Abstract
Background
The human bacterial pathogen Campylobacter jejuni contains two subspecies: C. jejuni subsp. jejuni (Cjj) and C. jejuni subsp. doylei (Cjd). Although Cjd strains are isolated infrequently in many parts of the world, they are obtained primarily from human clinical samples and result in an unusual clinical symptomatology in that, in addition to gastroenteritis, they are associated often with bacteremia. In this study, we describe a novel multiplex PCR method, based on the nitrate reductase (nap) locus, that can be used to unambiguously subspeciate C. jejuni isolates.
Results
Internal and flanking napA and napB primer sets were designed, based on existing C. jejuni and Campylobacter coli genome sequences to create two multiplex PCR primer sets, nap mpx1 and nap mpx2. Genomic DNA from 161 C. jejuni subsp. jejuni (Cjj) and 27 C. jejuni subsp. doylei (Cjd) strains were amplified with these multiplex primer sets. The Cjd strains could be distinguished clearly from the Cjj strains using either nap mpx1 or mpx2. In addition, combination of either nap multiplex method with an existing lpxA speciation multiplex method resulted in the unambiguous and simultaneous speciation and subspeciation of the thermophilic Campylobacters. The Cjd nap amplicons were also sequenced: all Cjd strains tested contained identical 2761 bp deletions in napA and several Cjd strains contained deletions in napB.
Conclusion
The nap multiplex PCR primer sets are robust and give a 100% discrimination of C. jejuni subspecies. The ability to rapidly subspeciate C. jejuni as well as speciate thermophilic Campylobacter species, most of which are pathogenic in humans, in a single amplification will be of value to clinical laboratories in strain identification and the determination of the environmental source of campylobacterioses caused by Cjd. Finally, the sequences of the Cjd napA and napB loci suggest that Cjd strains arose from a common ancestor, providing clues as to the potential evolutionary origin of Cjd.
Similar content being viewed by others
Background
Campylobacter spp. are a common cause of acute bacterial gastroenteritis in humans [1, 2]. The majority of campylobacterioses are caused by C. jejuni and are linked primarily to untreated water or consumption of poultry and raw milk [2]. C. jejuni has been divided into two subspecies: C. jejuni subsp. jejuni (Cjj) and C. jejuni subsp. doylei (Cjd). Cjd strains were isolated originally as "gastric campylobacter-like organisms type 2 (GCLO2)" from human gastric biopsies [4] and "nitrate-negative campylobacter-like organisms (NNC)" from Australian pediatric patients with gastroenteritis [5]. As the "NNC" designation suggests, the characteristic feature of Cjd, used commonly to distinguish Cjd strains from Cjj strains, is the inability to reduce nitrate. Other phenotypic characteristics, such as variable growth at 42°C [6], high susceptibility to cephalothin [6], and the absence of γ-glutamyl transferase (GGT) and L-arginine arylamidase activity [7] have been associated also with Cjd; however, like Cjj, all Cjd strains are hippuricase positive.
Cjd strains also differ from Cjj in two other clinically-related aspects: first, Cjd strains can be found throughout the gastrointestinal tract, leading to both gastritis [4, 8] and enteritis [6, 9–11], and are obtained often from pediatric patients [6, 9, 11, 12]. Second, in South Africa, unlike Cjj strains, Cjd strains are isolated more often from blood cultures than from stool cultures [3]; Cjd was isolated from 24% of the Campylobacter-positive blood cultures [3], in contrast to 7.7% of the Campylobacter-positive stool cultures, obtained at Red Cross Children's Hospital, Cape Town, during the years 1990–2005. Additionally, Morey reported that Cjd was isolated from 85.2% of Campylobacter/Helicobacter-related bacteremia cases in Australia during a five-year period [12].
In some parts of the world, notably South Africa, Cjd strains represent a significant proportion of the total campylobacters isolated from human clinical samples; 16% of the non-Cjj/coli Campylobacter strains isolated in Cape Town, South Africa were Cjd. However, despite the unusual clinical symptomatology and relatively high prevalence in certain parts of the world, Cjd is generally isolated infrequently and few strains exist (compared to Cjj) for this subspecies. One possible reason is that many clinical laboratories do not characterize Campylobacter isolates past the genus level, much less subspeciate C. jejuni isolates as Cjj or Cjd. It is also likely that both the susceptibility of Cjd to cephalothin and variable growth at 42°C prevents the isolation of a substantial number of Cjd strains under normal Cjj isolation conditions; the Cjd strains isolated in South Africa were obtained using the Cape Town Protocol [3] which uses passive filtration through a 0.65 μM membrane filter, growth at 37°C and no antibiotic selection.
Another factor in the lack of subspeciation characterization of C. jejuni strains is the absence of a rapid test to distinguish Cjj from Cjd. Phenotypic characterization of Cjd, based on the absence of nitrate reductase activity, remains the primary means of identification; however, such nitrate reductase assays require a large number of cells and can take 24–48 h. Additionally, nitrate reductase assays require pure cultures; mixtures of Cjj and Cjd cells would type as nitrate+/Cjj.
Many commonly used molecular-based Campylobacter detection methods cannot be used to subspeciate C. jejuni due to the high similarity between the two subspecies. Molecular-based methods that can subspeciate C. jejuni do exist, based on hybridization [7], amplified fragment length polymorphism fingerprinting [13, 14], or characterization of the 16S/23S rDNA internal spacer region [15]; however, these methods are lengthy or require specialized and costly equipment. In this study we present a simple, novel multiplex PCR method that can be used to unambiguously subspeciate C. jejuni.
Results
Development of the napAB multiplex subspeciation PCR
Preliminary results from C. jejuni DNA microarray experiments, using Cjd strain RM2095 as a tester strain, indicated that, in addition to other loci, the napA and napB genes in strain RM2095 were either absent or highly divergent with respect to Cjj napA and napB (Parker et al., submitted for publication). Since napA and napB encode the large and small subunits of nitrate reductase, respectively, this was not unexpected, given the absence of nitrate reductase activity in RM2095. Thus, the microarray results suggested that the nap phenotype in RM2095 was due most likely to deletions in napA and/or napB and that these results might be extended to Cjd in general; a loss of function in either subunit would result in a loss of enzyme activity. Therefore, an obvious target for a subspeciation multiplex PCR would be the nap locus. The nap locus in several Campylobacter species consists of six genes (in order): napA, napG, napH, napB, napL, and napD. Upstream of napA in Cjj and C. coli is the tpx gene, encoding a thiol peroxidase. The conservation of gene order and high nt identity between C. jejuni and C. coli at this locus suggested that primers designed to amplify both species should also amplify Cjd strains.
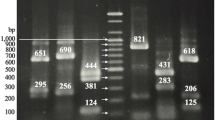
Therefore, the sequences of the nap regions (tpx to napD) of three strains (Cjj NCTC 11168, Cjj RM1221 and C. coli RM2228), obtained from the genome sequence for each strain [GenBank: AL111168, GenBank: CP000025, and GenBank: AAFL00000000, respectively], were aligned and four primer pairs were designed to regions of high conservation within the alignment: primers internal to and flanking napA, and primers internal to and flanking napB. The primers were organized into two multiplex primer sets: nap mpx1 and nap mpx2 (Table 1). A representative gel illustrating the nap mpx1 and nap mpx2 amplicons is presented in Fig. 1A.
Identification of C. jejuni subsp. doylei strains by multiplex PCR. A. napAB multiplex PCR. C. jejuni genomic DNAs were amplified with either the nap mpx 1 ("1") or mpx 2 ("2") primer sets; the PCRs were run on a 1% agarose gel. D1: Cjd strain RM2095; D2: Cjd strain 269.97; D3: Cjd strain RM2096; D4: Cjd strain CCUG 18266; J1: Cjj strain NCTC 11168; J2: Cjj strain RM1221. Band sizes in bp are indicated on the left side of the gel. B. Combination speciation and subspeciation PCR. Campylobacter genomic DNAs were amplified with the combined nap mpx 2 and thermophilic lpxA Campylobacter speciation primer sets; PCRs were run on a 2% agarose gel. Cj: Cjj strain NCTC 11168; D1: Cjd strain CCUG 18266; D2: Cjd strain 269.97; Cc: C. coli strain RM2228; Cl: C. lari strain RM2100; Cu: C. upsaliensis strain RM3195; Ch: C. helveticus strain ATCC 51209. Band sizes in bp are indicated on the right side of the gel. Bands resulting from the speciation primer sets are indicated with an asterisk (*).
Since napA is quite large (~2.8 kb), Cjj strains would not be expected to amplify with the napA flanking primers under standard PCR conditions; however, both Cjj strains did amplify with the napA internal primer set (1454 bp: Fig. 1A). The Cjd strains did not amplify with the napA internal primer set but did amplify with the napA flanking primers. The resulting amplicon (1240 bp: Fig 1A) was reduced in size from that predicted for Cjj by about 2.8 kb, indicating the presence of a deletion within napA; amplification with different sets of flanking primers indicated that this deletion extended into napG (data not shown). Noteworthy also was the fact that the Cjd strains could be divided into two classes, differentiated by an apparent presence or absence of napB. The "napA-napB+" strains (D3, D4: Fig 1A), termed Cjd1, amplified with the napB internal primer set (326 bp: Fig. 1A) and gave a full length amplicon (973 bp: Fig 1A) with the napB flanking primer set. The "napA-napB-" strains (D1, D2: Fig 1A), termed Cjd2, gave a reduced length amplicon (494 bp: Fig 1A) with the napB flanking primer set, indicating the presence of a napB deletion. Both classes would be unable to reduce nitrate, based on the common napA deletion.
Validation of the nap multiplex PCR using additional Campylobacter genomic DNAs
To test the specificity of the nap mpx primer sets, an additional 20 human clinical Cjd strains and 158 human clinical, animal and environmental Cjj strains were amplified using the nap mpx2 primer set. The PCR results were identical to those presented in Fig. 1A [Table S1, see additional file 1] with two exceptions. The HS:63 Penner serotype reference strain, listed originally as Cjj, amplified as a Cjd2 isolate; however, this serotype has been seen previously in Cjd isolates [16] and therefore this strain may have been misidentified as a Cjj. Also, one Cjd strain amplified as a Cjj isolate; a second nitrate reductase test on this strain indicated that the strain was mis-typed originally.
Genomic DNA from the C. coli strain RM2228 amplified also with the nap mpx2 primer set; the resulting banding pattern was identical to that of Cjj (data not shown). Consequently, representatives of the remaining non-jejuni/coli Campylobacter spp. were amplified using the nap mpx2 primer set. Genomic DNA from five of these species [C. upsaliensis (RM3195), C. lari (RM2100), C. helveticus (RM3228), C. sputorum (RM3237) and C. mucosalis (RM3234)] amplified, although the banding patterns were distinct from those in Fig. 1A [Table S1, see additional file 1]. Therefore, 107 additional strains of these five species and 10 additional C. coli strains were amplified using both primer sets. All of the C. coli strains and some of the C. upsaliensis strains produced both of the Cjj-characteristic 1454 bp and 1016 bp bands following amplification with nap mpx2 (11/11 and 23/72, respectively; [Table S1, see additional file 1]). Moreover, the banding patterns for many of these 34 C. coli and C. upsaliensis strains were similar to those seen in Fig. 1A for both multiplex primer sets; therefore, C. coli and C. upsaliensis strains could be confused potentially with Cjj isolates.
Combination of the nap and lpxA multiplex PCRs gives unambiguous speciation and subspeciation of thermophilic campylobacters
If the nap multiplex PCRs were performed on strains already speciated as C. jejuni, then the non-specificity of the nap PCR towards C. coli and C. upsaliensis would not be of concern. However, in many instances, food, animal and environmental samples contain multiple Campylobacter species, especially C. jejuni and C. coli. In this case, the non-specificity of the nap PCR would not permit accurate identification. A PCR speciation method for thermophilic Campylobacter spp, based on the lipid A gene lpxA, was published recently [17]. Therefore, to extend the discriminatory power of the nap subspeciation PCR method, we tested whether unambiguous speciation and subspeciation of thermophilic Campylobacter spp. would be feasible by combining the lpxA and nap mpx PCR methods. Genomic DNAs from five thermophilic Campylobacter species were amplified with the nine-primer nap/lpxA primer set. Unique banding patterns were obtained for all species/subspecies (Fig. 1B), indicating that unambiguous speciation and subspeciation of thermophilic campylobacters can be accomplished with only one amplification.
Characterization of the Cjd napB and napA loci
As described above, the Cjd strains were subdivided into two groups based on the apparent presence (Cjd1) or absence (Cjd2) of napB. To characterize the Cjd napB genes, the amplicons obtained with the napB flanking primers were sequenced. The napB loci of Cjd2 strains RM2095, RM3782 and SSI 5384 contained an approx. 520 bp deletion extending from nt -23 to nt 495 (data not shown). All three deletions had identical endpoints; however, single nucleotide polymorphisms present in the napB amplicon sequences indicated that all three strains were genetically distinct. Interestingly, three of the four Cjd1 strains contained an extra G at napB nt 15, resulting in a truncated NapB; thus, of the eight Cjd1 and Cjd2 strains, only RM2096 would be predicted to encode a full-length, and presumably functional, NapB protein.
Similarly, the amplicons obtained with the napA flanking primers were sequenced. As with napB, single nucleotide polymorphisms are present also in the amplicon sequences of five of the eight strains, indicating that these five strains are different. The remaining strains, ATCC 49350, ATCC 49351 and CCUG 18266, have identical napA sequences. As predicted, all eight Cjd napA genes contain 2761 bp deletions that extend from napA (nt 493) into napG; unexpectedly, however, all eight deletions have identical endpoints. Amplification and sequencing of the napA loci from an additional 19 Cjd isolates also revealed the presence of identical napA deletions.
Discussion
The apparent absence of the genes encoding nitrate reductase (napA and napB) in Cjd strain RM2095 correlated well with the nitrate reduction-negative phenotype characteristic of this subspecies. Thus, we reasoned that a potential subspeciation marker for C. jejuni would be the nap locus, specifically napA and napB, and we developed therefore a napA/B multiplex PCR method to subspeciate C. jejuni. Amplification results indicate that this nap multiplex PCR method, using internal and flanking primer sets for napA and napB, can be used to subspeciate unambiguously C. jejuni: Cjd strains (D1-4: Fig. 1A) can be distinguished readily from Cjj strains (J1-2: Fig. 1A). Moreover, different banding patterns were observed with Cjj and Cjd using nap mpx1 or nap mpx2, suggesting that either multiplex primer set is sufficient to subspeciate C. jejuni. The almost total concordance between the subspecies identification of 188 C. jejuni strains and the multiplex PCR results indicates that the new multiplex PCR is robust and can be used successfully to subspeciate C. jejuni. One strain, the Penner HS:63 reference strain, classified originally as Cjj, was identified as Cjd by our assay, suggesting a potential flaw in the PCR method. However, identification of a strain as Cjd by our method requires a deletion in napA and/or napB. Since a deletion in either of these two genes would lead necessarily to a loss of nitrate reductase activity, C. jejuni strains containing such defects would be, by definition Cjd, regardless of the original classification. Therefore, the Penner HS:63 reference strain was either not subspeciated or was subspeciated incorrectly. Finally, combination of the napA/B multiplex method with an lpxA multiplex method designed to speciate thermophilic Campylobacters permits the speciation and subspeciation of thermophilic Campylobacters with a single amplification reaction. Combination of the two methods is especially important when the species identification of a Campylobacter strain is unknown, as C. coli and C. upsaliensis strains may be confused potentially with Cjj (although not Cjd) strains if the nap multiplex method is used solely.
Currently, Cjd strains are subspeciated using the nitrate reductase assay, an assay used initially also in this study to identify Cjd strains. Although the nitrate reductase assay is relatively easy to perform, the multiplex PCR methods described here have several advantages over the nitrate reductase assay. First, subspeciation of C. jejuni strains, as well as the speciation/subspeciation of thermophilic Campylobacters, can be accomplished with this assay in a matter of hours, compared to the 2–3 days necessary for the nitrate reductase assay, and in a single amplification reaction. Second, the nitrate reductase assay requires a pure culture. Mixtures of Campylobacter strains of the same species or strains representing different species occur often during isolation [3, 18, 19] and would be problematic with regards to phenotypic assays; mixtures of Cjj and Cjd cells would type as Cjj. In contrast, the multiplex PCR method does not require a pure culture and would identify the species/subspecies comprising such a mixture. In fact, the multiplex speciation method identified successfully each species in a mixture of four thermophilic Campylobacter genomic DNAs (data not shown). Third, the nitrate reductase assay requires often more than a single colony for accurate identification. However, the multiplex PCR method requires at most only a single colony. Indeed, 19 of the Cjd genomic DNAs used in this study were obtained by boiling a single storage bead in 10 mM Tris pH 8.0, indicating that strains in storage at -80°C do not even have to be grown out to be characterized. Finally, the nitrate reductase assay does lead occasionally to false identification. One of the strains in this study, identified initially as Cjd, was determined by the multiplex PCR method to be Cjj. A repeat of the nitrate reductase assay on this strain indicated that it was, in fact, Cjj and that the original nitrate reductase identification was in error. Taken together, these results indicate that the napA/B multiplex PCR method described here is a valuable tool for clinical and research laboratories and can be used successfully to subspeciate rapidly C. jejuni strains and speciate thermophilic Campylobacters when combined with the lpxA multiplex PCR method.
An unexpected outcome of this study arose from the sequencing of the Cjd napA and napB amplicons. Some Cjd strains contained deletions in napB (Cjd2) and some contained apparently full-length napB genes (Cjd1), although point mutations in many of the Cjd1 napB genes would result in non-functional proteins; however, all 27 characterized Cjd strains contained identical napA deletions of 2761 bp. Based on the presence of these identical napA deletions, it appears that the Cjd strains, isolated on four continents over at least two decades, share a common ancestor. Furthermore, as the defining phenotype of Cjd is the absence of nitrate reduction, the evidence suggests strongly that Cjd arose from a single evolutionary event, i.e. the napA deletion, with the divergence at napB occurring somewhat later; deletion of the gene encoding the large subunit of nitrate reductase would remove the constraint maintaining a functional small subunit-encoding gene. Obviously, further experiments will be necessary to validate this hypothesis. Such experiments would entail the isolation and characterization of additional Cjd isolates, facilitated by the multiplex PCRs described above. Although the results presented here will need to be investigated further, they nevertheless provide intriguing insights into the origin of this subspecies.
Conclusion
Campylobacter jejuni subsp. doylei strains are isolated infrequently. They are isolated primarily from human clinical patients and often from blood cultures, suggesting that they may be more pathogenic than Cjj strains. However, little is known about the doylei subspecies, especially with regards to the environmental reservoirs for this organism; no animal host has been yet identified for Cjd [20]. Cjj strains are isolated often from avian hosts. The absence of Cjd from avian sources, such as poultry, may be due to the lower maximum growth temperature (37°C) for this subspecies. This lower growth temperature may restrict Cjd strains to other hosts with lower normal body temperatures, such as swine and domestic pets. It may influence also the isolation of Cjd at the clinical level since enrichment and selection of C. jejuni as a whole is normally performed at 42°C, precluding the isolation of most Cjd strains. Also, antibiotics used in typical C. jejuni isolation media may restrict the growth of some Cjd strains.
The multiplex PCR method presented in this study will enhance greatly the identification of Cjd strains from human clinical, veterinary and environmental samples. Since this PCR does not require large numbers of cells or pure cultures, Cjd strains can be identified even when they comprise only a small subset of the overall population. Removing the need for enrichment or selection would reduce the isolation bias that might minimize or eliminate detection of Cjd strains. Finally, we have demonstrated here that PCR identification of Cjd strains can be performed using only the cells coating bacterial storage beads from -80°C freezer stocks. Thus, strains identified only as C. jejuni can be re-assessed rapidly without the necessity of growing the strains out to purity. This was instrumental in identifying, for example, an HS:63 Penner serotype reference strain, identified initially as Cjj, as a Cjd strain; several other strains identified only as C. jejuni may be, in fact, C. jejuni subsp. doylei.
Methods
Bacterial strains, growth conditions and chemicals
Campylobacter strains used in this study are listed in Table 2. All Cjd strains were cultured routinely at 37°C under microaerophilic conditions (5% O2, 10% CO, and 85% N2) on Anaerobe basal agar (ABA; Oxoid, Basingstoke, UK) amended with 5% (v/v) laked horse blood (Hema Resource & Supply, Aurora, OR). All chemicals were purchased from Sigma-Aldrich (St. Louis, MO) or Fisher Scientific (Houston, TX). PCR enzymes and reagents were purchased from New England Biolabs (Beverly, MA) or Epicentre (Madison, WI). DNA sequencing chemicals and capillaries were purchased from Applied Biosystems (Foster City, CA). Sequencing and PCR oligonucleotides were purchased from MWG-Biotech (High Point, NC).
Phenotypic characterization of C. jejuni subsp. doylei strains
To determine nitrate reduction in C. jejuni, nitrate disks and anaerobic nitrate reagents A and B (Remel, Lenexa, KS) were used. Zinc dust was obtained from BioMérieux. Cjj strain NCTC 11168 and Cjd strain ATCC 49350 were used as controls for the procedure. Each Cjd strain was streaked onto ABA agar supplemented with 5% horse blood and grown for 48 h as described above. A nitrate disk was then placed on a thick zone of growth on the plate and incubated for an additional 24 h under the same conditions. The nitrate disk was removed subsequently and placed in a sterile tube. One drop of both reagents A and B were added to the disk. Reduction of nitrate is indicated by a color change (clear to red); if no color change was observed after 3 min, zinc dust was added. A color change prior to the addition of zinc is indicative of Cjj and a color change only after addition of zinc is indicative of Cjd.
Multiplex PCR
Genomic DNA from the Cjd strains in Table 2 was prepared using the Wizard Genomic DNA kit (Promega, Madison, WI) according to the manufacturer's protocols. Additional Cjd genomic DNAs were prepared by boiling single Microbank bacterial storage beads (Pro-Lab, Austin, TX) from freezer stocks for 5 min in 100 μl of 10 mM Tris pH 8.0. Genomic DNA from other Campylobacter strains was prepared as described [21]. nap multiplex PCRs were performed on a Tetrad thermocycler (Bio-Rad, Hercules, CA) with the following settings: 30 s at 94°C; 30 s at 53°C; 2 min at 72°C (30 cycles). Each amplification mixture contained 50 ng genomic DNA, 1× PCR buffer (Epicentre), 1× MasterAmp enhancer (Epicentre), 2.5 mM MgCl2, 250 μM each dNTP, 50 pmol each primer (Table 1), and 1 U Taq polymerase (New England Biolabs). nap/lpxA multiplex PCRs were performed under similar parameters, with a final MgCl2 concentration of 2.0 mM; additionally, 30 pmol of the KK2 primer (Table 1) and 10 pmol of the lpxAC. coli, lpxAC. jejuni, Cl0122 and lpxAC. upsaliensis primers (Table 1) were added to each reaction.
DNA sequencing
Cycle sequencing reactions were performed on a Tetrad thermocycler using the ABI PRISM BigDye terminator cycle sequencing kit (version 3.1) and standard protocols. All extension products were purified on DyeEx 96 well plates (Qiagen). DNA sequencing was performed on an ABI PRISM 3130XL Genetic Analyzer (Applied Biosystems) using the POP-7 polymer and ABI PRISM Genetic Analyzer Data Collection and ABI PRISM Genetic Analyzer Sequencing Analysis software.
GenBank accession numbers
The partial Cjd napA and napB sequences were submitted to GenBank and have the following accession numbers: ATCC 49350 napA: [GenBank:EF218724 ]; ATCC 49350 napB: [GenBank: EF218725]; ATCC 49351 napA: [GenBank: EF218726]; ATCC 49351 napB: [GenBank: EF218727]; RM2095 napA: [GenBank: EF218728]; RM2095 napB: [GenBank: EF218729]; RM2096 napA: [GenBank: EF218730]; RM2096 napB: [GenBank: EF218731]; RM3782 napA: [GenBank: EF218732]: RM3782 napB: [GenBank: EF218733]; SSI 5384 napA: [GenBank: EF218734]; SSI 5384 napB: [GenBank: EF218735]; CCUG 18266 napA: [GenBank: EF218736]; CCUG 18266 napB: [GenBank: EF218737]; 269.97 napA: [GenBank: EF218738]; 269.97 napB: [GenBank: EF218739].
References
Friedman CR, Neimann J, Wegener HC, Tauxe RV: Epidemiology of Campylobacter jejuni infections in the United States and other industrialized nations. Campylobacter. Edited by: Nachamkin I, Blaser MJ. 2000, Washington, DC: ASM Press, 121-138.
Miller WG, Mandrell RE: Prevalence of Campylobacter in the food and water supply: incidence, outbreaks, isolation and detection. Campylobacter: Molecular and Cellular Biology. Edited by: Konkel ME, Ketley JM. 2005, Norwich, UK: Horizon Scientific Press, 101-163.
Lastovica AJ: Emerging Campylobacter spp.: the Tip of the Iceberg. Clin Microbiol News. 2006, 28 (7): 49-56. 10.1016/j.clinmicnews.2006.03.004.
Kasper G, Dickgiesser N: Isolation from gastric epithelium of Campylobacter-like bacteria that are distinct from Campylobacter pyloridis". Lancet. 1985, 1 (8420): 111-112. 10.1016/S0140-6736(85)92005-7.
Steele TW, Lanser JA, Sangster N: Nitrate-negative campylobacter-like organisms. Lancet. 1985, 1 (8425): 394-10.1016/S0140-6736(85)91410-2.
Lastovica AJ, Skirrow MB: Clinical significance of Campylobacter and related species other than Campylobacter jejuni and C. coli. Campylobacter. Edited by: Nachamkin I, Blaser MJ. 2000, Washington, DC: ASM Press, 89-120.
Barros-Velazquez J, Jimenez A, Villa TG: Speciation of thermotolerant Campylobacter isolates involved in foodborne disease by means of DNA restriction analysis and molecular probes. J Agric Food Chem. 2002, 50 (22): 6563-6568. 10.1021/jf0204553.
Owen RJ, Beck A, Borman P: Restriction endonuclease digest patterns of chromosomal DNA from nitrate-negative Campylobacter jejuni-like organisms. Eur J Epidemiol. 1985, 1 (4): 281-287. 10.1007/BF00237103.
Taylor DN, Kiehlbauch JA, Tee W, Pitarangsi C, Echeverria P: Isolation of group 2 aerotolerant Campylobacter species from Thai children with diarrhea. J Infect Dis. 1991, 163 (5): 1062-1067.
Jimenez A, Velaquez JB, Rodriguez J, Chomon B, Villa TG: Biotyping of Campylobacter jejuni and Campylobacter coli infections in Spain. J Infect. 1994, 29 (3): 305-310. 10.1016/S0163-4453(94)91243-2.
Musmanno RA, Russi M, Figura N, Guglielmetti P, Zanchi A, Signori R, Rossolini A: Unusual species of campylobacters isolated in the Siena Tuscany area, Italy. New Microbiol. 1998, 21 (1): 15-22.
Morey F: Five years of Campylobacter bacteremia in central Australia. Campylobacters, Helicobacters, and Related Organisms. Edited by: Newell DG, Ketley JM, Feldman RA. 1996, New York, NY: Plenum Press, 491-494.
Duim B, Vandamme PA, Rigter A, Laevens S, Dijkstra JR, Wagenaar JA: Differentiation of Campylobacter species by AFLP fingerprinting. Microbiology. 2001, 147 (Pt 10): 2729-2737.
On SL, Harrington CS: Identification of taxonomic and epidemiological relationships among Campylobacter species by numerical analysis of AFLP profiles. FEMS Microbiol Lett. 2000, 193 (1): 161-169. 10.1111/j.1574-6968.2000.tb09419.x.
Christensen H, Jorgensen K, Olsen JE: Differentiation of Campylobacter coli and C. jejuni by length and DNA sequence of the 16S-23S rRNA internal spacer region. Microbiology. 1999, 145: 99-105.
Lorenz E, Lastovica A, Owen RJ: Subtyping of Campylobacter jejuni Penner serotypes 9, 38 and 63 from human infections, animals and water by pulsed field gel electrophoresis and flagellin gene analysis. Lett Appl Microbiol. 1998, 26: 179-182. 10.1046/j.1472-765X.1998.00311.x.
Klena JD, Parker CT, Knibb K, Ibbitt JC, Devane PM, Horn ST, Miller WG, Konkel ME: Differentiation of Campylobacter coli, Campylobacter jejuni, Campylobacter lari, and Campylobacter upsaliensis by a multiplex PCR developed from the nucleotide sequence of the lipid A gene lpxA. J Clin Microbiol. 2004, 42 (12): 5549-5557. 10.1128/JCM.42.12.5549-5557.2004.
Englen MD, Fedorka-Cray PJ: Evaluation of a commercial diagnostic PCR for the identification of Campylobacter jejuni and Campylobacter coli. Lett Appl Microbiol. 2002, 35 (4): 353-356. 10.1046/j.1472-765X.2002.01193.x.
Miller WG, Bates AH, Horn ST, Brandl MT, Wachtel MR, Mandrell RE: Detection on surfaces and in Caco-2 cells of Campylobacter jejuni cells transformed with new gfp, yfp, and cfp marker plasmids. Appl Environ Microbiol. 2000, 66 (12): 5426-5436. 10.1128/AEM.66.12.5426-5436.2000.
On SLW: Taxonomy, phylogeny, and methods for the identification of Campylobacter species. Campylobacter: Molecular and Cellular Biology. Edited by: Ketley JM, Konkel ME. 2005, Norfolk, UK: Horizon Scientific Press, 13-42.
Miller WG, Pearson BM, Wells JM, Parker CT, Kapitonov VV, Mandrell RE: Diversity within the Campylobacter jejuni type I restriction-modification loci. Microbiology. 2005, 151 (Pt 2): 337-351. 10.1099/mic.0.27327-0.
Parkhill J, Wren BW, Mungall K, Ketley JM, Churcher C, Basham D, Chillingworth T, Davies RM, Feltwell T, Holroyd S: The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature. 2000, 403 (6770): 665-668. 10.1038/35001088.
Black RE, Levine MM, Clements ML, Hughes TP, Blaser MJ: Experimental Campylobacter jejuni infection in humans. J Infect Dis. 1988, 157 (3): 472-479.
Korlath JA, Osterholm MT, Judy LA, Forfang JC, Robinson RA: A point-source outbreak of campylobacteriosis associated with consumption of raw milk. J Infect Dis. 1985, 152 (3): 592-596.
Steele TW, Owen RJ: Campylobacter jejuni subsp. doylei subsp. nov., a subspecies of nitrate-negative campylobacters isolated from human clinical specimens. Int J Syst Bacteriol. 1988, 38: 316-318.
Fouts DE, Mongodin EF, Mandrell RE, Miller WG, Rasko DA, Ravel J, Brinkac LM, Deboy RT, Parker CT, Daugherty SC: Major structural differences and novel potential virulence mechanisms from the genomes of multiple campylobacter species. PLoS Biol. 2005, 3 (1): e15-10.1371/journal.pbio.0030015.
Acknowledgements
This work was supported by the United States Department of Agriculture, Agricultural Research Service CRIS project 5325-42000-045, and supports a U.S. collaboration in the European Commission Fifth Framework Project QLK1-CT-2002-0220, "CAMPYCHECK". AJL is indebted to the South African Medical Research Council for financial assistance.
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
WGM and AJL designed the research project. WGM designed the nap mpx1 and mpx2 multiplex primer sets and was the principal author of the manuscript. CTP constructed the C. jejuni DNA microarray and performed the microarray experiments and analysis. SH performed all of the multiplex amplifications and sequenced all of the napA and napB amplicons. AJL collected the 21 South African Cjd strains and performed the initial nitrate reductase assays on these isolates. All authors approved and read the final manuscript.
Electronic supplementary material
12866_2006_328_MOESM1_ESM.pdf
Additional File 1: Table S1. Campylobacter strains used to validate the nap multiplex PCR assay. The 321 Campylobacter strains used in the nap multiplex PCR validation are briefly described, including source/location of isolation and serotype, where available. Additionally, for each strain, the amplicon sizes (in bp, when present) for the nap and lpxA multiplex PCRs are provided. (PDF 51 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Miller, W.G., Parker, C.T., Heath, S. et al. Identification of genomic differences between Campylobacter jejuni subsp. jejuni and C. jejuni subsp. doylei at the nap locus leads to the development of a C. jejuni subspeciation multiplex PCR method. BMC Microbiol 7, 11 (2007). https://doi.org/10.1186/1471-2180-7-11
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2180-7-11