Abstract
Background
The selection pressure imposed by the parasite has a functional consequence on the immune genes, leading to altered immune function in which regulatory T cells (Tregs) induced by parasites during infectious challenges modulate or thwart T effector cell mechanism.
Methods
We identified and investigated regulatory polymorphisms in the immune gene IL2 and its receptor IL2R alpha (also known as CD25) in Gabonese individuals exposed to plentiful parasitic infections.
Results
We identified two reported variants each for IL2 and its receptor IL2R alpha gene loci. Also identified were two novel variants, -83 /-84 CT deletions (ss410961576) for IL2 and -409C/T (ss410961577) for IL2R alpha. We further validated all identified promoter variants for their allelic gene expression using transient transfection assays. Three promoter variants of the IL2 locus revealed no significant expression of the reporter gene. The identified novel variant (ss410961577C/T) of the IL2R alpha revealed a significant higher expression of the reporter gene in comparison to the major allele (P<0.05). In addition, the rs12722616C/T variant of the IL2R alpha locus altered the transcription factor binding site TBP (TATA box binding protein) and C/EBP beta (CCAAT/enhancer binding protein beta) that are believed to regulate the Treg function.
Conclusions
The identification and validation of such regulatory polymorphisms in the immune genes may provide a basis for future studies on parasite susceptibility in a population where T cell functions are compromised.
Similar content being viewed by others
Background
Parasites increase their survival rate in the host by means of a complex interaction with the host immune system. However understanding such interaction on the part of the host and parasite during infections still remains a fundamental issue. In such processes, it is believed that the host regulatory T (Tregs) cells play an essential role [1, 2]. The parasite induces a regulatory T cell (Tregs) population that can modulate the magnitude of effector T cell functions thereby leading to a subtle immune response during infections [3]. The regulatory T cell populations remain diverse; a few of them are induced during infections while the others are considered to be natural Tregs vitally implicated in averting autoimmunity [4]. Tregs are believed to influence host inflammatory and immune responses via mechanisms of cell-to-cell contact, inhibitory cytokines and cytokine deprivation [3]. Pathogen driven selection operating on the host immune genes can impose a nucleotide variation in the primed sequence whereby substantial changes in gene expression is directed [2]. Human gene expression is a controlled transcriptional process in the promoter region of a given gene and is regulated by cis-acting DNA sequence elements. Any nucleotide alteration in the promoter region is likely to alter the gene expression, reflecting the level of susceptibility to a parasitic infection as well as Treg expression [5]. A number of loci are known to be associated with Treg activity. Genes such as IL10, IL13, STAT6, TNFRSF18, TLRs and FOXP3 have been as key players in regulating Tregs [2, 4–7]. One such gene of interest is the interleukin 2 (IL2) and its receptor IL2R alpha (CD25) that are known to modulate the proliferation and differentiation of T cells and are essential for peripheral homeostasis of the CD4+CD25+ Tregs [8].
The human IL2 is located on the q arm of the chromosome 4 (specifically 4q27) and its receptor IL2R alpha (CD25) maps to the p arm of the chromosome 10 (specifically 10p15.1). The human IL-2 is primarily produced by T cells in response to antigenic stimulation and is a major mediator of the immune response [9]. Studies have demonstrated that IL-2 is essential for the proliferation and maintenance of Tregs and can disrupt Treg homeostasis [10, 11], whereas the IL-2R plays a significant role in Treg differentiation and proliferation [12]. Tregs have been shown to constitutively express IL-2R (CD25), allowing Tregs to respond to low levels of IL-2 produced by conventional CD4+ T cells [10, 11]. The removal of IL-2 from activated T cells can lead to a deprivation of cells, which is indicated by studies on IL2-deficient mice [13]. The IL-2 receptor has three chains, α, β and γ, which constitute the high affinity IL2 receptor. The IL2R alpha (CD25) is responsible for activating the IL2 signaling complex and regulates the signal transduction [14, 15]. The IL2-R alpha subunit forms the largest of the three IL-2/IL2-R interfaces. Association of α chain with the β and γ heterodimer creates a receptor with a much higher affinity for IL-2 than the β and γ chains receptor [16]. Antigen recognition by the T cell receptor induces the synthesis or activation of the transcription factors such as NFAT, AP-1, and NFκB, which are located in the promoter region of the IL2 gene and are essential for activating its transcription [17]. Studies have pointed to the single nucleotide polymorphisms (SNPs) located within the upstream −10 kb of the IL2 gene that includes the promoter region, and possibly even beyond, thereby contributing to IL2 transcriptional properties in vivo[18]. The inhibition of IL2R alpha (CD25) during thymocyte differentiation is related to IL2R alpha promoter after response to pre-TCR signals and is essential for the specific response of mature T cells later on [19]. Additionally studies have shown that SNPs within IL2R alpha are associated with both Grave’s disease and Type1 diabetes [20, 21]. Reports have indicated that polymorphisms in the genes encoding IL2 are associated with ulcerative colitis, inflammatory bowel disorder, rheumatoid arthritis and Behcet’s disease [22–25], whereas the receptor of IL2, the IL2R alpha variants were associated with type I diabetes and multiple sclerosis [21, 26, 27].
In the current study, our goal is to identify regulatory single nucleotide polymorphisms (SNPs) in the promoter region of the IL2 and its receptor IL2R alpha (CD25) gene loci in a Sub-Saharan African population exposed to a wide array of parasitic diseases. In this study, with a view to identifying regulatory SNPs in the promoter regions of the IL2 and its receptor IL2R alpha (CD25), we sequenced the promoter region of such genes as were upstream of the transcriptional start site, using samples from 40 unrelated Gabonese individuals. The identified regulatory SNPs were further validated for their allelic gene expression, which may possibly be correlated with various physiological responses.
Methods
Genomic DNA Isolation
Forty DNA samples were collected from unrelated Gabonese individuals and informed written consent for participation in the study was obtained from all participants. Blood samples were collected from adult male patients with uncomplicated malaria at the Medical Research Unit of the Albert Schweitzer Hospital, Lambaréné, Gabon, between August and November 2004 [28]. All uncomplicated malaria individuals were male adults living in malaria-endemic countries; usually such individuals will not show symptoms of malaria although carrying parasites in their blood. The study subjects represent another cohort where we investigated novel chemotherapy against malaria [29]. To avoid complications arising from unnoticed pregnancy, only males were chosen. The study was approved by the local ethics committee of the International Foundation of the Albert Schweitzer Hospital. Genomic DNA from whole blood was isolated using QIAamp DNA Blood Mini kit (Qiagen, Hilden, Germany).
Sequencing and SNP identification
For purposes of sequencing analysis, gene and genomic sequences of the IL2 (NM_000586) and IL2R alpha (CD25) (NM_000417) were obtained from the SNPper database [http://snpper.chip.org/]. PCR primers were designed to amplify the promoter region of the gene using the PRIMER3 Software [http:// www genome.wi.mit.edu/cgi-bin/primer/primer3_http://www.cgi]. The promoter regions of the human IL2 and IL2R alpha (CD25) genes were amplified by polymerase chain reaction (PCR). The primer pairs employed for amplifying promoter regions of the IL2 were IL2F: 5′-TAAATAAGGCCATAGAATGG-3′ and IL2R: 5′- GTTACATTAGCCCACACTTA -3′. The primer pairs employed to amplify promoter regions of IL2R alpha were IL2RF: 5′- GATCCACCCACCTTGGTCTA -3′ and IL2RR: 5′-GGCAGCCAGGCACCATGATGAAC -3′ (MWG Operon, Germany). In brief: PCR were carried out in a 20 μl reaction volumes with 5ng of genomic DNA, 1x PCR buffer (20 mM Tris–HCl pH 8.4, 50 mM KCl, 1.5 mM of MgCl2; Qiagen), 0.125mM of dNTPs, 0.5mM of each primer and 1 U Taq DNA polymerase (Qiagen, Hilden, Germany) on a PTC-200 Thermal cycler (MJ Research, USA). Thermal cycling parameters for the amplification of both IL2 and IL2R alpha were as follows: initial denaturation at 94°C for 5min, followed by 40 cycles of 15 sec at 94°C denaturation, 60 sec at 60°C annealing temperature, 60 sec at 72°C extension, followed by a final extension of 10 min at 72°C. PCR products were cleaned up using Exo-SAP-IT (USB, Affymetrix, USA) and 1μl of the purified product was directly used as templates for sequencing, using the BigDye terminator v. 2.0 cycle sequencing kit (Applied Biosystems, USA) on an ABI 3130 XL DNA sequencer, according to the manufacturer’s instructions. Polymorphisms in the promoter regions were identified by assembling the sequences, the respective reference sequences being obtained from SNPper database (http://snpper.chip.org) using Codon code Aligner 4.0 software (http://www.codoncode.com/) and were then reconfirmed visually from their respective electropherograms.
Cloning and construct preparation
The SNPs identified in the promoter regions had their polymorphism status reconfirmed for their polymorphism status by subsequent cloning procedures. Those genomic DNA sequences identified as having SNPs in the promoter regions of the IL2 and IL2R alpha were amplified with infusion primers (flanks a 15bp homology to the linearised pGL3 vector) and were then cloned to a linearised pGL3 basic vector. In brief, PCR amplifications were carried out in 50 μl reaction volumes under the same program conditions as mentioned above. The amplified PCR products was analyzed by electrophoresis in 1.5% agarose gels, using a 100 bp DNA ladder molecular size marker (Invitrogen, Karlsruhe, Germany) and PCR -products were gel eluted and purified using a Nucleospin kit (Macherey-Nagel, Düren, Germany) before being cloned to the pGL3 basic vector using Infusion advantage PCR cloning kit (Clonetech, Mounatain view, CA). Plasmids were isolated using QIAprep® Spin Miniprep Kit (Qiagen, Hilden, Germany). To ensure accuracy of the sequenced promoter regions, several independent plasmids containing inserts were sequenced in both directions using appropriate primer pairs. The plasmid exhibiting the confirmed polymorphism was transformed into one shot E. coli (Invitrogen, Karlsruhe, Germany). Two independent colonies were selected from these transformations and maxi prep was performed using Endofree plasmid maxi kit (Qiagen, Hilden, Germany).
Transient transfection assays
We tested the activities of the observed polymorphic promoters for both IL2 and IL2R alpha using Jurkat T cell lines. Basically, four independent transfection experiments for each construct in duplicates were performed with Jurkat T cells (DSMZ, Braunschweig, Germany). Jurkat T cells (0.8X10 6 cells/μl) were grown in a RPMI 1640 (Sigma-Aldrich, Hamburg, Germany) supplemented medium containing 10% FBS, 2mM L-Glutamine and 1% Streptamycin-pencillin substrate (Invitrogen, Karlsruhe, Germany). Jurkat cells (0.8X10 6 cells/μl) were transfected with TransIT reagent (Mirus Bio, Madison, USA) as recommended by the manufacturer. In brief: 120μl of TransIT reagent was added to 3ml of RPMI 1640 serum free medium (Sigma-Aldrich, Hamburg Germany), this being then incubated for 20 min at room temperature. Each of the 24 well plates was then seeded with 0.5ml (0.8X10 6 cells/μl) of Jurkat T cells along with 500ng of plasmid DNA constructs and 20ng of Renilla, before being allowed to incubate for 20 minutes. After incubation, 52μl of TransIT+RPMI serum free medium mix were suspended across each well. The whole procedure was performed in 2X24 well plate formats. After 24 hours, one plate was induced with 20ng/ml Phorbol 12-myristate 13-acetate (PMA, Sigma-Aldrich, Hamburg, Germany) as well as 25μg/ml Concanavalin A (Sigma-Aldrich, Hamburg, Germany). After 24 hours cells were harvested by centrifugation, washed twice with phosphate-buffered saline and lysed in 100μl of 1x passive lysis buffer (Promega, Mannheim, Germany). After incubation for 20 min at room temperature on a rocking platform, 10μl of the lysate was used to measure luciferase activity in the SIRIUS luminometer (Berthold detection system, Pforzheim, Germany). We employed the dual luciferase reporter assay system (Promega, Mannheim, Germany). For each experiment, a plasmid expressing constitutively Renilla luciferase in low amounts was used as a positive control [30]; while a promoterless plasmid (pGL3 basic) was integrated as a negative control. Each construct was measured 8 times, then both stimulated and non stimulated with two different DNA preparations. Relative luciferase activity was calculated as luciferase firefly/luciferase Renilla multiplied by 1000.
Transcription factor binding search
An extensive search for transcription factor binding sites for the observed SNPs in the promoter region was performed using a TF-Search online tool (http://www.gene-regulation.com/pub/programs/alibaba2) that utilizes TRANSFAC 4.0.
Statistical analysis
Data were normalized and have been analyzed by StatView (http://www.statview.com). The mean ratio (Luciferase/Renilla) across all measurements was considered for purposes of calculating relative luciferase activity. The luciferase activity of the two different IL2 and IL2R alpha (CD25) promoter variants was comparatively analyzed by a t-test (before and after stimulation). In addition, each constructs activity was compared to the activity of the major allele (common alleles) in both an induced and a non induced state. The statistically significant level was set as 0.05.
Results
All forty subjects were sequenced for the entire promoter region of the IL2 and IL2R alpha (CD25) and were investigated for described and novel SNPs. For the IL2 promoter region, two predescribed SNPs (rs2069762T/G and rs2067006T/A) were observed. In addition we observed a novel CT deletion at the position −83/-84 in one individual. This novel SNP was submitted to the SNPper database and a corresponding ID was obtained (#ss410961576). The corresponding references SNP ID (#rs); observed allele frequencies from investigated IL2 gene are summarized in Table 1. In order to characterize these variants in terms of function, a transient transfection assay were performed using Jurkat T cells. The activity of the three different IL2 promoter variants analyzed by luciferase activities is compared in Figure 1. The p-values of the two observed SNP constructs of the IL2 variant remained insignificant in comparison to the major allele before and after stimulation (P > 0.05). When explicitly examined for a transcription factor binding site in the observed variants of IL2, the rs2067006T/A variant was found to be positioned on the transcription site ETS-1, which is a member of the ETS (E-twenty six) family of transcription factors that function as transcriptional activators or repressors in numerous genes believed to be involved in stem cell development, cell senescence, and death. The other two variants (#ss410961576CTdel and rs2069762T/G) do not alter any putative transcription factor binding site.
Comparison of the activity of the three identified IL2 promoter variants analyzed by luciferase activities. The ratio of the relative light units (firefly/renilla) is given. P values are calculated by t-Test from four different experiments performed in duplicates with two different DNA preparations. Each construct was induced by PMA and ConA. The letters indicate the presence of mutations T/G, CT del and T/A.
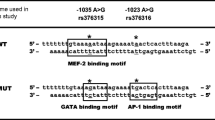
For the IL2R alpha (CD25), two described variants in the promoter region (rs12722617C/T and rs12722616C/T) were observed. In addition we observed a novel C/T variant at the position −409 in two individuals. This novel SNP was submitted to the SNPper database and a corresponding ID was obtained (#ss410961577). The corresponding references SNP ID (#rs); observed allele frequencies from investigated IL2R alpha (CD25) are summarized in Table 2. The activity of the three different IL2 promoter variants analyzed by luciferase activities is compared in Figure 2. The variant rs12722617C/T and rs12722616C/T were observed to be in absolute linkage in all individuals analyzed. The p-values of the SNP construct (rs12722617C/T + rs12722616C/T) of the IL2R alpha variant remained insignificant in comparison to the major allele before and after induction (P >0.05), but remained significant when compared to the new variant in a stimulated state (P<0.0005). The observed novel variant #ss410961577C/T showed significantly increased activity in comparison to the major allele (P<0.05). When looked in explicit for transcription factor binding site in the observed variants of IL2R alpha (CD25) variants, two variants (rs12722617C/T and #ss410961577C/T) were found not to alter any putative transcription factor binding site. However the SNP variant (rs12722616C/T) which was in linkage disequilibrium (LD) with rs12722617C/T is positioned at the transcription factor binding site TBP (TATA box binding protein) and C/EBP beta (CCAAT/enhancer binding protein beta).
The activity of the three identified IL2R alpha promoter variants as compared by luciferase activities. The ratio of the relative light units (firefly/renilla) is given. P values are calculated by t-Test in four different experiments performed in duplicates with two different DNA preparations. Each construct was induced by PMA and ConA. The letters indicate the presence of polymorphic variants.
Discussion
The underlying idea of the current study is to understand how regulatory SNPs in populations that are naturally exposed to array of parasites contribute to immune outcome. A significant impact on the diversity among immune gene families is believed to be attributable to invasion maneuvers performed by the parasites. Therefore variations in the promoter regions of these immune system genes can potentially amend the gene expression levels either by changing specificity of transcription binding sites or by altering the kinetics of transcription initiation [7]. In our current study, we identified two known SNPs (rs2069762T/G and rs2067006T/A) and one novel CT deletion (#ss410961576) at the position −83/-84 when screened for IL2 promoter region. None of the investigated SNP constructs revealed differential luciferase activity compared to the major allele. In the current investigation the IL2 variant (rs2067006T/A) was considered to be the major allele, as all the individuals were found to harbor this particular variant. When compared to the NCBI HapMap database, the frequencies of the observed SNP variants (rs2067006T/A and rs2069762T/G of IL2) were in accordance with Yoruba population of Nigeria which represents a Sub Saharan African group. The rs2067006T/A variant is positioned on to the transcription site ETS-1, a member of the ETS (E-twenty six) family of transcription factors. The ETS-1 DNA-binding domain recognizes the core consensus DNA in target genes and acts either as transcriptional activators or repressors [31]. Recent studies have demonstrated that ETS-1 belongs to a large protein complex which binds to the Treg-specific demethylated region (TSDR) in the FOXP3 locus, thereby restricting the stable FOXP3 expression in the Tregs [32]. FOXP3 is described as a master regulator of natural Tregs development and function [4], while mature regulatory T cells expressing a non-functional fusion protein of FOXP3 lack any suppressor function [33]. Since all the investigated individuals inherited this variant, we believe that the studied population may have a restricted FOXP3 expression leading to a subtle T effector cell function.
In our study we also identified two known SNPs (rs12722617C/T and rs12722616C/T) along with one novel C/T variant (#ss410961577) at the position −516 when screened for the IL2R alpha (CD25) promoter region of the IL2R alpha. Both the predescribed SNP variants of the IL2R alpha (rs12722617C/T and rs12722616C/T) were observed in linkage in all studied individuals. We did not observe a differential luciferase activity compared to the major allele for the construct (rs12722617C/T + rs12722616C/T). However the identified novel variant (#ss410961577C/T) revealed significant increased activity in comparison to the constructs with the major allele. This particular novel variant did not alter any putative transcription factor binding site. However the variant rs12722616C/T which was in LD with rs12722617C/T altered the transcription factor binding site TBP (TATA box binding protein) and C/EBP beta (CCAAT/enhancer binding protein beta). The TATA box is a type of promoter sequence that indicates the transcriptional start site whereby a genetic sequence can be read and decoded. It is named after the conserved DNA sequence TATAAA. TBP (TATA binding protein) together with TATA associated factors (TAFS) make the transcription factor TFIID which binds the TATA box in combination with other transcription factors (TFIIB, TFIIA, TFIIE, TFIIF, TFIIH) [34]. All the above transcription factors along with RNA polymerase II enzyme form a transcription initiation complex. Therefore it is possible that the TATA box in the construct rs12722616C/T ma contributes to the transcription initiation in vitro in the Jurkat cells. The other transcription factor C/EBP beta (CCAAT/enhancer binding protein beta) is believed to modulate inflammatory processes. A recent study has demonstrated in an experimental autoimmune encephalomyelitis model that C/EBP expression by dendritic cells (DC) influences Th17 versus Treg differentiation but has little or no impact upon Th1 development [35]. Examining the HapMap database, all the frequencies of reported variants of the IL2R alpha loci were found to be in accordance with the frequencies in the Yoruba population.
Human IL2 and its receptor IL2R alpha are described as constituting a potent T cell growth factor and are mainly produced by activated CD4+ T cells, though also by naïve CD8+ T cells, dendritic cells, and thymic cells [36]. Human IL-2 is considered to be vital for the development of CD4+CD25+ regulatory T cells [37]. Also studies have demonstrated that IL-2 also potentially plays a role in the thymic development of Tregs [38, 39]. Studies have shown that polymorphisms in the IL2 are associated with many diseases including auto immune diseases. Recent studies have demonstrated that the studied IL2 promoter variant (rs2069762T/G) is associated with Helicobacter pylori infection [40], multiple sclerosis [41], pathogenesis of childhood lymphoma [42], pathogenesis of new-onset diabetes after transplantation (NODAT) [43], measles-specific cellular immunity [44], a higher risk of acute rejection episodes during kidney transplantation [45] and in allergic disorders [46]. However, the role of the described and novel variants of IL2 and IL2R alpha in this study needs to be validated in terms of the specific role these play in different parasitic diseases.
Conclusions
In summary, the regulatory SNPs identified in this current study will provide useful information for understanding the relevance of sequence polymorphisms in populations exposed to many parasitic diseases and may serve as a basis for backup studies examining disease susceptibility.
References
Belkaid Y, Rouse BT: Natural regulatory T cells in infectious disease. Nat Immunol. 2005, 6: 353-360.
Maizels RM: Parasite immunomodulation and polymorphisms of the immune system. J Biol. 2009, 8: 62-10.1186/jbiol166.
Velavan TP, Ojurongbe O: Regulatory T cells and parasites. J Biomed Biotechnol. 2011, 201 (1): 520940-
Hanel SA, Velavan TP, Kremsner PG, Kun JF: Novel and functional regulatory SNPs in the promoter region of FOXP3 gene in a Gabonese population. Immunogenetics. 2011, 63: 409-415. 10.1007/s00251-011-0524-x.
Velavan TP, Buyukyazici B, Kremsner PG, Kun JF: Combined promoter haplotypes of the IL10R genes are associated with protection against severe malaria in Gabonese children. Immunogenetics. 2012, 64: 87-95. 10.1007/s00251-011-0563-3.
Velavan TP, Bechlars S, Huang X, Kremsner PG, Kun JF: Novel regulatory SNPs in the promoter region of the TNFRSF18 gene in a Gabonese population. Braz J Med Biol Res. 2011, 44: 418-420. 10.1590/S0100-879X2011007500036.
Velavan TP, Bechlars S, Tomiuk J, Kremsner PG, Kun JF: Molecular characterization of regulatory polymorphisms in the promoter region of the STAT6 gene in a Gabonese population. Mem Inst Oswaldo Cruz. 2011, 106: 65-69. 10.1590/S0074-02762011000100011.
Bayer AL, Yu A, Adeegbe D, Malek TR: Essential role for interleukin-2 for CD4(+)CD25(+) T regulatory cell development during the neonatal period. J Exp Med. 2005, 201: 769-777. 10.1084/jem.20041179.
Zou T, Satake A, Corbo-Rodgers E, Schmidt AM, Farrar MA, Maltzman JS, et al: Cutting edge: IL-2 signals determine the degree of TCR signaling necessary to support regulatory T cell proliferation in vivo. J Immunol. 2012, 189: 28-32. 10.4049/jimmunol.1200507.
Cheng G, Yu A, Malek TR: T-cell tolerance and the multi-functional role of IL-2R signaling in T-regulatory cells. Immunol Rev. 2011, 241: 63-76. 10.1111/j.1600-065X.2011.01004.x.
Setoguchi R, Hori S, Takahashi T, Sakaguchi S: Homeostatic maintenance of natural Foxp3(+) CD25(+) CD4(+) regulatory T cells by interleukin (IL)-2 and induction of autoimmune disease by IL-2 neutralization. J Exp Med. 2005, 201: 723-735. 10.1084/jem.20041982.
Webster KE, Walters S, Kohler RE, Mrkvan T, Boyman O, Surh CD, et al: In vivo expansion of T reg cells with IL-2-mAb complexes: induction of resistance to EAE and long-term acceptance of islet allografts without immunosuppression. J Exp Med. 2009, 206: 751-760. 10.1084/jem.20082824.
Reya T, Bassiri H, Biancaniello R, Carding SR: Thymic stromal-cell abnormalities and dysregulated T-cell development in IL-2-deficient mice. Dev Immunol. 1998, 5: 287-302. 10.1155/1998/19567.
Jonuleit H, Schmitt E, Stassen M, Tuettenberg A, Knop J, Enk AH: Identification and functional characterization of human CD4(+)CD25(+) T cells with regulatory properties isolated from peripheral blood. J Exp Med. 2001, 193: 1285-1294. 10.1084/jem.193.11.1285.
Sakaguchi S, Sakaguchi N, Asano M, Itoh M, Toda M: Immunologic self-tolerance maintained by activated T cells expressing IL-2 receptor alpha-chains (CD25). Breakdown of a single mechanism of self-tolerance causes various autoimmune diseases. J Immunol. 1995, 155: 1151-1164.
Grant AJ, Roessler E, Ju G, Tsudo M, Sugamura K, Waldmann TA: The interleukin 2 receptor (IL-2R): the IL-2R alpha subunit alters the function of the IL-2R beta subunit to enhance IL-2 binding and signaling by mechanisms that do not require binding of IL-2 to IL-2R alpha subunit. Proc Natl Acad Sci U S A. 1992, 89: 2165-2169. 10.1073/pnas.89.6.2165.
Attema JL, Reeves R, Murray V, Levichkin I, Temple MD, Tremethick DJ, et al: The human IL-2 gene promoter can assemble a positioned nucleosome that becomes remodeled upon T cell activation. J Immunol. 2002, 169: 2466-2476.
Wang J, Wicker LS, Santamaria P: IL-2 and its high-affinity receptor: genetic control of immunoregulation and autoimmunity. Semin Immunol. 2009, 21: 363-371. 10.1016/j.smim.2009.04.004.
Yeh JH, Spicuglia S, Kumar S, Sanchez-Sevilla A, Ferrier P, Imbert J: Control of IL-2Ralpha gene expression: structural changes within the proximal enhancer/core promoter during T-cell development. Nucleic Acids Res. 2002, 30: 1944-1951. 10.1093/nar/30.9.1944.
Brand OJ, Lowe CE, Heward JM, Franklyn JA, Cooper JD, Todd JA, et al: Association of the interleukin-2 receptor alpha (IL-2Ralpha)/CD25 gene region with Graves' disease using a multilocus test and tag SNPs. Clin Endocrinol (Oxf). 2007, 66: 508-512.
Lowe CE, Cooper JD, Brusko T, Walker NM, Smyth DJ, Bailey R, et al: Large-scale genetic fine mapping and genotype-phenotype associations implicate polymorphism in the IL2RA region in type 1 diabetes. Nat Genet. 2007, 39: 1074-1082. 10.1038/ng2102.
Bowes J, Ho P, Flynn E, Ali F, Marzo-Ortega H, Coates LC, et al: Comprehensive assessment of rheumatoid arthritis susceptibility loci in a large psoriatic arthritis cohort. Ann Rheum Dis. 2012, 71 (8): 1350-1354. 10.1136/annrheumdis-2011-200802.
Festen EA, Goyette P, Scott R, Annese V, Zhernakova A, Lian J, et al: Genetic variants in the region harbouring IL2/IL21 associated with ulcerative colitis. Gut. 2009, 58: 799-804. 10.1136/gut.2008.166918.
Marquez A, Orozco G, Martinez A, Palomino-Morales R, Fernandez-Arquero M, Mendoza JL, et al: Novel association of the interleukin 2-interleukin 21 region with inflammatory bowel disease. Am J Gastroenterol. 2009, 104: 1968-1975. 10.1038/ajg.2009.224.
Shahram F, Nikoopour E, Rezaei N, Saeedfar K, Ziaei N, Davatchi F, et al: Association of interleukin-2, interleukin-4 and transforming growth factor-beta gene polymorphisms with Behcet’s disease. Clin Exp Rheumatol. 2011, 29: S28-S31.
Alcina A, Fedetz M, Ndagire D, Fernandez O, Leyva L, Guerrero M, et al: IL2RA/CD25 gene polymorphisms: uneven association with multiple sclerosis (MS) and type 1 diabetes (T1D). PLoS One. 2009, 4: e4137-10.1371/journal.pone.0004137.
Klinker MW, Schiller JJ, Magnuson VL, Wang T, Basken J, Veth K, et al: Single-nucleotide polymorphisms in the IL2RA gene are associated with age at diagnosis in late-onset Finnish type 1 diabetes subjects. Immunogenetics. 2010, 62: 101-107. 10.1007/s00251-009-0417-4.
Kreidenweiss A, Kremsner PG, Dietz K, Mordmuller B: In vitro activity of ferroquine (SAR97193) is independent of chloroquine resistance in Plasmodium falciparum. AmJTrop Med Hyg. 2006, 75: 1178-1181.
Supan C, Mombo-Ngoma G, Dal-Bianco MP, Ospina Salazar CL, Issifou S, Mazuir F, et al: Pharmacokinetics of ferroquine, a novel 4-aminoquinoline, in asymptomatic carriers of Plasmodium falciparum infections. Antimicrob Agents Chemother. 2012, 56: 3165-3173. 10.1128/AAC.05359-11.
Juliger S, Bongartz M, Luty AJ, Kremsner PG, Kun JF: Functional analysis of a promoter variant of the gene encoding the interferon-gamma receptor chain I. Immunogenetics. 2003, 54: 675-680.
Shaikhibrahim Z, Wernert N: ETS transcription factors and prostate cancer: the role of the family prototype ETS-1 (review). Int J Oncol. 2012, 40: 1748-1754.
Polansky JK, Schreiber L, Thelemann C, Ludwig L, Kruger M, Baumgrass R, et al: Methylation matters: binding of Ets-1 to the demethylated Foxp3 gene contributes to the stabilization of Foxp3 expression in regulatory T cells. J Mol Med (Berl). 2010, 88: 1029-1040. 10.1007/s00109-010-0642-1.
Lin W, Haribhai D, Relland LM, Truong N, Carlson MR, Williams CB, et al: Regulatory T cell development in the absence of functional Foxp3. Nat Immunol. 2007, 8: 359-368. 10.1038/ni1445.
Maldonado E, Ha I, Cortes P, Weis L, Reinberg D: Factors involved in specific transcription by mammalian RNA polymerase II: role of transcription factors IIA, IID, and IIB during formation of a transcription-competent complex. Mol Cell Biol. 1990, 10: 6335-6347.
Tsai VW, Mohammad MG, Tolhurst O, Breit SN, Sawchenko PE, Brown DA: CCAAT/enhancer binding protein-delta expression by dendritic cells regulates CNS autoimmune inflammatory disease. J Neurosci. 2011, 31: 17612-17621. 10.1523/JNEUROSCI.3449-11.2011.
Nelson BH: IL-2, regulatory T cells, and tolerance. J Immunol. 2004, 172: 3983-3988.
Cheng G, Yuan X, Tsai MS, Podack ER, Yu A, Malek TR: IL-2 receptor signaling is essential for the development of Klrg1+ terminally differentiated T regulatory cells. J Immunol. 2012, 189 (4): 1780-1791. 10.4049/jimmunol.1103768.
Malek TR, Porter BO, Codias EK, Scibelli P, Yu A: Normal lymphoid homeostasis and lack of lethal autoimmunity in mice containing mature T cells with severely impaired IL-2 receptors. J Immunol. 2000, 164: 2905-2914.
Malek TR, Yu A, Vincek V, Scibelli P, Kong L: CD4 regulatory T cells prevent lethal autoimmunity in IL-2Rbeta-deficient mice. Implications for the nonredundant function of IL-2. Immunity. 2002, 17: 167-178. 10.1016/S1074-7613(02)00367-9.
Schmidt HM, Ha DM, Taylor EF, Kovach Z, Goh KL, Fock KM, et al: Variation in human genetic polymorphisms, their association with Helicobacter pylori acquisition and gastric cancer in a multi-ethnic country. J Gastroenterol Hepatol. 2011, 26: 1725-1732. 10.1111/j.1440-1746.2011.06799.x.
Fedetz M, Ndagire D, Fernandez O, Leyva L, Guerrero M, Arnal C, et al: Multiple sclerosis association study with the TENR-IL2-IL21 region in a Spanish population. Tissue Antigens. 2009, 74: 244-247. 10.1111/j.1399-0039.2009.01298.x.
Song N, Han S, Lee KM, Choi JY, Park SK, Jeon S, et al: Genetic Variants in Interleukin-2 and Risk of Lymphoma among Children in Korea. Asian Pac J Cancer Prev. 2012, 13: 621-623. 10.7314/APJCP.2012.13.2.621.
Kim YG, Ihm CG, Lee TW, Lee SH, Jeong KH, Moon JY, et al: Association of genetic polymorphisms of interleukins with new-onset diabetes after transplantation in renal transplantation. Transplantation. 2012, 93: 900-907. 10.1097/TP.0b013e3182497534.
Haralambieva IH, Ovsyannikova IG, Kennedy RB, Vierkant RA, Pankratz VS, Jacobson RM, et al: Associations between single nucleotide polymorphisms and haplotypes in cytokine and cytokine receptor genes and immunity to measles vaccination. Vaccine. 2011, 29: 7883-7895. 10.1016/j.vaccine.2011.08.083.
Park SJ, Yoon YC, Kang SW, Kim TH, Kim YW, Joo H, et al: Impact of IL2 and IL2RB genetic polymorphisms in kidney transplantation. Transplant Proc. 2011, 43: 2383-2387. 10.1016/j.transproceed.2011.06.014.
Braegger CP, Spencer J, MacDonald TT: Ontogenetic aspects of the intestinal immune system in man. Int J Clin Lab Res. 1992, 22: 1-4. 10.1007/BF02591385.
Pre-publication history
The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1471-2350/13/117/prepub
Acknowledgements
We thank Andrea Weierich and Velia Grummes for technical assistance during the study period. The project received funding from a grant of the EU commission –TRANCHI (INCO-CT-2006-032436).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no conflicting interests.
Authors’ contributions
BL designed the field study; VTP and JFJ Kun designed and supervised the experiments; XH and VK performed the experiments; PTS and VTP analyzed the data; BL and VTP contributed materials/analysis tools; XH and VTP wrote the paper. All authors read and approved the final manuscript.
Xiangsheng Huang, Vera Kühne contributed equally to this work.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Huang, X., Kühne, V., Kun, J.F.J. et al. In-vitro characterization of novel and functional regulatory SNPs in the promoter region of IL2 and IL2R alpha in a Gabonese population. BMC Med Genet 13, 117 (2012). https://doi.org/10.1186/1471-2350-13-117
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2350-13-117