Abstract
Background
Little is known about the role of most asthma susceptibility genes during human lung development. Genetic determinants for normal lung development are not only important early in life, but also for later lung function.
Objective
To investigate the role of expression patterns of well-defined asthma susceptibility genes during human and murine lung development. We hypothesized that genes influencing normal airways development would be over-represented by genes associated with asthma.
Methods
Asthma genes were first identified via comprehensive search of the current literature. Next, we analyzed their expression patterns in the developing human lung during the pseudoglandular (gestational age, 7-16 weeks) and canalicular (17-26 weeks) stages of development, and in the complete developing lung time series of 3 mouse strains: A/J, SW, C57BL6.
Results
In total, 96 genes with association to asthma in at least two human populations were identified in the literature. Overall, there was no significant over-representation of the asthma genes among genes differentially expressed during lung development, although trends were seen in the human (Odds ratio, OR 1.22, confidence interval, CI 0.90-1.62) and C57BL6 mouse (OR 1.41, CI 0.92-2.11) data. However, differential expression of some asthma genes was consistent in both developing human and murine lung, e.g. NOD1, EDN1, CCL5, RORA and HLA-G. Among the asthma genes identified in genome wide association studies, ROBO1, RORA, HLA-DQB1, IL2RB and PDE10A were differentially expressed during human lung development.
Conclusions
Our data provide insight about the role of asthma susceptibility genes during lung development and suggest common mechanisms underlying lung morphogenesis and pathogenesis of respiratory diseases.
Similar content being viewed by others
Introduction
There is good evidence that genetic factors strongly influence the risk of asthma, and associations between numerous genes and asthma have been evaluated in the past decades [1, 2]. Recent genome wide association studies (GWAS) of asthma have identified several additional asthma susceptibility genes [3–10]. Little is known about the role of most asthma susceptibility genes during human lung development.
The "developmental origins" hypothesis [11] proposes that specific in utero events at critical periods during organogenesis and maturation result in long-term physiological or metabolic changes, ultimately contributing to disease in later life [12, 13]. Our group previously showed that Wnt signaling genes that were differentially expressed during fetal lung development were associated with impaired lung function in two cohorts of school-aged asthmatic children [14]. These results suggest the importance of early life events in determining lung function. They also highlight the benefit of integrating gene expression and genetic association data to connect transcriptomic events in the early developing lung to genetic associations of lung function in later life.
Asthma is a disease characterized by both airway inflammation and smooth muscle contraction, leading to airway obstruction. Dendritic cells, mast cells, and T-lymphocytes, as well as airway smooth muscle cells, all begin to appear within the lung parenchyma during the pseudoglandular stage of lung development. We therefore hypothesized that genes influencing normal airways development, especially during the branching morphogenesis stage of human lung development, would be over-represented by genes associated with asthma. To test this hypothesis, we investigated the role of a well-defined set of asthma susceptibility genes during human and murine lung development. 96 asthma genes were first identified via comprehensive search of the current literature. Next, we analyzed their expression patterns in the developing human lung during the pseudoglandular (gestational age, 7-16 weeks) and canalicular (17-27 weeks) stages of development, and in the complete developing lung time series of 3 mouse strains: A/J, SW and C57BL6.
We show that overall, there was no over-representation of the asthma genes among genes differentially expressed during lung development, which may reflect the diverse ontological contexts of the asthma genes. However, some genes showed a consistent pattern of differential expression in all developing lung data sets, e.g. NOD1, EDN1, RORA, CCL5 and HLA-G, which suggests that these genes play a fundamental role in normal lung development.
Methods
Tissue samples
The human fetal lung tissues were obtained from National Institute of Child Health and Human Development supported tissue databases and microarray profiled as previously described [14, 15]. Creation of the tissue repository was approved by the University of Missouri-Kansas City Pediatric Institutional Review Board. 38 RNA samples from 38 subjects (estimated gestational age 7-22 weeks or 53-154 days post conception) were included in the analysis (Table 1). The murine data have previously been described and their microarray data are available at NCBI Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo); A/J [16], n = 24 samples; SW [17], n = 11; and C57BL6 mice [18], n = 5, Table 1.
Microarray analysis
The developing human lung time series data is available at NCBI Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo), GSE14334 (Affymetrix Human Genome GeneChip U133 Plus 2.0 microarray platform). Expression values were extracted and normalized from .CEL files using the Affy package and the Robust Multi-array Average (RMA) method in R/BioConductor (http://www.bioconductor.org) which returns the measured expression signal of each micrroarray gene probe in logarithmic base 2 scale. Validation of the human microarray analysis by qPCR for genes differentially expressed during lung development has been performed earlier and this demonstrated that 83% of individual gene expression trajectories could be replicated [15]. The developing whole mouse lung transcriptome data from three different mouse strains were extracted and normalized, separately, using RMA in R/BioConductor; 24 samples from A/J (Affymetrix Mu74Av2 platform); 11 samples from SW (Affymetrix Mu11K A and B platforms); and 5 samples from C57BL6 (Affymetrix Mouse 430 Plus 2.0 platform).
Literature search
A PubMed (http://www.ncbi.nlm.nih.gov/pubmed) search was performed on March 8, 2010 using the terms 1) "asthma" together with 2) "genetic association" or "case control" in order to cover all published papers between July 1, 2008 and December 31, 2009. We applied the following inclusion criteria for an asthma gene: 1) significant association with asthma affection status in at least two populations and 2) at least one significant association study with no fewer than 150 cases and 150 controls or 150 trios. Genes identified through three earlier literature searches based on papers published before July 1, 2008 were also included if they met our two predefined criteria [1, 19, 20]. In addition, all GWAS of asthma published through September 2010, were also evaluated and asthma genes were included if our criteria were met. Please see Supplemental data for details about the asthma genes included in our analyses. Mouse orthologues of human genes were identified using NCBI's HomoloGene database (http://www.ncbi.nlm.nih.gov/homologene).
Statistical analysis
Differential gene expression analysis relative to gestational age was performed using a linear regression model (lmFit) as implemented in the Limma package in R/BioConductor. Each microarray gene probe's logarithmic base 2 expression signal was regressed against the gestational age as a continuous variable representing days of the developing lung. We adjusted for multiple testing using the Benjamini and Hochberg method, which controls the false discovery rate (i.e. the expected proportion of false discoveries amongst the rejected hypotheses), and the adjusted p-values were used to declare a significant gene expression pattern over age [21]. "Differentially expressed" refers to an adjusted p-value of <0.05 in the linear regression model. Fisher's exact test was next performed in Stata Statistical Software (Collage Station, Tx) to test whether microrarray probes representing predefined asthma genes were over-represented among differentially expressed probes relative to probes representing "non-asthma genes". This analysis was restricted to microarray probes that were gene annotated because the asthma gene probes were all annotated. The same analysis steps were performed in human and murine data sets. Gene ontology (GO) enrichment analysis was performed using DAVID (The Database for Annotation, Visualization and Integrated Discovery) [22, 23].
Results
In total, 96 asthma susceptibility genes were identified in the literature (Additional file 1, Table E1 [1, 3–10, 24–96]). All genes show significant association with asthma in at least two human populations, one of which has no fewer than 150 cases and 150 controls or 150 trios. The 96 genes were represented by 220 probes on the human microarray (Table 1). Not all human genes have a mouse orthologue and the mouse microarray data sets have slightly lower numbers of asthma genes and their corresponding microarray probes.
We found that 28% of all microarray probes in the human data set were differentially expressed during the analyzed lung development period (human estimated gestational age 7-22 weeks), Table 2. A similar figure was seen in the A/J mouse and somewhat lower figures in the SW and C57BL6 mouse strains. Gene ontology (GO) enrichment analysis using DAVID of the human list of differentially expressed genes returned 879 significant GO terms, of which 6 terms pertain directly to the lung development. Among the asthma gene probes, 32% were differentially expressed during early human lung development. While there was a trend towards over-representation (Odds ratio, OR 1.22, CI 0.90-1.62) this was not statistically significant in comparison to the non-asthma gene probes (28%). In agreement with the human data, no over-representation of asthma gene probes was found among probes differentially expressed during lung development in mice strains, although there was a trend in the C57BL6 strain (OR 1.41, CI 0.92-2.11), Table 2.
Although asthma genes as a group was not differentially expressed more than non-asthma genes during early lung development, some genes were consistently differential expressed, as listed in Table 3 (see full list in Additional file 1, Table E2). Expression of NOD1, EDN1 and IL4R were positively correlated with gestational age in the human data, whereas ROBO1 and PLAUR were negatively correlated (i.e. lower expression levels the higher gestational age). Among the asthma genes identified in GWAS, ROBO1, RORA, HLA-DQB1, IL2RB and PDE10A showed most significant evidence of involvement in lung development (all adjusted p < 0.001 for differential expression). Analyses were also done comparing gene expression patterns between the pseudoglandular (primary branching morphogenesis stage) and canalicular stages (with 112 days post conception as the dividing time point between the 2 stages). The list of top genes differentially expressed between these two stages (Additional file 1, Table E3 and Figure E1) corresponds well with the list of top genes using time as a continuous variable (Table 3).
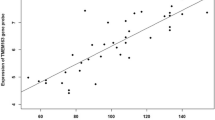
Next, we evaluated all differentially expressed asthma genes in the human data set to see which genes showed a consistent expression pattern across human and murine data sets. Table 4 shows all genes with at least one significant probe per gene in the human data and at least one significant probe in a mouse data set (n = 19 with adjusted p-value <0.05). Eight genes had one or more significant probes in all data sets, with NOD1, EDN1, CCL5, RORA and HLA-G showing the most consistent expression patterns across human and mouse (see detailed EDN1/Edn1 expression over time in human and mouse lung tissue; Figure 1 and 2). In terms of bio-ontologic enrichment, the 19 asthma genes consistently differentially expressed in human and mouse lung development were enriched for ontological attributes "Regulation of cytokine production" (IRAK3, CD86, NOD1, TNF, IL18, SCGB1A1) and "Regulation of cell activation" (STAT6, CD86, IL18, IL4R, RORA, SCGB1A1) (Additional file 1, Table E4.) In terms of gene product characteristics, "Disulfide bond", "Secreted" and "Signal peptide" are attributes of a majority of the genes. 15 of the 19 genes in Table 4 have been extensively studied in human and murine experiments that support their involvement in asthma pathogenesis (Additional file 1, Table E5).
Expression of Edn1 over time in mouse whole lung tissue in relation to time (days post conception). Solid circles represent the A/J data (p = 0.007 for differential expression), open squares represent the C57BL6 data (p = 0.02) and solid triangles represent the SW data (p = 0.001). The fitted line through each data set represents the beta coefficient from linear regression analysis.
In order to disentangle pre- and postnatal expression patterns in the murine data sets, separate pre- and postnatal analyses were attempted. However, this subgroup analysis was not meaningful for the SW and C56BL6 data sets because of substantially reduced sample size. The A/J data contains two prenatal time points (day 11 and 17), each with 4 unique samples and Table E6 shows overlapping results for human and prenatal A/J data. Eight of the previously identified 19 genes with consistent expression pattern across human and murine data sets (Table 4) were also identified when prenatal A/J data was used (including Edn1).
Discussion
Little is known about the role of most asthma susceptibility genes during human lung development. Here we present a thorough evaluation of gene expression patterns of current published asthma genes in the developing human and murine lung. While there was no general over-representation of asthma genes among differentially expressed genes, some asthma genes were consistently differentially expressed in multiple developing lung transcriptomes, e.g. NOD1, EDN1, CCL5, RORA and HLA-G suggesting key functional roles in lung development.
Determinants for a normal lung development are critical not only early in life, but also for later lung function. Longitudinal studies have shown that infants with reduced lung function have an increased risk of developing asthma and respiratory illness later in life [97, 98]. Shared genetic factors for reduced lung function in children with asthma and adults who smoke (e.g. MMP12 variants) emphasize the role of genetics on long term lung function [99]. Wnt signaling genes (e.g. Wif1, Wisp1) were not identified as asthma genes in our literature search, and were thus not included in our analyses. In our previous article by Sharma et al, Wif1 and Wisp1 were differentially expressed during fetal lung development and polymorphisms in these genes also showed association with lung function measured as FEV1 and FVC, but association to asthma per se was not tested [14].
The transcriptional control of lung morphogenesis is key for normal development from primordium to a fully differentiated, functioning organ [100, 101]. Human lung growth has historically been categorised into five stages based on histological and anatomical characteristics: embryonic (26 days to 5 weeks), pseudoglandular (5-16 weeks), canalicular (16-26 weeks), saccular (26 weeks to birth), and alveolar (birth to 6 months) [100]. Additional "molecular" phases within the pseudoglandular stage have been observed, which extends our knowledge of lung development beyond traditional embryology [15].
GWAS have contributed to important knowledge about underlying functional genetics in many complex diseases [102]. The majority of trait associated SNPs show weak to moderate effect sizes, which supports earlier evidence that complex diseases result from several genetic and, often, environmental factors. Evidence of a functional role is also lacking for most identified genes. In order to increase our understanding of the mechanism and potential function of asthma susceptibility genes identified in published GWAS and "classic" asthma candidate genes, we evaluated their gene expression patterns in the developing human lung. Comparative analyses also showed that many of the differentially expressed genes in the human data set were also differentially expressed during murine lung development. Among the GWAS asthma genes, ROBO1, RORA, HLA-DQB1, IL2RB and PDE10A were differentially expressed in the human data. These genes represent a wide range of structural and ontological families with different assumed functions, but their potential involvement in lung development has previously not been thoroughly evaluated. Regulation of cytokine production and cell activation were the most significant bio-ontologic attributes to genes differentialy expressed during lung development.
Using the murine data sets for comparative analyses, RORA, which encodes for a nuclear hormone receptor, showed the most consistent expression pattern (expression positively correlated with gestational age in all data sets). ROBO1 expression was on the other hand negatively correlated with gestational age in all tested data sets (albeit significant in only 2/3 sets), which indicates an important effect early in the developing lung and then a diminishing effect over time. The ROBO1 protein is involved in axon guidance and neuronal precursor cell migration. PTGDR, WDR36, PRNP, DENND1B, PDE4D, TLE4 and TSLP also showed weak evidence of differential expression in the human data using adjusted p < 0.05 as cut off (Additional file 1, Table E2), but none showed consistent gene expression patterns in the murine data sets.
NOD1 showed the strongest evidence for differential expression in the human data and this pattern was consistent in the C57BL6 strain. However, Nod1 was not represented on the platforms used for analyses on the A/J and SW strains and could thus not be evaluated in these data sets (also true for another asthma gene with consistent expression patterns, PCDH1 [52]). NOD1 encodes for a cytosolic protein which contains an N-terminal caspase recruitment domain (CARD) and plays an important role for recognition of bacterial compounds and initiation of the innate immune response [103]. Little is known about the role of NOD1 during lung development and our findings indicate that NOD1 could have important contribution.
EDN1 was the second most differentially expressed asthma gene in the human data set and very consistent expression patterns were found in all murine data sets. Also for the embryonic stage analyses (pseudoglandular vs canalicular), EDN1 was among the most highly differentially expressed genes. In general, embryonic stage results were very similar to the results using time as a continuous variable. EDN1 belong to a family of secreted peptides produced by vascular endothelial cells with multiple effects on cardiovascular, neural, pulmonary and renal physiology [104, 105]. EDN1 shows involvement in pulmonary hypertension, fibrosis, obstructive diseases and acute lung injury, and is also required for the normal development of several tissues. Mice lacking the Edn1 gene die of respiratory failure at birth and show severe craniofacial abnormalities, as well as cardiovascular defects [106, 107]. Transgenic mice with lung-specific over-expression of the human EDN1 gene develop, on the other hand, chronic lung inflammation and fibrosis [108]. Edn1 heterozygous knockout mice also show increased bronchial responsiveness and these result link EDN1 functionally to asthma and obstructive diseases [72]. To date, three studies report significant association between EDN1 and asthma [41, 109, 110]. Our data, as well as previous studies, point to an important role for EDN1 in normal lung development, which warrants further studies.
Our study has several limitations. Our 38 human lung tissue samples were restricted to the pseudoglandular and canalicular stages. Information about key exposures that could influence gene expression patterns, such as maternal smoking, residential area, and parental allergy is not available. Thirty-eight samples are a relatively small sample size for expression analyses due to human biological variation and fetal lung tissue during the later stages of gestation was not available. It is possible that some asthma genes are important for human lung development during the later stages of gestation, but we were not able to evaluate this with our current data set. To complement the human data, we analysed expression patterns from early gestational to postnatal stages of lung development in three different murine strains. We used this murine data to replicate, in silico, the human results in the early stages and to infer human gene expression pattern in the later stages of the developing lung. Also, the microarray platforms used in the included data sets do not entirely cover the human (and murine) transcriptome and important genes may have been missed (e.g. GPRA/NPSR1 [111] is not represented on the U133 Plus 2.0 microarray chip and could not be evaluated). Protein analyses could provide a better view to understand specific gene functions and the post-transcriptional regulation level, but such data was not available in our study. Our asthma gene list represents genes that met our predefined criteria for asthma association, and some genes genes may have been missed (e.g. those only captured by the search terms "family based study" AND "asthma"). Given the rapid rate at which novel asthma susceptibility loci are being discovered, some of the most recent asthma genes may have been missed. These may introduce a potential null bias in the analysis.
Conclusions
We have evaluated gene expression patterns of asthma susceptibility genes identified via a comprehensive literature search of candidate gene studies and GWAS published to date. We found strong and consistent evidence of differential expression of several asthma genes in the developing human and murine lung. Among genes identified in asthma GWAS, ROBO1, RORA, HLA-DQB1, IL2RB and PDE10A showed most consistent expression patterns and from asthma candidate genes, e.g. NOD1, EDN1, CCL5 and HLA-G were identified. Our analyses provide functional insight about asthma susceptibility genes during normal lung development, which improves our understanding about normal and pathological processes related to respiratory diseases in children and adults.
Abbreviations
- CI:
-
Confidence interval
- DAVID:
-
Database for Annotation, Visualization and Integrated Discovery
- GEO:
-
Gene Expression Omnibus
- GO:
-
Gene ontology
- GWAS:
-
Genome wide association studies
- NCBI:
-
National Center for Biotechnology Information
- OR:
-
Odds ratio
- qPCR:
-
Quantitative real time polymerase chain reaction.
References
Ober C, Hoffjan S: Asthma genetics 2006: the long and winding road to gene discovery. Genes Immun. 2006, 7 (2): 95-100. 10.1038/sj.gene.6364284.
Rogers AJ, Raby BA, Lasky-Su JA, Murphy A, Lazarus R, Klanderman BJ, Sylvia JS, Ziniti JP, Lange C, Celedon JC, Silverman EK, Weiss ST: Assessing the reproducibility of asthma candidate gene associations, using genome-wide data. Am J Respir Crit Care Med. 2009, 179 (12): 1084-1090. 10.1164/rccm.200812-1860OC.
Gudbjartsson DF, Bjornsdottir US, Halapi E, Helgadottir A, Sulem P, Jonsdottir GM, Thorleifsson G, Helgadottir H, Steinthorsdottir V, Stefansson H, Williams C, Hui J, Beilby J, Warrington NM, James A, Palmer LJ, Koppelman GH, Heinzmann A, Krueger M, Boezen HM, Wheatley A, Altmuller J, Shin HD, Uh ST, Cheong HS, Jonsdottir B, Gislason D, Park CS, Rasmussen LM, Porsbjerg C, et al: Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat Genet. 2009, 41 (3): 342-347. 10.1038/ng.323.
Hancock DB, Romieu I, Shi M, Sienra-Monge JJ, Wu H, Chiu GY, Li H, del Rio-Navarro BE, Willis-Owens SA, Weiss ST, Raby BA, Gao H, Eng C, Chapela R, Burchard EG, Tang H, Sullivan PF, London SJ: Genome-wide association study implicates chromosome 9q21.31 as a susceptibility locus for asthma in mexican children. PLoS Genet. 2009, 5 (8): e1000623-10.1371/journal.pgen.1000623.
Himes BE, Hunninghake GM, Baurley JW, Rafaels NM, Sleiman P, Strachan DP, Wilk JB, Willis-Owen SA, Klanderman B, Lasky-Su J, Lazarus R, Murphy AJ, Soto-Quiros ME, Avila L, Beaty T, Mathias RA, Ruczinski I, Barnes KC, Celedon JC, Cookson WO, Gauderman WJ, Gilliland FD, Hakonarson H, Lange C, Moffatt MF, O'Connor GT, Raby BA, Silverman EK, Weiss ST: Genome-wide association analysis identifies PDE4D as an asthma-susceptibility gene. Am J Hum Genet. 2009, 84 (5): 581-593. 10.1016/j.ajhg.2009.04.006.
Li X, Howard TD, Zheng SL, Haselkorn T, Peters SP, Meyers DA, Bleecker ER: Genome-wide association study of asthma identifies RAD50-IL13 and HLA-DR/DQ regions. J Allergy Clin Immunol. 2010, 125 (2): 328-335. 10.1016/j.jaci.2009.11.018. e311
Mathias RA, Grant AV, Rafaels N, Hand T, Gao L, Vergara C, Tsai YJ, Yang M, Campbell M, Foster C, Gao P, Togias A, Hansel NN, Diette G, Adkinson NF, Liu MC, Faruque M, Dunston GM, Watson HR, Bracken MB, Hoh J, Maul P, Maul T, Jedlicka AE, Murray T, Hetmanski JB, Ashworth R, Ongaco CM, Hetrick KN, Doheny KF, et al: A genome-wide association study on African-ancestry populations for asthma. J Allergy Clin Immunol. 2009
Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, Heath S, von Mutius E, Farrall M, Lathrop M, Cookson WO, consortium G: A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010, 363 (13): 1211-1221. 10.1056/NEJMoa0906312.
Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, Depner M, von Berg A, Bufe A, Rietschel E, Heinzmann A, Simma B, Frischer T, Willis-Owen SA, Wong KC, Illig T, Vogelberg C, Weiland SK, von Mutius E, Abecasis GR, Farrall M, Gut IG, Lathrop GM, Cookson WO: Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007, 448 (7152): 470-473. 10.1038/nature06014.
Sleiman PM, Flory J, Imielinski M, Bradfield JP, Annaiah K, Willis-Owen SA, Wang K, Rafaels NM, Michel S, Bonnelykke K, Zhang H, Kim CE, Frackelton EC, Glessner JT, Hou C, Otieno FG, Santa E, Thomas K, Smith RM, Glaberson WR, Garris M, Chiavacci RM, Beaty TH, Ruczinski I, Orange JM, Allen J, Spergel JM, Grundmeier R, Mathias RA, Christie JD, et al: Variants of DENND1B associated with asthma in children. N Engl J Med. 2010, 362 (1): 36-44. 10.1056/NEJMoa0901867.
Barker DJ, Martyn CN: The maternal and fetal origins of cardiovascular disease. J Epidemiol Community Health. 1992, 46 (1): 8-11. 10.1136/jech.46.1.8.
Barker DJ: In utero programming of chronic disease. Clin Sci (Lond). 1998, 95 (2): 115-128. 10.1042/CS19980019.
Stick S: Pediatric origins of adult lung disease. 1. The contribution of airway development to paediatric and adult lung disease. Thorax. 2000, 55 (7): 587-594. 10.1136/thorax.55.7.587.
Sharma S, Tantisira K, Carey V, Murphy AJ, Lasky-Su J, Celedon JC, Lazarus R, Klanderman B, Rogers A, Soto-Quiros M, Avila L, Mariani T, Gaedigk R, Leeder S, Torday J, Warburton D, Raby B, Weiss ST: A Role for WNT-Signaling Genes in the Pathogenesis of Impaired Lung Function in Asthma. Am J Respir Crit Care Med. 2009
Kho AT, Bhattacharya S, Tantisira KG, Carey VJ, Gaedigk R, Leeder JS, Kohane IS, Weiss ST, Mariani TJ: Transcriptomic analysis of human lung development. Am J Respir Crit Care Med. 2009, 181 (1): 54-63.
Bonner AE, Lemon WJ, You M: Gene expression signatures identify novel regulatory pathways during murine lung development: implications for lung tumorigenesis. J Med Genet. 2003, 40 (6): 408-417. 10.1136/jmg.40.6.408.
Mariani TJ, Reed JJ, Shapiro SD: Expression profiling of the developing mouse lung: insights into the establishment of the extracellular matrix. Am J Respir Cell Mol Biol. 2002, 26 (5): 541-548.
Naxerova K, Bult CJ, Peaston A, Fancher K, Knowles BB, Kasif S, Kohane IS: Analysis of gene expression in a developmental context emphasizes distinct biological leitmotifs in human cancers. Genome Biol. 2008, 9 (7): R108-10.1186/gb-2008-9-7-r108.
Weiss ST, Raby BA, Rogers A: Asthma genetics and genomics 2009. Curr Opin Genet Dev. 2009, 19 (3): 279-282. 10.1016/j.gde.2009.05.001.
Vercelli D: Discovering susceptibility genes for asthma and allergy. Nat Rev Immunol. 2008, 8 (3): 169-182. 10.1038/nri2257.
Benjamini Y, Hochberg Y: Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995, 57: 289-300.
Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA: DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003, 4 (5): P3-10.1186/gb-2003-4-5-p3.
Huang da W, Sherman BT, Lempicki RA: Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009, 4 (1): 44-57.
Bailey MT, Kierstein S, Sharma S, Spaits M, Kinsey SG, Tliba O, Amrani Y, Sheridan JF, Panettieri RA, Haczku A: Social stress enhances allergen-induced airway inflammation in mice and inhibits corticosteroid responsiveness of cytokine production. J Immunol. 2009, 182 (12): 7888-7896. 10.4049/jimmunol.0800891.
Barton SJ, Koppelman GH, Vonk JM, Browning CA, Nolte IM, Stewart CE, Bainbridge S, Mutch S, Rose-Zerilli MJ, Postma DS, Maniatis N, Henry AP, Hall IP, Holgate ST, Tighe P, Holloway JW, Sayers I: PLAUR polymorphisms are associated with asthma, PLAUR levels, and lung function decline. J Allergy Clin Immunol. 2009, 123 (6): 1391-1400. 10.1016/j.jaci.2009.03.014. e1317
Batra J, Pratap Singh T, Mabalirajan U, Sinha A, Prasad R, Ghosh B: Association of inducible nitric oxide synthase with asthma severity, total serum immunoglobulin E and blood eosinophil levels. Thorax. 2007, 62 (1): 16-22. 10.1136/thx.2006.057935.
Berlin AA, Lincoln P, Tomkinson A, Lukacs NW: Inhibition of stem cell factor reduces pulmonary cytokine levels during allergic airway responses. Clin Exp Immunol. 2004, 136 (1): 15-20. 10.1111/j.1365-2249.2004.02404.x.
Berry MA, Hargadon B, Shelley M, Parker D, Shaw DE, Green RH, Bradding P, Brightling CE, Wardlaw AJ, Pavord ID: Evidence of a role of tumor necrosis factor alpha in refractory asthma. N Engl J Med. 2006, 354 (7): 697-708. 10.1056/NEJMoa050580.
Bierbaum S, Nickel R, Koch A, Lau S, Deichmann KA, Wahn U, Superti-Furga A, Heinzmann A: Polymorphisms and haplotypes of acid mammalian chitinase are associated with bronchial asthma. Am J Respir Crit Care Med. 2005, 172 (12): 1505-1509. 10.1164/rccm.200506-890OC.
Bosse Y, Lemire M, Poon AH, Daley D, He JQ, Sandford A, White JH, James AL, Musk AW, Palmer LJ, Raby BA, Weiss ST, Kozyrskyj AL, Becker A, Hudson TJ, Laprise C: Asthma and genes encoding components of the vitamin D pathway. Respir Res. 2009, 10: 98-10.1186/1465-9921-10-98.
Buckova D, Izakovicova Holla L, Vacha J: Polymorphism 4G/5G in the plasminogen activator inhibitor-1 (PAI-1) gene is associated with IgE-mediated allergic diseases and asthma in the Czech population. Allergy. 2002, 57 (5): 446-448. 10.1034/j.1398-9995.2002.03582.x.
Cameron L, Depner M, Kormann M, Klopp N, Illig T, von Mutius E, Kabesch M: Genetic variation in CRTh2 influences development of allergic phenotypes. Allergy. 2009, 64 (10): 1478-1485. 10.1111/j.1398-9995.2009.02053.x.
Castro-Giner F, Kunzli N, Jacquemin B, Forsberg B, de Cid R, Sunyer J, Jarvis D, Briggs D, Vienneau D, Norback D, Gonzalez JR, Guerra S, Janson C, Anto JM, Wjst M, Heinrich J, Estivill X, Kogevinas M: Traffic-related air pollution, oxidative stress genes, and asthma (ECHRS). Environ Health Perspect. 2009, 117 (12): 1919-1924.
Chatila TA: Interleukin-4 receptor signaling pathways in asthma pathogenesis. Trends Mol Med. 2004, 10 (10): 493-499. 10.1016/j.molmed.2004.08.004.
Chatterjee R, Batra J, Das S, Sharma SK, Ghosh B: Genetic association of acidic mammalian chitinase with atopic asthma and serum total IgE levels. J Allergy Clin Immunol. 2008, 122 (1): 202-208. 10.1016/j.jaci.2008.04.030. 208 e201-207
Chelbi H, Ghadiri A, Lacheb J, Ghandil P, Hamzaoui K, Hamzaoui A, Combadiere C: A polymorphism in the CCL2 chemokine gene is associated with asthma risk: a case-control and a family study in Tunisia. Genes Immun. 2008, 9 (7): 575-581. 10.1038/gene.2008.50.
Chen YQ, Shi HZ: CD28/CTLA-4--CD80/CD86 and ICOS--B7RP-1 costimulatory pathway in bronchial asthma. Allergy. 2006, 61 (1): 15-26. 10.1111/j.1398-9995.2006.01008.x.
Cho SH, Hall IP, Wheatley A, Dewar J, Abraha D, Del Mundo J, Lee H, Oh CK: Possible role of the 4G/5G polymorphism of the plasminogen activator inhibitor 1 gene in the development of asthma. J Allergy Clin Immunol. 2001, 108 (2): 212-214. 10.1067/mai.2001.117260.
Chu EK, Cheng J, Foley JS, Mecham BH, Owen CA, Haley KJ, Mariani TJ, Kohane IS, Tschumperlin DJ, Drazen JM: Induction of the plasminogen activator system by mechanical stimulation of human bronchial epithelial cells. Am J Respir Cell Mol Biol. 2006, 35 (6): 628-638. 10.1165/rcmb.2006-0040OC.
Crosby JR, Guha M, Tung D, Miller DA, Bender B, Condon TP, York-DeFalco C, Geary RS, Monia BP, Karras JG, Gregory SA: Inhaled CD86 antisense oligonucleotide suppresses pulmonary inflammation and airway hyper-responsiveness in allergic mice. J Pharmacol Exp Ther. 2007, 321 (3): 938-946. 10.1124/jpet.106.119214.
Daley D, Lemire M, Akhabir L, Chan-Yeung M, He JQ, McDonald T, Sandford A, Stefanowicz D, Tripp B, Zamar D, Bosse Y, Ferretti V, Montpetit A, Tessier MC, Becker A, Kozyrskyj AL, Beilby J, McCaskie PA, Musk B, Warrington N, James A, Laprise C, Palmer LJ, Pare PD, Hudson TJ: Analyses of associations with asthma in four asthma population samples from Canada and Australia. Hum Genet. 2009, 125 (4): 445-459. 10.1007/s00439-009-0643-8.
Goenka S, Kaplan MH: Transcriptional regulation by STAT6. Immunol Res. 2011, 50 (1): 87-96. 10.1007/s12026-011-8205-2.
Hattori T, Konno S, Hizawa N, Isada A, Takahashi A, Shimizu K, Gao P, Beaty TH, Barnes KC, Huang SK, Nishimura M: Genetic variants in the mannose receptor gene (MRC1) are associated with asthma in two independent populations. Immunogenetics. 2009, 61 (11-12): 731-738. 10.1007/s00251-009-0403-x.
Higa S, Hirano T, Mayumi M, Hiraoka M, Ohshima Y, Nambu M, Yamaguchi E, Hizawa N, Kondo N, Matsui E, Katada Y, Miyatake A, Kawase I, Tanaka T: Association between interleukin-18 gene polymorphism 105A/C and asthma. Clin Exp Allergy. 2003, 33 (8): 1097-1102. 10.1046/j.1365-2222.2003.01739.x.
Hong X, Zhou H, Tsai HJ, Wang X, Liu X, Wang B, Xu X, Xu X: Cysteinyl leukotriene receptor 1 gene variation and risk of asthma. Eur Respir J. 2009, 33 (1): 42-48. 10.1183/09031936.00057708.
Hossny EM, Amr NH, Elsayed SB, Nasr RA, Ibraheim EM: Association of polymorphisms in the mast cell chymase gene promoter region (-1903 g/A) and (TG)n(GA)m repeat downstream of the gene with bronchial asthma in children. J Investig Allergol Clin Immunol. 2008, 18 (5): 376-381.
Imada Y, Fujimoto M, Hirata K, Hirota T, Suzuki Y, Saito H, Matsumoto K, Akazawa A, Katsunuma T, Yoshihara S, Ebisawa M, Shibasaki M, Arinami T, Tamari M, Noguchi E: Large scale genotyping study for asthma in the Japanese population. BMC Res Notes. 2009, 2: 54-10.1186/1756-0500-2-54.
Imboden M, Nicod L, Nieters A, Glaus E, Matyas G, Bircher AJ, Ackermann-Liebrich U, Berger W, Probst-Hensch NM: The common G-allele of interleukin-18 single-nucleotide polymorphism is a genetic risk factor for atopic asthma. The SAPALDIA Cohort Study. Clin Exp Allergy. 2006, 36 (2): 211-218. 10.1111/j.1365-2222.2006.02424.x.
Islam T, Breton C, Salam MT, McConnell R, Wenten M, Gauderman WJ, Conti D, Van Den Berg D, Peters JM, Gilliland FD: Role of inducible nitric oxide synthase in asthma risk and lung function growth during adolescence. Thorax. 2009, 65 (2): 139-145.
Jaradat M, Stapleton C, Tilley SL, Dixon D, Erikson CJ, McCaskill JG, Kang HS, Angers M, Liao G, Collins J, Grissom S, Jetten AM: Modulatory role for retinoid-related orphan receptor alpha in allergen-induced lung inflammation. Am J Respir Crit Care Med. 2006, 174 (12): 1299-1309. 10.1164/rccm.200510-1672OC.
Jetten AM: Retinoid-related orphan receptors (RORs): critical roles in development, immunity, circadian rhythm, and cellular metabolism. Nucl Recept Signal. 2009, 7: e003-
Koppelman GH, Meyers DA, Howard TD, Zheng SL, Hawkins GA, Ampleford EJ, Xu J, Koning H, Bruinenberg M, Nolte IM, van Diemen CC, Boezen HM, Timens W, Whittaker PA, Stine OC, Barton SJ, Holloway JW, Holgate ST, Graves PE, Martinez FD, van Oosterhout AJ, Bleecker ER, Postma DS: Identification of PCDH1 as a novel susceptibility gene for bronchial hyperresponsiveness. Am J Respir Crit Care Med. 2009, 180 (10): 929-935. 10.1164/rccm.200810-1621OC.
Kowal K, Bodzenta-Lukaszyk A, Pampuch A, Szmitkowski M, Zukowski S, Donati MB, Iacoviello L: Analysis of -675 4 g/5 G SERPINE1 and C-159T CD14 polymorphisms in house dust mite-allergic asthma patients. J Investig Allergol Clin Immunol. 2008, 18 (4): 284-292.
Kowal K, Moniuszko M, Zukowski S, Bodzenta-Lukaszyk A: Concentrations of plasminogen activator inhibitor-1 (PAI-1) and urokinase plasminogen activator (uPA) in induced sputum of asthma patients after allergen challenge. Folia Histochem Cytobiol. 2010, 48 (4): 518-523. 10.2478/v10042-010-0075-2.
Kucharewicz I, Mogielnicki A, Kasacka I, Buczko W, Bodzenta-Lukaszyk A: Plasmin system regulation in an ovalbumin-induced rat model of asthma. Int Arch Allergy Immunol. 2008, 147 (3): 190-196. 10.1159/000142041.
Lee CC, Lin WY, Wan L, Tsai Y, Tsai CH, Huang CM, Chen CP, Tsai FJ: Association of interleukin-18 gene polymorphism with asthma in Chinese patients. J Clin Lab Anal. 2008, 22 (1): 39-44. 10.1002/jcla.20218.
Lee JH, Moore JH, Park SW, Jang AS, Uh ST, Kim YH, Park CS, Park BL, Shin HD: Genetic interactions model among Eotaxin gene polymorphisms in asthma. J Hum Genet. 2008, 53 (10): 867-875. 10.1007/s10038-008-0314-y.
Li H, Romieu I, Sienra-Monge JJ, Ramirez-Aguilar M, Estela Del Rio-Navarro B, Kistner EO, Gjessing HK, Lara-Sanchez Idel C, Chiu GY, London SJ: Genetic polymorphisms in arginase I and II and childhood asthma and atopy. J Allergy Clin Immunol. 2006, 117 (1): 119-126. 10.1016/j.jaci.2005.09.026.
Li YF, Tseng PJ, Lin CC, Hung CL, Lin SC, Su WC, Huang YL, Sung FC, Tai CK: NAD(P)H: Quinone oxidoreductase 1, glutathione S-transferase M1, environmental tobacco smoke exposure, and childhood asthma. Mutat Res. 2009, 678 (1): 53-58.
Liu Q, Xia Y, Zhang W, Li J, Wang P, Li H, Wei C, Gong Y: A functional polymorphism in the SPINK5 gene is associated with asthma in a Chinese Han Population. BMC Med Genet. 2009, 10: 59-
Madore AM, Perron S, Turmel V, Laviolette M, Bissonnette EY, Laprise C: Alveolar macrophages in allergic asthma: an expression signature characterized by heat shock protein pathways. Hum Immunol. 2010, 71 (2): 144-150. 10.1016/j.humimm.2009.11.005.
Martinez B, Barrios K, Vergara C, Mercado D, Jimenez S, Gusmao L, Caraballo L: A NOS1 gene polymorphism associated with asthma and specific immunoglobulin E response to mite allergens in a Colombian population. Int Arch Allergy Immunol. 2007, 144 (2): 105-113. 10.1159/000103221.
Masumoto J, Yang K, Varambally S, Hasegawa M, Tomlins SA, Qiu S, Fujimoto Y, Kawasaki A, Foster SJ, Horie Y, Mak TW, Nunez G, Chinnaiyan AM, Fukase K, Inohara N: Nod1 acts as an intracellular receptor to stimulate chemokine production and neutrophil recruitment in vivo. J Exp Med. 2006, 203 (1): 203-213. 10.1084/jem.20051229.
Matsuzaki S, Ishizuka T, Hisada T, Aoki H, Komachi M, Ichimonji I, Utsugi M, Ono A, Koga Y, Dobashi K, Kurose H, Tomura H, Mori M, Okajima F: Lysophosphatidic acid inhibits CC chemokine ligand 5/RANTES production by blocking IRF-1-mediated gene transcription in human bronchial epithelial cells. J Immunol. 2010, 185 (8): 4863-4872. 10.4049/jimmunol.1000904.
McKay A, Komai-Koma M, MacLeod KJ, Campbell CC, Kitson SM, Chaudhuri R, Thomson L, McSharry C, Liew FY, Thomson NC: Interleukin-18 levels in induced sputum are reduced in asthmatic and normal smokers. Clin Exp Allergy. 2004, 34 (6): 904-910. 10.1111/j.1365-2222.2004.01973.x.
Millstein JCD, Gilliland FD, Gauderman WJ: A testing framework for identifying suseptibility genes in the presence of epistasis. Am J Hum Genet. 2006, 78: 15-27. 10.1086/498850.
Minelli C, Granell R, Newson R, Rose-Zerilli MJ, Torrent M, Ring SM, Holloway JW, Shaheen SO, Henderson JA: Glutathione-S-transferase genes and asthma phenotypes: a Human Genome Epidemiology (HuGE) systematic review and meta-analysis including unpublished data. Int J Epidemiol. 2010, 39 (2): 539-562. 10.1093/ije/dyp337.
Moller-Larsen S, Nyegaard M, Haagerup A, Vestbo J, Kruse TA, Borglum AD: Association analysis identifies TLR7 and TLR8 as novel risk genes in asthma and related disorders. Thorax. 2008, 63 (12): 1064-1069. 10.1136/thx.2007.094128.
Movahedi M, Moin M, Gharagozlou M, Aghamohammadi A, Dianat S, Moradi B, Nicknam MH, Nikbin B, Amirzargar A: Association of HLA class II alleles with childhood asthma and Total IgE levels. Iran J Allergy Asthma Immunol. 2008, 7 (4): 215-220.
Munthe-Kaas MC, Carlsen KH, Haland G, Devulapalli CS, Gervin K, Egeland T, Carlsen KL, Undlien D: T cell-specific T-box transcription factor haplotype is associated with allergic asthma in children. J Allergy Clin Immunol. 2008, 121 (1): 51-56. 10.1016/j.jaci.2007.07.068.
Munthe-Kaas MC, Carlsen KL, Carlsen KH, Egeland T, Haland G, Devulapalli CS, Akselsen H, Undlien D: HLA Dr-Dq haplotypes and the TNFA-308 polymorphism: associations with asthma and allergy. Allergy. 2007, 62 (9): 991-998. 10.1111/j.1398-9995.2007.01377.x.
Nagase T, Kurihara H, Kurihara Y, Aoki-Nagase T, Nagai R, Ouchi Y: Disruption of ET-1 gene enhances pulmonary responses to methacholine via functional mechanism in knockout mice. J Appl Physiol. 1999, 87 (6): 2020-2024.
Nicolae D, Cox NJ, Lester LA, Schneider D, Tan Z, Billstrand C, Kuldanek S, Donfack J, Kogut P, Patel NM, Goodenbour J, Howard T, Wolf R, Koppelman GH, White SR, Parry R, Postma DS, Meyers D, Bleecker ER, Hunt JS, Solway J, Ober C: Fine mapping and positional candidate studies identify HLA-G as an asthma susceptibility gene on chromosome 6p21. Am J Hum Genet. 2005, 76 (2): 349-357. 10.1086/427763.
Pampuch A, Kowal K, Bodzenta-Lukaszyk A, Di Castelnuovo A, Chyczewski L, Donati MB, Iacoviello L: The -675 4G/5G plasminogen activator inhibitor-1 promoter polymorphism in house dust mite-sensitive allergic asthma patients. Allergy. 2006, 61 (2): 234-238. 10.1111/j.1398-9995.2005.00948.x.
Pegorier S, Arouche N, Dombret MC, Aubier M, Pretolani M: Augmented epithelial endothelin-1 expression in refractory asthma. J Allergy Clin Immunol. 2007, 120 (6): 1301-1307. 10.1016/j.jaci.2007.09.023.
Raby BA, Hwang ES, Van Steen K, Tantisira K, Peng S, Litonjua A, Lazarus R, Giallourakis C, Rioux JD, Sparrow D, Silverman EK, Glimcher LH, Weiss ST: T-bet polymorphisms are associated with asthma and airway hyperresponsiveness. Am J Respir Crit Care Med. 2006, 173 (1): 64-70. 10.1164/rccm.200503-505OC.
Ray R, Choi M, Zhang Z, Silverman GA, Askew D, Mukherjee AB: Uteroglobin suppresses SCCA gene expression associated with allergic asthma. J Biol Chem. 2005, 280 (11): 9761-9764. 10.1074/jbc.C400581200.
Saadi A, Gao G, Li H, Wei C, Gong Y, Liu Q: Association study between vitamin D receptor gene polymorphisms and asthma in the Chinese Han population: a case-control study. BMC Med Genet. 2009, 10: 71-
Salam MT, Islam T, Gauderman WJ, Gilliland FD: Roles of arginase variants, atopy, and ozone in childhood asthma. J Allergy Clin Immunol. 2009, 123 (3): 596-602. 10.1016/j.jaci.2008.12.020. 602 e591-598
Sanz C, Isidro-Garcia M, Davila I, Moreno E, Laffond E, Lorente F: Analysis of 927T > C CYSLTRI and -444A > C LTC4S polymorphisms in patients with asthma. J Investig Allergol Clin Immunol. 2006, 16 (6): 331-337.
Seibold MA, Reese TA, Choudhry S, Salam MT, Beckman K, Eng C, Atakilit A, Meade K, Lenoir M, Watson HG, Thyne S, Kumar R, Weiss KB, Grammer LC, Avila P, Schleimer RP, Fahy JV, Rodriguez-Santana J, Rodriguez-Cintron W, Boot RG, Sheppard D, Gilliland FD, Locksley RM, Burchard EG: Differential enzymatic activity of common haplotypic versions of the human acidic Mammalian chitinase protein. J Biol Chem. 2009, 284 (29): 19650-19658. 10.1074/jbc.M109.012443.
Suttner K, Rosenstiel P, Depner M, Schedel M, Pinto LA, Ruether A, Adamski J, Klopp N, Illig T, Vogelberg C, Schreiber S, von Mutius E, Kabesch M: TBX21 gene variants increase childhood asthma risk in combination with HLX1 variants. J Allergy Clin Immunol. 2009, 123 (5): 1062-1068. 10.1016/j.jaci.2009.02.025. 1068 e1061-1068
Szalai C, Kozma GT, Nagy A, Bojszko A, Krikovszky D, Szabo T, Falus A: Polymorphism in the gene regulatory region of MCP-1 is associated with asthma susceptibility and severity. J Allergy Clin Immunol. 2001, 108 (3): 375-381. 10.1067/mai.2001.117930.
Szczepankiewicz A, Breborowicz A, Skibinska M, Wilkosc M, Tomaszewska M, Hauser J: Association analysis of brain-derived neurotrophic factor gene polymorphisms in asthmatic children. Pediatr Allergy Immunol. 2007, 18 (4): 293-297. 10.1111/j.1399-3038.2007.00525.x.
Szczepankiewicz A, Rose-Zerilli MJ, Barton SJ, Holgate ST, Holloway JW: Association analysis of brain-derived neurotrophic factor gene polymorphisms in asthmatic families. Int Arch Allergy Immunol. 2009, 149 (4): 343-349. 10.1159/000205580.
van den Oord RA, Sheikh A: Filaggrin gene defects and risk of developing allergic sensitisation and allergic disorders: systematic review and meta-analysis. Bmj. 2009, 339: b2433-10.1136/bmj.b2433.
Wang J, Xu Y, Zhao H, Sui H, Liang H, Jiang X: Genetic variations in chemoattractant receptor expressed on Th2 cells (CRTH2) is associated with asthma susceptibility in Chinese children. Mol Biol Rep. 2009, 36 (6): 1549-1553. 10.1007/s11033-008-9349-6.
Wang JY, Shyur SD, Wang WH, Liou YH, Lin CG, Wu YJ, Wu LS: The polymorphisms of interleukin 17A (IL17A) gene and its association with pediatric asthma in Taiwanese population. Allergy. 2009, 64 (7): 1056-1060. 10.1111/j.1398-9995.2009.01950.x.
Vergara C, Tsai YJ, Grant AV, Rafaels N, Gao L, Hand T, Stockton M, Campbell M, Mercado D, Faruque M, Dunston G, Beaty TH, Oliveira RR, Ponte EV, Cruz AA, Carvalho E, Araujo MI, Watson H, Schleimer RP, Caraballo L, Nickel RG, Mathias RA, Barnes KC: Gene encoding Duffy antigen/receptor for chemokines is associated with asthma and IgE in three populations. Am J Respir Crit Care Med. 2008, 178 (10): 1017-1022. 10.1164/rccm.200801-182OC.
White JH, Chiano M, Wigglesworth M, Geske R, Riley J, White N, Hall S, Zhu G, Maurio F, Savage T, Anderson W, Cordy J, Ducceschi M, Vestbo J, Pillai SG: Identification of a novel asthma susceptibility gene on chromosome 1qter and its functional evaluation. Hum Mol Genet. 2008, 17 (13): 1890-1903. 10.1093/hmg/ddn087.
Yamagata S, Tomita K, Sato R, Niwa A, Higashino H, Tohda Y: Interleukin-18-deficient mice exhibit diminished chronic inflammation and airway remodelling in ovalbumin-induced asthma model. Clin Exp Immunol. 2008, 154 (3): 295-304. 10.1111/j.1365-2249.2008.03772.x.
Yang CJ, Liu YK, Liu CL, Shen CN, Kuo ML, Su CC, Tseng CP, Yen TC, Shen CR: Inhibition of acidic mammalian chitinase by RNA interference suppresses ovalbumin-sensitized allergic asthma. Hum Gene Ther. 2009, 20 (12): 1597-1606. 10.1089/hum.2008.092.
Ye Q, Fujita M, Ouchi H, Inoshima I, Maeyama T, Kuwano K, Horiuchi Y, Hara N, Nakanishi Y: Serum CC-10 in inflammatory lung diseases. Respiration. 2004, 71 (5): 505-510. 10.1159/000080636.
Zeilinger S, Pinto LA, Nockher WA, Depner M, Klopp N, Illig T, von Mutius E, Renz H, Kabesch M: The effect of BDNF gene variants on asthma in German children. Allergy. 2009, 64 (12): 1790-1794. 10.1111/j.1398-9995.2009.02131.x.
Zheng XQ, Li CC, Xu DP, Lin A, Bao WG, Yang GS, Yan WH: Analysis of the plasma soluble human leukocyte antigen-G and interleukin-10 levels in childhood atopic asthma. Hum Immunol. 2010, 71 (10): 982-987. 10.1016/j.humimm.2010.06.018.
Zhou H, Hong X, Jiang S, Dong H, Xu X: Analyses of associations between three positionally cloned asthma candidate genes and asthma or asthma-related phenotypes in a Chinese population. BMC Med Genet. 2009, 10: 123-10.1186/1471-2350-10-123.
Haland G, Carlsen KC, Sandvik L, Devulapalli CS, Munthe-Kaas MC, Pettersen M, Carlsen KH: Reduced lung function at birth and the risk of asthma at 10 years of age. N Engl J Med. 2006, 355 (16): 1682-1689. 10.1056/NEJMoa052885.
Martinez FD, Wright AL, Taussig LM, Holberg CJ, Halonen M, Morgan WJ: Asthma and wheezing in the first six years of life. The Group Health Medical Associates. N Engl J Med. 1995, 332 (3): 133-138. 10.1056/NEJM199501193320301.
Hunninghake GM, Cho MH, Tesfaigzi Y, Soto-Quiros ME, Avila L, Lasky-Su J, Stidley C, Melen E, Soderhall C, Hallberg J, Kull I, Kere J, Svartengren M, Pershagen G, Wickman M, Lange C, Demeo DL, Hersh CP, Klanderman BJ, Raby BA, Sparrow D, Shapiro SD, Silverman EK, Litonjua AA, Weiss ST, Celedon JC: MMP12, lung function, and COPD in high-risk populations. N Engl J Med. 2009, 361 (27): 2599-2608. 10.1056/NEJMoa0904006.
Maeda Y, Dave V, Whitsett JA: Transcriptional control of lung morphogenesis. Physiol Rev. 2007, 87 (1): 219-244. 10.1152/physrev.00028.2006.
Franzdottir SR, Axelsson IT, Arason AJ, Baldursson O, Gudjonsson T, Magnusson MK: Airway branching morphogenesis in three dimensional culture. Respir Res. 2010, 11: 162-10.1186/1465-9921-11-162.
Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA: Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA. 2009, 106 (23): 9362-9367. 10.1073/pnas.0903103106.
Ting JP, Duncan JA, Lei Y: How the noninflammasome NLRs function in the innate immune system. Science. 2010, 327 (5963): 286-290. 10.1126/science.1184004.
Fagan KA, McMurtry IF, Rodman DM: Role of endothelin-1 in lung disease. Respir Res. 2001, 2 (2): 90-101. 10.1186/rr44.
Stow LR, Jacobs ME, Wingo CS, Cain BD: Endothelin-1 gene regulation. FASEB J. 2011, 25 (1): 16-28. 10.1096/fj.10-161612.
Kurihara Y, Kurihara H, Oda H, Maemura K, Nagai R, Ishikawa T, Yazaki Y: Aortic arch malformations and ventricular septal defect in mice deficient in endothelin-1. J Clin Invest. 1995, 96 (1): 293-300. 10.1172/JCI118033.
Kurihara Y, Kurihara H, Suzuki H, Kodama T, Maemura K, Nagai R, Oda H, Kuwaki T, Cao WH, Kamada N, et al: Elevated blood pressure and craniofacial abnormalities in mice deficient in endothelin-1. Nature. 1994, 368 (6473): 703-710. 10.1038/368703a0.
Hocher B, Schwarz A, Fagan KA, Thone-Reineke C, El-Hag K, Kusserow H, Elitok S, Bauer C, Neumayer HH, Rodman DM, Theuring F: Pulmonary fibrosis and chronic lung inflammation in ET-1 transgenic mice. Am J Respir Cell Mol Biol. 2000, 23 (1): 19-26.
Immervoll T, Loesgen S, Dutsch G, Gohlke H, Herbon N, Klugbauer S, Dempfle A, Bickeboller H, Becker-Follmann J, Ruschendorf F, Saar K, Reis A, Wichmann HE, Wjst M: Fine mapping and single nucleotide polymorphism association results of candidate genes for asthma and related phenotypes. Hum Mutat. 2001, 18 (4): 327-336. 10.1002/humu.1194.
Zhu G, Carlsen K, Carlsen KH, Lenney W, Silverman M, Whyte MK, Hosking L, Helms P, Roses AD, Hay DW, Barnes MR, Anderson WH, Pillai SG: Polymorphisms in the endothelin-1 (EDN1) are associated with asthma in two populations. Genes Immun. 2008, 9 (1): 23-29. 10.1038/sj.gene.6364441.
Melen E, Bruce S, Doekes G, Kabesch M, Laitinen T, Lauener R, Lindgren CM, Riedler J, Scheynius A, van Hage-Hamsten M, Kere J, Pershagen G, Wickman M, Nyberg F: Haplotypes of G protein-coupled receptor 154 are associated with childhood allergy and asthma. Am J Respir Crit Care Med. 2005, 171 (10): 1089-1095. 10.1164/rccm.200410-1317OC.
Acknowledgements
The authors wish to thank Dr. Weiliang Qiu, Channing Laboratory, Brigham and Women's Hospital and Harvard Medical School, for a valuable statistical review. Financial support: Supported by National Institutes of Health grants K25 HL91124, R01 HL88028, and P50 NS40828 (ATK.); R01 ES10855 (JSL); R01 HL097144 (STW), U01 HL65899 (STW and KGT); EM is supported by post doc grants from the Swedish Heart Lung Foundation, the Swedish Fulbright Commission, Centre for Allergy Research, Karolinska Institutet and Riksbankens Jubileumsfond, Erik Rönnberg's scholarship for research on early childhood diseases.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
All authors declare no competing interests and no support from any organisation for the submitted work; no financial relationships with any organisations that might have an interest in the submitted work in the previous 3 years; no other relationships or activities that could appear to have influenced the submitted work.
Authors' contributions
EM carried out the literature search and the statistical analyses together with ATK, SS and VJC. EM, RG, JSL, TJM, STW and KGT participated in the design and planning of the study. EM, ATK and KGT drafted the manuscript. All authors read and approved the final manuscript.
Electronic supplementary material
12931_2011_1111_MOESM1_ESM.DOC
Additional file 1: Supplementary Tables and Figure. Expression analysis of asthma candidate genes during human and murine lung development. (DOC 972 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Melén, E., Kho, A.T., Sharma, S. et al. Expression analysis of asthma candidate genes during human and murine lung development. Respir Res 12, 86 (2011). https://doi.org/10.1186/1465-9921-12-86
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1465-9921-12-86