdk5 activity to regulate hippocampal synaptic strength. Nat. Commun. 6, 8665 (2015).
Sen, N. et al. GOSPEL: A neuroprotective protein that binds to GAPDH upon S-nitrosylation. Neuron 63, 81–91 (2009).
Marozkina, N. V. & Gaston, B. S-nitrosylation signaling regulates cellular protein interactions. Bba-Gen. Subj. 1820, 722–729 (2012).
Zhang, P. et al. S-nitrosylation of cyclin-dependent kinase 5 (Cdk5) regulates its kinase activity and dendrite growth during neuronal development. J. Neurosci. 30, 14366–14370 (2010).
Kim, S., Wing, S. S. & Ponka, P. S-nitrosylation of IRP2 regulates its stability via the ubiquitin-proteasome pathway. Mol. Cell Biol. 24, 330–337 (2004).
Qu, J. et al. S-nitrosylation activates Cdk5 and contributes to synaptic spine loss induced by β-amyloid peptide. Proc. Natl Acad. Sci. USA 108, 14330–14335 (2011).
Cho, D. H. et al. S-nitrosylation of Drp1 mediates β-amyloid-related mitochondrial fission and neuronal injury. Science 324, 102–105 (2009).
Jaffrey, S. R., Erdjument-Bromage, H., Ferris, C. D., Tempst, P. & Snyder, S. H. Protein S-nitrosylation: A physiological signal for neuronal nitric oxide. Nat. Cell Biol. 3, 193–197 (2001).
Nakamura, T. et al. Aberrant protein S-nitrosylation in neurodegenerative diseases. Neuron 78, 596–614 (2013).
Stykel, M. G. & Ryan, S. D. Nitrosative stress in Parkinson’s disease. Npj Parkinsons Dis. 8, 104 (2022).
Wijasa, T. S. et al. Quantitative proteomics of synaptosome S-nitrosylation in Alzheimer’s disease. J. Neurochem 152, 710–726 (2020).
Fu, A. K. Y. et al. Blockade of EphA4 signaling ameliorates hippocampal synaptic dysfunctions in mouse models of Alzheimer’s disease. P Natl Acad. Sci. USA 111, 9959–9964 (2014).
Lau, S. F. et al. The VCAM1-ApoE pathway directs microglial chemotaxis and alleviates Alzheimer’s disease pathology. Nat. Aging 3, 1219 (2023).
Knopman, D. S. et al. Alzheimer disease. Nat. Rev. Dis. Primers 7, 33 (2021).
Graff-Radford, J. et al. New insights into atypical Alzheimer’s disease in the era of biomarkers. Lancet Neurol. 20, 222–234 (2021).
Zhang, K., Han, Y. F., Zhang, P., Zheng, Y. Q. & Cheng, A. B. Comparison of fluorescence biosensors and whole-cell patch clamp recording in detecting ACh, NE, and 5-HT. Front. Cell. Neurosci. 17, 1166480 (2023).
Nakamura, T. & Lipton, S. A. Protein S-nitrosylation as a therapeutic target for neurodegenerative diseases. Trends Pharm. Sci. 37, 73–84 (2016).
Seneviratne, U. et al. -nitrosation of proteins relevant to Alzheimer’s disease during early stages of neurodegeneration. Proc. Natl Acad. Sci. USA 113, 4152–4157 (2016).
Zhao, Q. F., Yu, J. T. & Tan, L. S-nitrosylation in Alzheimer’s disease. Mol. Neurobiol. 51, 268–280 (2015).
Dyer, R. R., Ford, K. I. & Robinson, R. A. S. The roles of S-nitrosylation and S-glutathionylation in Alzheimer’s disease. Method Enzymol. 626, 499–538 (2019).
Ramesh, M., Gopinath, P. & Govindaraju, T. Role of post-translational modifications in Alzheimer’s disease. Chembiochem 21, 1052–1079 (2020).
Humbert, S., Manier, L. M. & Tsai, L. H. Synaptic localization of p39, a neuronal activator of cdk5. Neuroreport 11, 2213–2216 (2000).
Sattler, R. et al. Specific coupling of NMDA receptor activation to nitric oxide neurotoxicity by PSD-95 protein. Science 284, 1845–1848 (1999).
Kasai, H., Ziv, N. E., Okazaki, H., Yagishita, S. & Toyoizumi, T. Spine dynamics in the brain, mental disorders and artificial neural networks. Nat. Rev. Neurosci. 22, 407–422 (2021).
Lai, K. O. et al. TrkB phosphorylation by Cdk5 is required for activity-dependent structural plasticity and spatial memory. Nat. Neurosci. 15, 1506–1515 (2012).
Norrholm, S. D. et al. Cocaine-induced proliferation of dendritic spines in nucleus accumbens is dependent on the activity of cyclin-dependent kinase-5. Neuroscience 116, 19–22 (2003).
Fu, W. Y. et al. Cdk5 regulates EphA4-mediated dendritic spine retraction through an ephexin1-dependent mechanism. Nat. Neurosci. 10, 67–76 (2007).
Kim, Y. et al. Phosphorylation of WAVE1 regulates actin polymerization and dendritic spine morphology. Nature 442, 814–817 (2006).
Liang, Z. Y. et al. The pseudokinase CaMKv is required for the activity-dependent maintenance of dendritic spines. Nat. Commun. 7, 13282 (2016).
Huang, H. Q. et al. Cdk5-dependent phosphorylation of liprinα1 mediates neuronal activity-dependent synapse development. P Natl Acad. Sci. USA 114, E6992–E7001 (2017).
Montero-Calle, A. et al. Proteomics analysis of prefrontal cortex of Alzheimer’s disease patients revealed dysregulated proteins in the disease and novel proteins associated with amyloid-β pathology. Cell Mol. Life Sci. 80, 141 (2023).
Fukasawa, J. T. et al. CDK5 and MAPT gene expression in Alzheimer’s disease brain samples. Curr. Alzheimer Res 15, 182–186 (2018).
Nikhil, K., Viccaro, K. & Shah, K. Multifaceted regulation of ALDH1A1 by Cdk5 in Alzheimer’s disease pathogenesis. Mol. Neurobiol. 56, 1366–1390 (2019).
Ryder, J. et al. Divergent roles of GSK3 and CDK5 in APP processing. Biochem. Biophys. Res. Co. 312, 922–929 (2003).
Wang, Y. L. et al. Metformin ameliorates synaptic defects in a mouse model of AD by inhibiting Cdk5 activity. Front. Cell. Neurosci. 14, 170 (2020).
Sheng, Y. H., Zhang, L., Su, S. C., Tsai, L. H. & Zhu, J. J. Cdk5 is a new rapid synaptic homeostasis regulator capable of initiating the early Alzheimer-like pathology. Cereb. Cortex 26, 2937–2951 (2016).
Shukla, V. et al. TFP5, a peptide inhibitor of aberrant and hyperactive Cdk5/p25, attenuates pathological phenotypes and restores synaptic function in CK-p25Tg Mice. J. Alzheimers Dis. 56, 335–349 (2017).
Valerio, A. et al. Gene expression profile activated by the chemokine CCL5/RANTES in human neuronal cells. J. Neurosci. Res. 78, 371–382 (2004).
Ajoy, R. et al. CCL5 promotion of bioenergy metabolism is crucial for hippocampal synapse complex and memory formation. Mol. Psychiatr. 26, 6451–6468 (2021).
Nakamura, T. et al. Noncanonical transnitrosylation network contributes to synapse loss in Alzheimer’s disease. Science 371, 253 (2021).
Asada, A. et al. Myristoylation of p39 and p35 is a determinant of cytoplasmic or nuclear localization of active cycline-dependent kinase 5 complexes. J. Neurochem 106, 1325–1336 (2008).
Shen, Y. et al. Stimulation of the hippocampal POMC/MC4R circuit alleviates synaptic plasticity impairment in an Alzheimer’s disease model. Cell Rep. 17, 1819–1831 (2016).
Acknowledgements
The research received support from various funding sources, including the Program of National Natural Science Foundation of China (82102184), the Shanghai high-level local university construction project (PT21011), Shanghai 2023 “Science and Technology Innovation Action Plan” Natural Science Foundation Project (23ZR1436800), NATCM’s Project of High-level Construction of Key TCM Disciplines (ZYYZDXK-2023070), Alzheimer’s Association Research Fellowship (AARF-19-619387), and the Junior Thousand Talents Program of China.
Author information
Authors and Affiliations
Contributions
P.Z., H.C. and A.C. conceived and supervised the project. P.Z., A.C., J.W. (Jingyi Wang), J.L., J.W. (Jie Wang) and M.X. performed the experiments. P.Z., H.C. and A.C. wrote the manuscript with input and help from other members from P.Z. and H.C. lab. All authors contributed to data interpretation and data analysis.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Abhishek Kumar Singh and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Ibrahim Javed and Benjamin Bessieres. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Cheng, A., Wang, J., Li, J. et al. S-Nitrosylation of p39 promotes its degradation and contributes to synaptic dysfunction induced by β-amyloid peptide. Commun Biol 7, 1113 (2024). https://doi.org/10.1038/s42003-024-06832-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-024-06832-z
- Springer Nature Limited