Abstract
Background
The selection of individuals based on their predicted breeding values and mating of related individuals can increase the proportion of identical-by-descent alleles. In this context, the objectives of this study were to estimate inbreeding coefficients based on alternative metrics and data sources such as pedigree (FPED), hybrid genomic relationship matrix H (FH), and ROH of different length (FROH); and calculate Pearson correlations between the different metrics in a closed Nellore cattle population selected for body weight adjusted to 378 days of age (W378). In addition to total FROH (all classes) coefficients were also estimated based on the size class of the ROH segments: FROH1 (1–2 Mb), FROH2 (2–4 Mb), FROH3 (4–8 Mb), FROH4 (8–16 Mb), and FROH5 (> 16 Mb), and for each chromosome (FROH_CHR). Furthermore, we assessed the effect of each inbreeding metric on birth weight (BW), body weights adjusted to 210 (W210) and W378, scrotal circumference (SC), and residual feed intake (RFI). We also evaluated the chromosome-specific effects of inbreeding on growth traits.
Results
The correlation between FPED and FROH was 0.60 while between FH and FROH and FH and FPED were 0.69 and 0.61, respectively. The annual rate of inbreeding was 0.16% for FPED, 0.02% for FH, and 0.16% for FROH. A 1% increase in FROH5 resulted in a reduction of up to -1.327 ± 0.495 kg in W210 and W378. Four inbreeding coefficients (FPED, FH, FROH2, and FROH5) had a significant effect on W378, with reductions of up to -3.810 ± 1.753 kg per 1% increase in FROH2. There was an unfavorable effect of FPED on RFI (0.01 ± 0.0002 kg dry matter/day) and of FROH on SC (-0.056 ± 0.022 cm). The FROH_CHR coefficients calculated for BTA3, BTA5, and BTA8 significantly affected the growth traits.
Conclusions
Inbreeding depression was observed for all traits evaluated. However, these effects were greater for the criterion used for selection of the animals (i.e., W378). The increase in the genomic inbreeding was associated with a higher inbreeding depression on the traits evaluated when compared to pedigree-based inbreeding. Genomic information should be used as a tool during mating to optimize control of inbreeding and, consequently, minimize inbreeding depression in Nellore cattle.
Similar content being viewed by others
Background
The selection of breeding candidates based on their breeding values predicted by the Best Linear Unbiased Predictor (BLUP) method [1], has had major impacts on animal productivity [2]. The inclusion of information from relatives combined with individual phenotypic records can increase the accuracy prediction of estimated breeding values (EBVs) but also the probability of co-selection of collateral relatives (families). Consequently, BLUP-based selection can lead to a reduction in genetic variability within populations and an increase in the overall level of inbreeding if genetic relationships are not properly taken into account during the selection and mating processes [3]. In addition, high selection intensity and the use of a small number of breeding animals combined with reproductive biotechnologies (e.g., artificial insemination) can reduce the genetic variability within populations and hence increase the proportion of identical-by-descent alleles [4, 5].
Over the past decades, inbreeding coefficients in commercial populations have been calculated based on pedigree records [6,7,8,9]. The more recent availability of genomic information on a large number of individuals and the genome-wide coverage from genotyping platforms have enabled the calculation of more accurate inbreeding [10]. Genomic inbreeding coefficients can be simultaneously calculated for genotyped and non-genotyped animals based on the diagonal elements of the hybrid genomic relationship matrix (H) [11]. Furthermore, the identification of large homozygous segments likely inherited from a common ancestor, called runs of homozygosity (ROH), can also contribute to the assessment of genomic inbreeding in genotyped animals [12,13,14]. An increasing number of studies have identified ROH in cattle breeds [5, 14,15,16,17,18], as this approach enables the differentiation between old and recent inbreeding events that occurred in populations [19].
The negative effect of increased inbreeding on complex traits (i.e., inbreeding depression) such as growth and fertility, calculated based on pedigree [9, 20] and, more recently, using genomic data [14, 21, 22], has been widely reported in the livestock literature. Inbreeding depression can be estimated based on the regression of phenotypic values for a certain trait on the inbreeding coefficient of the animals [20]. However, this approach estimates the average effect of inbreeding based only on the linear relationship between the inbreeding coefficients and the complex traits and it does not directly account for the allelic content shared among individuals. Thus, inbreeding depression can be more reliably estimated by fitting the inbreeding coefficients as linear covariates in the animal models [14, 22, 23].
With a cattle herd estimated at 202.8 million heads and a slaughter of 42.31 million heads, Brazil is responsible for 27.7% of world beef exports, generating 12.97 billion dollars [24], what highlighting the impact of the beef agro-industrial system in Brazil and its importance to the world. In this scenario, Nellore cattle, originating from India and arriving in Brazil through imports between 1920 and 1963 [25], established themselves as one of the most important breeds for beef cattle farming in Brazil [26].Various studies in Nellore cattle have characterized the distribution of ROH and reported genomic inbreeding coefficients [15, 27], evaluated the population structure and genetic diversity in the breed [28] and assessed the level of pedigree-based inbreeding depression on economically traits of interest in Nellore breeding programs interest [9]. In recent decades, the Institute of Animal Sciences (IZ, Sertaozinho, SP, Brazil) has developed a unique genetic resource: a closed herd composed of three lines of Nellore cattle. The Control line is selected for average W378 (phenotypic), the Selection line is selected for higher W378 (phenotypic), and the Traditional line was selected until 2008 for higher W378 and, from 2008 onwards, for higher EBV for W378 and lower EBV for residual feed intake (RFI) [29,30,31].
Therefore, the primary objectives of this study were to identify and characterize ROH and to estimate inbreeding coefficients based on pedigree (FPED), matrix H (FH), and ROH (FROH) in a closed herd Nellore (Bos taurus indicus) population. In addition, we estimated inbreeding depression on growth, fertility, and feed efficiency traits, as well as the chromosome-specific effects of inbreeding on growth traits in the same population.
Results
Pedigree structure
For the “Total” dataset, a maximum number of 17 generations was established based on the pedigree (Table 1). The minimum values of the maximum number of ancestral generations, ECG, and PCI observed in the “Total” dataset are related to founder animals. Among all animals in the pedigree, 230 sires and 1,072 dams are progenitors of the genotyped animals. The pedigree structure of the genotyped animals indicates a relatively deep and complete pedigree.
Runs of homozygosity (ROH)
Table 2 shows the descriptive statistics of ROH segments on the Nellore population.
Figure 1 shows the distribution of the number of ROH segments per chromosome according to ROH length class. The mean number of ROH segments per chromosome was 5,464 ± 1,897 (CV = 34%). The total number of ROH segments ranged from 9,277 on BTA5 to 2,615 on BTA25, corresponding to 5.8% and 1.6% of the total number of segments, respectively. The five chromosomes with the largest number of ROH segments (BTA5, BTA7, BTA3, BTA12, and BTA1) contained 26% of the total number of ROH segments found in the studied population. The largest number of segments (n = 67,266) was observed for the shortest ROH length class (1–2 Mb). The number of ROH segments found decreased at a rate of 42.64% as the length class increased. In addition, 6,291 ROH segments larger than 16 Mb were detected in 89% of the individuals, which corresponds to about 4% of the total number of ROH segments found.
Inbreeding coefficients
Figure 2 shows the distribution of the inbreeding coefficients of genotyped animals. The average inbreeding of the genotyped animals was 0.039 ± 0.019 for FPED (CV = 48%), 0.041 ± 0.044 for FH (CV = 107%), 0.015 ± 0.005 for FROH1 (CV = 33%), 0.019 ± 0.007 for FROH2 (CV = 36%), 0.030 ± 0.009 for FROH3 (CV = 30%), 0.034 ± 0.014 for FROH4 (CV = 41%), 0.030 ± 0.020 for FROH5 (CV = 66%), and 0.128 ± 0.031 for FROH (CV = 24%). The minimum value for FPED corresponds to founder animals and/or individuals with unknown parents. Regarding FROH by length class, the average values tended to increase with increasing length class.
Distribution of inbreeding coefficients of genotyped animals calculated based on pedigree (FPED), H matrix (FH), runs of homozygosity (ROH) segments of 1–2 Mb (FROH1), 2–4 Mb (FROH2), 4–8 Mb (FROH3), 8–16 Mb (FROH4) and > 16 Mb (FROH5), and ROH segments of all length classes (FROH). The vertical red line indicates the mean value of the distributions
The pedigree-based inbreeding coefficients exhibited the lowest variability among genotyped individuals when compared to the distribution of FH and FROH. Although the mean was close to FPED, the FH values showed greater variability, with a right-skewed unimodal distribution, including negative values. Among the coefficients evaluated, the highest mean value was observed for FROH, which, like FPED, does not present negative values, in addition to a left-skewed distribution related to positive values of high magnitude.
Pearson’s correlations between the eight inbreeding coefficients estimated in genotyped animals are illustrated in Fig. 3. The correlations ranged from − 0.28 between FROH1 and FROH4 to 0.74 between FROH and FROH5. The correlation between FPED and FROH increased with increasing size of the ROH segments, reaching 0.60 between FPED and FROH. A similar trend was observed for the correlations between total FROH and the coefficients calculated according to length class, reaching 0.74 between FROH and FROH5. Furthermore, FH showed a moderate correlation with FPED (0.61) and FROH (0.69).
Pearson’s correlations between the inbreeding coefficients of genotyped animals calculated based on pedigree (FPED), H matrix (FH), runs of homozygosity (ROH) segments of 1–2 Mb (FROH1), 2–4 Mb (FROH2), 4–8 Mb (FROH3), 8–16 Mb (FROH4) and > 16 Mb (FROH5), and ROH segments of all length classes (FROH)
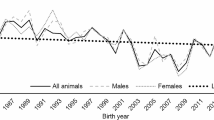
Inbreeding coefficients increased from 2000 onwards (the year from which there was a greater number of genotyped animals), with significant annual rates (P < 0.001) of 0.16% for FPED, 0.02% for FH, and 0.16% for FROH (Fig. 4).
Effect of inbreeding
Table 3 shows the effect of each inbreeding coefficient on the growth, fertility, and feed efficiency traits evaluated only in genotyped animals. At least one inbreeding coefficient had a significant effect on all traits, except RFI. All significant values were negative, indicating a reduction in the phenotypic values of the traits as the inbreeding coefficient of the animal increased (inbreeding depression). Each 1% increase in recent inbreeding (e.g., FROH5) was associated with a decrease of -0.087 ± 0.041 kg in BW. For the W210 trait, inbreeding depression based on FPED, FH, FROH5, and FROH was statistically significant, ranging from − 0.966 ± 0.452 to -0.457 ± 0.183 kg. The selection criterion applied to the population studied, W378, was significantly affected by four inbreeding estimators due to recent (e.g., FROH5) and older (e.g., FROH2) inbreeding, in addition to FROH and FH, with the effect ranging from − 0.966 ± 0.294 to -3.581 ± 1.554 kg. A 1% increase in FROH was associated with − 0.046 ± 0.022 cm reduction in SC.
To maximize the use of existing phenotypic records and increase the statistical power of detection of inbreeding depression on the traits evaluated, the relationship matrices consistent with the inbreeding coefficient metrics were calculated and used in the animal model analyses. This approach enabled the identification of 50% more significant effects of inbreeding coefficients on the traits (Table 4) compared to the previous adjustment method (Table 3).
Five inbreeding coefficients (FPED, FH, FROH2, FROH5 and FROH) had a significant effect on W378, consistently resulting in a mean reduction of 1.891 kg in this trait per 1% increase in inbreeding. The greatest inbreeding depression on W378 (-3.81 ± 1.753 kg) was caused by a 1% increase in FROH2. Furthermore, there was an unfavorable effect of FPED on RFI, with an increase of 0.01 ± 0.0002 kg/DM/day per 1% increase in FPED. Lastly, an unfavorable effect of -0.056 ± 0.022 cm per 1% increase in FROH was observed for SC.
Effect of inbreeding at the chromosome level
Considering chromosomes with the largest number of SNPs within the regions defined as ROH segments (BTA5, BTA1, BTA2, BTA3, BTA7), there was a significant negative effect of FROH_CHR3 and FROH_CHR5 on W210. Furthermore, the inbreeding coefficient from BTA5 negatively affected W378 (Table 5).
Considering the chromosomes with the highest proportion of ROH segments longer than 8 Mb (BTA9, BTA2, BTA8, BTA20, BTA1), there were negative effects of FROH_CHR8 on BW and W210. Among chromosomes with the highest proportions of segments shorter than 8 Mb (BTA5, BTA7, BTA3, BTA12, BTA1), only FROH_CHR3 and FROH_CHR5 presented significant effects on W210 and W378. These two chromosomes were also classified in the group containing the largest number of SNPs within the regions identified as ROH segments.
Discussion
Pedigree structure and runs of homozygosity (ROH)
Although the studied population is closed and was formed from a considerably small number of founder animals [29] the effective population size (Ne) estimated based on the “Total” pedigree is similar to that reported by Santana et al. [28]. These authors found a Ne equal to 100 based on pedigree containing more than 4 million Nellore animals born between 2005 and 2012, with an average of 6.03 complete generations. Intense selection in populations with small Ne contributes to increasing the frequency of long homozygous segments across the genome [32]. In the present study, the mean number of ROH segments per animal was 70.24 ± 14.25, with a mean length of 4.43 ± 5.27 Mb. Peripolli et al. [15], assessing imputed genotypes for HD level (777,962 SNPs), of more than 7,000 Nellore animals, found 55.15 ± 13.01 segments per animal, with a mean length of 3.24 Mb. Studying a different population of the same breed, Zavarez et al. [27] reported a mean ROH length of 4.79 Mb. Although the identification of ROH segments is sensitive to the criteria used for this purpose, the number and length of ROH segments per animal observed in the present study are similar to those reported in studies from other Nellore populations.
The prevalence of shorter ROH segments obtained in the present study (Fig. 1) corroborates with most ROH studies in cattle [17, 21, 33, 34]. It is expected that, due to the various events of meiosis and gene recombination [35], shorter segments are inherited from a distant common ancestor, while longer segments derive from more recent common ancestor [36]. ROH segments longer than 16 Mb were identified in 89% of the animals, indicating recent inbreeding events that occurred approximately three generations ago. Evaluating a Nellore population without restrictions on mating with individuals from other populations, Zavarez et al. [27] reported ROH segments longer than 10 Mb in 74% of the animals. The high incidence of animals with ROH segments longer than 16 Mb observed in the present study reflects the impacts of maintaining a closed population despite the control of kinship between parents during mating and the use of sires for only two breeding seasons (2 years).
Inbreeding coefficients
The mean values of inbreeding vary across populations for different reasons [37], a fact that impairs comparison between studies. However, in most of these studies, genomic inbreeding coefficients were higher than the pedigree-based coefficient [14, 22, 34, 38,39,40]. One exception was reported by Sumreddee et al. [21] for a closed herd of Montana Line 1 Hereford cattle, whose FPED (0.292 ± 0.053) was higher than the FROH (0.229 ± 0.051), probably because of the small number of genotyped animals (n = 797) in proportion to the total number of animals in the pedigree (n = 10,186).
In the present study, compared to the genomic coefficients, FPED showed the lowest variability (Fig. 2) because of the assumption of the pedigree-based estimator that animals with the same degree of kinship share the same proportion of alleles identical by descent [41]. Considering the finite size of the genome, pedigree-based coefficients represent only an estimate of true inbreeding [42, 43] and the combination of pedigree and genomic information enables better prediction of future inbreeding during matings [44, 45].
As shown in Fig. 2, the distribution of FH was right-skewed, with negative values being associated with animals that are more heterozygous than the population average [46]. Although the mean FH (0.041) was similar to the mean FPED (0.039), FH and FROH showed greater variability. Comprehensive genomic coverage of genetic markers enables the prediction of inbreeding using the true degree of identical-by-descent sharing between relatives [10]. Thus, animals with the same degree of kinship can have different inbreeding coefficients. Recent literature reviews indicated FROH as the main approach to estimate inbreeding in cattle [13, 47]. The mean FROH per length class (Fig. 2) slightly increased with increasing segment length, as also reported by Peripolli et al. [15] for a larger Nellore population (n = 7,729).
It is worth highlighting that genomic inbreeding based on relationship matrices are dependent on the method used for constructing and scaling the matrix elements. These methods are also based on identical-by-state alleles (IBS) as opposed to other genomic inbreeding methods such as those based on ROH that capture more haplotypic segments that may be identical-by-descent (IBD) as also assumed by the pedigree-based inbreeding metric. As discussed by Meuwissen et al. [48], inbreeding metrics based on different methods such as (1) homozygosity/heterozygosity based (e.g., molecular kinship matrix), (2) genetic drift-based, i.e., changes of allele frequencies; or (3) IBD-based, i.e., SNPs are used in linkage analyses to identify IBD segments, can differ substantially and therefore, the basis of each method should be considered when interpreting and comparing inbreeding results.
FPED was moderately correlated with FH (0.61) and FROH (0.60). The correlation between FPED and FROH increased with increasing size of the ROH segments (Fig. 3), which is in agreement with reports by Peripolli et al. [15] and Lozada-Soto et al. [22]. The correlation of FPED with FROH measures ranged from 0.13 (with FROH1) to 0.39 (with FROH5). This trend of the correlations is expected since the length of the ROH segments follows an exponential distribution, given by 1/2g, where g is the number of generations that connect the individual evaluated and the common ancestor [49]. Thus, segments shorter than 2 Mb, which were used to obtain FROH1, are associated with inbreeding events that occurred more than 25 generations ago [50], which are likely not included in the recorded pedigree datasets.
In the Nellore population studied, inbreeding coefficients increased. The annual inbreeding rate was calculated for animals born after 2004, due to the small number of genotyped animals born before 2004, it was 0.16% for FPED, 0.02% for FH and 0.16% for FROH. (Fig. 4). According to the guidelines of the Food and Agriculture Organization of the United Nations [51], the acceptable average rate of inbreeding in commercial populations for the appropriate conservation of genetic resources is up to 2% per year. Thus, the annual increase in inbreeding observed in the population studied is lower than that recommended for commercial herds. This indicates that measures aimed at minimizing kinship between individuals for the definition of mating and the use of sires for a maximum period of two consecutive breeding seasons [29, 30] were effective. Sumreddee et al. [21] reported an even smaller annual increase in inbreeding (0.05% per year in FPED) in the Line 1 Hereford cattle, a highly inbred closed experimental herd (FPED = 0.292 ± 0.053). On the other hand, Lozada-Soto et al. [22], who evaluated the database of the North American Angus Association, found an increase in inbreeding of 0.14–0.22% per year for FPED and of 0.14–0.23% for FROH. These results indicate that in cattle populations known to be small and inbred, such as experimental herds, mating control promoted rates of increase in inbreeding similar to those in large commercial populations of purebred cattle.
Effect of inbreeding
Fitting the H matrix (instead of the A matrix) for FH based analysis, coupled with a larger number of animals in the analysis of inbreeding depression based on FPED and FH resulted in greater statistical power for detecting the effect of inbreeding on the traits (Tables 3 and 4).
A 1% increase in FPED resulted in a significant effect on BW (-0.086 ± 0.029 kg) and on W210 (-0.575 ± 0.188 kg) (Table 4). Based on a larger Nellore database (n = 892,199), Pereira et al. [9] reported an effect of -0.38 ± 0.01 kg in W210 using FPED (ABLUP), which is in agreement with the present findings. Animals born lighter have a reduction in vigor at birth, increased failure of the first milk feeding event, and an increase in early mortality and mortality until weaning [52, 53]. Regardless of the method used (ABLUP or GBLUP), there was a significant effect of the increase in FROH5 on the growth traits (BW, W210, and W378). This result suggests that long segments resulting from recent inbreeding events exert greater unfavorable effects on growth traits than shorter segments. This corroborates with the hypothesis that recent inbreeding tends to have more negative effect on phenotypic performance than older inbreeding.
The FPED, FH, FROH2, FROH5, and FROH significantly affected W378, with an average effect of -1.891 kg per 1% increase in inbreeding (Table 4). Unfavorable effects of increased inbreeding on yearling weight has been widely reported in beef cattle populations [9, 22, 23]. As mentioned earlier, W378 was used exclusively (NeS and NeC herds) or partially (NeT herd) as the selection criterion of the herds that make up the studied population. Similarly, Sumreddee et al. [21], evaluating Line 1 Hereford cattle, observed significant unfavorable effects on ADG after weaning (selection criterion of the population studied) for all inbreeding coefficients evaluated.
The FROH2 exerted effects of greater magnitude on W378 when modeled by A matrix (-3.581 ± 1.554 kg) or G matrix (-3.810 ± 1.753). The magnitude and significance of the effect of FROH2 are associated with the establishment of the herds that make up the population studied, reported by Mercadante et al. [29]. Since this population comprises a maximum number of 17 generations, the equation proposed by Fisher [49] can be used to estimate the time interval during which the inbreeding events represented by FROH2 occurred. If 1 Morgan is equivalent to 100 Mb, FROH2 represents inbreeding events that occurred between 25 and 12.5 generations ago, comprising the generations during which the herds were formed.
Despite the negative effects of inbreeding on W378, Benfica et al. [54] reported a genetic gain of 2.51 kg per year for the same population. Similarly, despite the detection of unfavorable effects of increased inbreeding on growth traits in Angus cattle, Lozada-Soto et al. [22] observed annual genetic gains in weaning weight and yearling weight. This finding suggests that, within the dynamics between the unfavorable effect of increased inbreeding and the favorable effect caused by genetic gain through selection, the latter more intensely affected the regions of the genome related to the traits evaluated.
Except for some niche markets, meat producers are paid by the industry based on carcass weight [55]. The carcass weight, in turn, shows a strong genetic and phenotypic correlation with the animal’s yearling weight in taurine breeds (0.89 and 0.53) [56] and in Nellore cattle (0.55 and 0.72) [57]. We may therefore infer that the decrease in W378 observed in the present study in response to the increase in inbreeding affects the profitability of the beef industry.
Higher RFI values indicate less efficient individuals, while lower and negative values are related to more efficient individuals that consume less feed at the same levels of average daily gain (ADG). There was an unfavorable effect of FPED on RFI (0.01 ± 0.0002 kg DM/day) (Table 4), suggesting that increased inbreeding is associated with less efficient animals that consume a greater amount of feed for the same ADG.
A 1% increase in FROH was associated with a change of -0.056 ± 0.022 cm in SC (Table 4). Evaluating Nellore animals, Pereira et al. [9] found an even greater effect of FPED (-0.07 ± 0.01 cm) on SC. Results of lesser magnitude have been reported by Forneris et al. [23] who evaluated the effect of FPED (-0.023 ± 0.009 cm) and FH (-0.023 ± 0.01 cm) on SC. Specifically regarding bull fertility, an increase in inbreeding has been reported to be associated with a decrease in sperm concentration and motility in Holstein cattle [58] and with a reduced conception rate (bull effect) in Brown Swiss cattle [59].
Effect of inbreeding at chromosome level
Considering that ROH segments are non-uniformly distributed across the genome (Fig. 1) and that they do not affect phenotypes with the same intensity [60], it is important to calculate inbreeding at the chromosome level (FROH_CHR) to understand the impact of certain ROH segments across the genome on economically important traits [21, 60, 61]. Among the chromosomes harboring the largest number of SNPs in regions defined as ROH and those showing correlations of higher magnitude with FROH (Supplementary Material: Figure S1), FROH_CHR3 and FROH_CHR5 were associated with significant reduction in W210 of -3.430 ± 1.203 kg and − 4.354 ± 1.628 kg, respectively (Table 5). Furthermore, a 1% increase in FROH_CHR5 was associated with a change of -5.758 ± 2.755 kg in W378.
According to Purfield et al. [62], the negative effect of inbreeding on a phenotype is mainly due to recessive variants of deleterious alleles. The proportion of deleterious genes is higher in homozygous segments than in other regions of the genome. Studies in humans reported a higher proportion of deleterious genes in long segments [63, 64], while this proportion was higher within short and medium segments (< 3 Mb) in cattle studies [61].
Considering chromosomes with the highest proportions of segments longer than 8 Mb, there was a significant effect of FROH_CHR8 on BW (-0.698 ± 0.329 kg) and W210 (-3.016 ± 1.476 kg) (Table 5). Sumreddee et al. [21] found effects of similar magnitude on BW for FROH_CHR12 (-0.676 ± 0.289) and FROH_CHR27 (-0.855 ± 0.306) and on weaning weight for FROH_CHR17 (-6.827 ± 3.011). Among chromosomes harboring the highest proportions of segments shorter than 8 Mb, only the coefficients calculated based on BTA3 and BTA5, whose participation in FROH was already evaluated, had significant inbreeding depression. Nevertheless, contrary to Pilon et al. [60], we found no apparent association between the length of the chromosome covered by SNPs (number of SNPs) and inbreeding depression (Supplementary Material: Figure S2).
We highlight the need for future studies aimed at investigating regions located on chromosomes that exhibited significant effects on the traits evaluated and at identifying candidate genes and relevant metabolic pathways. Furthermore, studies investigating how recessive alleles with deleterious effects are eliminated over generations in this population are needed since this process is directly influenced by the ratio between the effect due to deleterious recessive alleles [65] and the effect caused by favorable gene combinations in heterozygosity, which are not subject to elimination [66]. These approaches will provide a more comprehensive understanding of the genetic architecture of economically important traits and how inbreeding affects them. In general, increased inbreeding had a significant negative impact on the studied traits.
Although most studies reported in the literature have evaluated imputed genotypes [14, 15, 21, 22, 67, 68], the imputation process can significantly affect the accuracy and robustness of genomic inbreeding estimates [69], which is a limitation of the present study. Another limitation is the lack of a specific reference genome for Bos taurus indicus animals. The genome used in the present study was developed based on the Hereford breed (Bos taurus taurus) [70].
Conclusions
We used data from a closed population of Nellore cattle with a relatively complete pedigree to evaluate different inbreeding metrics and inbreeding depression on growth, fertility, and feed efficiency traits. Regardless of the inbreeding metric used, the averages annual inbreeding rates were low as a result of the control of mating and the restricted use of sires. Pedigree depth directly affected the correlation between FPED and the coefficients calculated based on the length classes of ROH (FROH). Greater inbreeding depression was observed for the criterion used to select the breeding animals in the closed herd (W378). Significant negative effects on the growth traits were found for the FROH_CHR approach. The increase in genomic inbreeding was associated with unfavorable effects on the most frequently evaluated traits compared to the traditional pedigree-based method. Therefore, the use of genomic inbreeding coefficients during mating is valuable for better controlling inbreeding in Nellore cattle.
Materials and methods
All information used in this study was derived from pre-existing databases. The animal records were obtained in strict accordance with the Guidelines on Animal Welfare and Humane Slaughter of the State of São Paulo (Law No. 11.977).
Animals and genotypes
The Nellore animals that make up the database used belong to three lines established in 1980 at the Beef Cattle Research Center, Institute of Animal Science (IZ, Sertãozinho, SP, Brazil). These animals have been selected for yearling weight (W378). The control line (NeC) has been maintained under stabilizing selection and the animals have been selected for average W378 obtained after performance testing within line x year of birth. In the selection (NeS) and traditional (NeT) lines, the animals were selected for higher W378 within line x year of birth. From 2008 onwards, NeT animals were selected based on higher EBVs for W378 and lower EBVs for residual feed intake (RFI) [29, 30, 54].
Mating in this population was carried out in a way to minimize the level of pedigree-based inbreeding of the resulting offspring [29]. Furthermore, the selected sires were used for a maximum period of two consecutive breeding seasons to increase genetic gain and minimize inbreeding. The pedigree file contained 12,568 animals, which are offspring of 432 sires and 2,884 dams. The optiSel package of the R software [71] was used to calculate the following parameters: equivalent complete generations (ECG) [72], pedigree completeness index (PCI) [73], and effective population size [74, 75].
The genotyped animals (2,256) were born from 1977 (2 animals) to 2020 (176 animals); however, the number of genotyped animals increased after 2004. Over the years, the animals were genotyped using panels of three different densities: 770 animals with the Illumina BovineHD BeadChip panel (770k, Illumina Inc. San Diego, CA, USA); 1,328 animals with the GeneSeek Genomic Profiler HDi 75 K panel (GeneSeek Inc. San Diego, CA, USA), and 158 animals with the GeneSeek Genomic Profiler HDi 50 K panel (GeneSeek Inc. San Diego, CA, USA).
The positions of the single nucleotide polymorphisms (SNPs) were updated for the ARS-UCD 1.2 reference genome [70]. Animals genotyped with medium-density panels were imputed to the HD panel using the FImpute v.3 software [76]. Quality control of the genotypes consisted of excluding SNPs without a known genomic position or located on the non-autosomal chromosomes and SNPs with an extreme departure from Hardy-Weinberg equilibrium (P < 10 − 5), and SNP call rate lower than 92%. Samples with call rate lower than 85% were also removed, with 612,154 SNPs remaining at the end of the process.
Runs of homozygosity (ROH)
The PLINK v1.9 software [77] was used to identify ROH segments based on the following criteria: (i) sliding window of 50 SNPs across the genome; (ii) proportion of homozygous overlapping windows of 0.05; (iii) minimum number of 100 consecutive SNPs included in one ROH; vi) minimum length of one ROH set to 1 Mb; v) maximum gap between consecutive homozygous SNPs of 500 kb; vi) density of one SNP per 50 kb, and vii) up to one heterozygous genotype within a ROH. After ROH identification, the ROH segments were divided into the following categories: 1–2 Mb, 2–4 Mb, 4–8 Mb, 4–16 Mb, and > 16 Mb.
Estimation of inbreeding coefficients
Three general methods were used to calculate the inbreeding coefficients. FPED was calculated for all animals using the BLUPF90 + family of programs [78] according to the method proposed by Meuwissen and Luo [79]. To construct the H matrix, the pedigree-genomic relationship matrix, the G matrix was obtained based on the first method proposed by Van Raden [42]. FH was calculated for all animals using method 3 of Legarra et al. [46] by subtracting one from the value present in the diagonal of matrix H. The H matrix was generated using preGSf90 software [78]. FROH was estimated for genotyped animals according to McQquillan et al. [80]:
where \(\:{{L}_{ROH}}_{j},\) is the total length of segments in class j, and \(\:{L}_{TOTAL}\) is the total size of the genome covered by markers. The yearly rate of inbreeding was calculated for FPED, FH and FROH by regressing the inbreeding coefficient of the natural logarithm of (1 - F) on the year of birth of the animal [81, 82].
For each animal, in addition to FROH, coefficients were also calculated based on the size class of the segments: FROH1 (1–2 Mb), FROH2 (2–4 Mb), FROH3 (4–8 Mb), FROH4 (8–16 Mb), and FROH5 (> 16 Mb). One FROH was calculated for each chromosome (FROH_CHRx) regardless of size class, except for the sex chromosomes, by adapting the equation proposed by [80] as follows:
where \(\:x\) is the number of chromosomes, \(\:{{L}_{ROH\_CHR}}_{j}\) is the total length of the segments present on each chromosome for a given class, and \(\:{L}_{TOTAL}\) is the total size of the genome covered by markers.
Effect of inbreeding
The phenotypic records used to estimate the effects of inbreeding on complex traits were collected from animals born from 1978 to 2020. The phenotypes were divided into two databases: “Total”, which contains phenotypes from the entire population (genotyped and non-genotyped animals); and “Genotyped”, which contains only phenotypes from genotyped animals born from 2004 to 2020 (Table 6).
Growth until weaning was assessed based on birth weight (BW) and weaning weight adjusted to 210 days of age (W210) [31]. After weaning, males were submitted to a feedlot weight gain test lasting 168 days, which provided W378 [29, 30]. On the other hand, females were reared on pasture and their weights were measured and adjusted to 550 days of age (W550). Like W378, scrotal circumference (SC) was obtained at the end of the weight gain test and was measured horizontally in the middle portion corresponding to the maximum circumference of the scrotum [83]. Residual feed intake (RFI) was estimated during the weight gain test and post-weaning feed efficiency test as the residual of the linear regression equation of dry matter (DM) intake on average daily gain (ADG) and metabolic body weight (BW0.75) [84] as described by Benfica et al. [54].
The contemporary groups (CG) for the traits were formed by concatenating birth year, selection line, and sex, except for RFI where CG were created based on birth year, selection line, sex, and test group. CGs with fewer than four animals were removed for subsequent analyses.
To estimate the effects of inbreeding coefficients calculated only for the genotyped animals (i.e., FROH metrics), only phenotypic records from animals born after 2004 were included in the analyses due to the small number of genotyped animals born prior to 2004. Inbreeding depression was calculated by fitting the inbreeding coefficients one by one as a linear covariate in the models (linear effect). The analyses were performed in a two-trait animal model (BW x W210, W378 x W550, W210 x SC, and W210 x RFI) using the BLUPF90 + family of programs [78]. The (co)variance components were the same as those used for the annual genetic evaluation of the selection lines [85]. The significance of the regression coefficients (β) of the inbreeding estimators for the traits was assessed by the Student t-test (β/standard error). The general model can be written in matrix form as:
,
where \(\:\varvec{y}\) is the vector of phenotypic records for each trait; \(\:\mathbf{b}\) is the vector of fixed effects including CG and month of birth, and age of the animal at the measurement of the trait (linear effect, except for BW) and age of cow in days (linear and quadratic effects) as covariates; \(\:\varvec{\upbeta\:}\) is the linear regression coefficient related to each inbreeding coefficient; \(\:\mathbf{F}\) is the vector containing each inbreeding coefficient (FPED, FH, FROH, FROH1, FROH2, FROH3, FROH4, FROH5, and FROH_CHR); \(\:\mathbf{a}\) is the vector of direct additive genetic effects, and \(\:\mathbf{e}\) is the vector of residual effects. \(\:\mathbf{X}\) and \(\:\mathbf{Z}\) are the incidence matrices relating \(\:\varvec{b}\), \(\:\varvec{a}\) and \(\:\varvec{e}\) to the vector of phenotypic observations. In this case, the covariance matrix was defined as
where \(\:{\mathbf{S}}_{\mathbf{a}}\) is the additive genetic covariance matrix; \(\:{\mathbf{S}}_{\mathbf{e}}\) is the residual covariance matrix; A is the genetic relationship matrix (pedigree), and I is an identity matrix. The model used for the analyses of the effect of inbreeding on BW, W210 and W378 also included the maternal additive effect and the maternal permanent environmental effect:
where \(\:\mathbf{y}\) is the vector of the traits observed; \(\:\varvec{b}\) is the vector of fixed effects (same as described above); \(\:\mathbf{a}\:\)is the vector of direct additive genetic effects; \(\:\mathbf{m}\) is the vector of maternal additive genetic effects, \(\:\mathbf{c}\) is the vector of maternal permanent environmental effects and \(\:\mathbf{e}\) is the vector of residual effects. \(\:\mathbf{X},\)\(\:{\mathbf{Z}}_{1}\), \(\:{\mathbf{Z}}_{2}\) and \(\:{\mathbf{Z}}_{3}\) are incidence matrices related to \(\:\mathbf{b}\), \(\:\mathbf{a}\), \(\:\mathbf{m}\) and \(\:\mathbf{c}\). Here the covariance matrix was defined as
where \(\:{\mathbf{S}}_{\mathbf{m}}\) is the maternal additive genetic covariance matrix, \(\:{\:\mathbf{s}}_{\mathbf{a}\mathbf{m}}\) is the covariance between the direct and maternal genetic effects and \(\:{\varvec{S}}_{\varvec{c}}\) is the matrix of maternal permanent environmental variance.
First, to evaluate the impact of the methods used for calculating the inbreeding coefficients, only genotyped animals (“Genotyped” dataset) were included in the analyses using the A relationship matrix (ABLUP). Next, we also used the “Total” dataset to evaluate the effect of FPED (ABLUP) and FH (HBLUP) on the traits included in the study. For this purpose, the variance of the random effects was assumed to be \(\:v\text{a}\text{r}\:\left(\mathbf{a}\right)=\mathbf{A}\otimes\:{\mathbf{S}}_{\mathbf{a}}\) and \(\:v\text{a}\text{r}\:\left(\mathbf{m}\right)=\mathbf{A}\otimes\:{\mathbf{S}}_{\mathbf{m}}\) for ABLUP (FPED), \(\:\text{v}\text{a}\text{r}\:\left(\mathbf{a}\right)=\mathbf{H}\otimes\:{\mathbf{S}}_{\mathbf{a}}\) and \(\:v\text{a}\text{r}\:\left(\mathbf{m}\right)=\mathbf{H}\otimes\:{\mathbf{S}}_{\mathbf{m}}\) for HBLUP (FH). To estimate the effect of FROH and FROH_CHR on the traits, only phenotypes of genotyped animals (Genotyped) were included in the analyses. The assumptions of the variance of the random effects were \(\:\text{v}\text{a}\text{r}\:\left(\mathbf{a}\right)=\mathbf{G}\otimes\:{\mathbf{S}}_{\mathbf{a}}\:\text{a}\text{n}\text{d}\:v\text{a}\text{r}\:\left(\mathbf{m}\right)=\mathbf{G}\otimes\:{\mathbf{S}}_{\mathbf{m}}\:\)for GBLUP (FROH and FROH_CHR).
The effect of FROH_CHR on the growth traits was estimated for different sets of chromosomes. First, the five chromosomes with the greatest contribution to FROH were considered (BTA5, BTA1, BTA2, BTA3, BTA7), i.e., chromosomes harboring the largest number of SNPs within regions defined as an ROH segment. Next, the five chromosomes containing the highest proportions of ROH segments longer than 8 Mb concerning all segments identified were analyzed (BTA9, BTA2, BTA8, BTA20, BTA1), excluding BTA1 and BTA2, which had been analyzed in the previous step. Finally, chromosomes containing the highest proportions of segments < 8 Mb were analyzed (BTA5, BTA7, BTA3, BTA12, BTA1). These chromosomes had smaller contributions to the FROH of individuals. Again, BTA1, BTA3, BTA5, and BTA7, which had been evaluated in the previous steps, were excluded from this last analysis (Supplementary Material: Table S1).
Data availability
The data supporting this study’s findings belongs to an experimental animal breeding program, and restrictions are applied to the availability of data. However, data are available by contacting the corresponding authors upon reasonable request and with permission of the program (contacting the researcher mezmercadante@gmail.com).
References
Henderson CR. Best Linear unbiased estimation and prediction under a selection model. Biometrics. 1975;31:423–47.
Brito LF, Bedere N, Douhard F, Oliveira HR, Arnal M, Peñagaricano F, et al. Review: genetic selection of high-yielding dairy cattle toward sustainable farming systems in a rapidly changing world. Animal. 2021;1:100292.
Woolliams JA, Berg P, Dagnachew BS, Meuwissen THE. Genetic contributions and their optimization. J Anim Breed Genet. 2015;132:89–99.
VanRaden PM. Accounting for inbreeding and crossbreeding in genetic evaluation of large populations. J Dairy Sci. 1992;75:3136–44.
Forutan M, Ansari Mahyari S, Baes C, Melzer N, Schenkel FS, Sargolzaei M. Inbreeding and runs of homozygosity before and after genomic selection in North American Holstein cattle. BMC Genomics. 2018;19.
Mehrabani-Yeganeh H, Gibson J, Schaeffer LR. Using recent versus complete pedigree data in genetic evaluation of a closed nucleus broiler line. Poult Sci. 1999;78(7):937–41.
Tang GQ, Xue J, Lian MJ, Yang RF, Liu TF, Zeng ZY, et al. Inbreeding and genetic diversity in three imported swine breeds in China using pedigree data. Asian-Australas J Anim Sci. 2013;26(6):755–65.
Stachowicz K, Brito LF, Oliveira HR, Miller SP, Schenkel FS, Stachowicz K, et al. Assessing genetic diversity of various Canadian sheep breeds through pedigree analyses. Can J Anim Sci. 2018;98:741–9.
Pereira RJ, Santana ML, Ayres DR, Bignardi AB, Menezes GRO, Silva LOC, et al. Inbreeding depression in Zebu cattle traits. J Anim Breed Genet. 2016;133:523–33.
Visscher PM, Medland SE, Ferreira MAR, Morley KI, Zhu G, Cornes BK, et al. Assumption-free estimation of heritability from genome-wide identity-by-descent sharing between Fullsibs. PLoS Genet. 2005;2:1–10.
Legarra A, Aguilar I, Misztal I. A relationship matrix including full pedigree and genomic information. J Dairy Sci. 2009;92:4656–63.
Curik I, Ferenčaković M, Sölkner J. Invited Review. Genomic dissection of inbreeding depression: a gate to new opportunities. J Anim Sci. 2017;46:773–82.
Cole JB, Perspective. Can we actually do anything about inbreeding? J Dairy Sci. 2024;107:643–8.
Nishio M, Inoue K, Ogawa S, Ichinoseki K, Arakawa A, Fukuzawa Y et al. Comparing pedigree and genomic inbreeding coefficients, and inbreeding depression of reproductive traits in Japanese black cattle. BMC Genomics. 2023;24.
Peripolli E, Stafuzza NB, Munari DP, Lima ALF, Irgang R, Machado MA et al. Assessment of runs of homozygosity islands and estimates of genomic inbreeding in Gyr (Bos indicus) dairy cattle. BMC Genomics. 2018;19.
Doublet AC, Croiseau P, Fritz S, Michenet A, Hozé C, Danchin-Burge C et al. The impact of genomic selection on genetic diversity and genetic gain in three French dairy cattle breeds. Genet Sel Evol. 2019;51.
Mulim HA, Brito LF, Pinto LFB, Ferraz JBS, Grigoletto L, Silva MR et al. Characterization of runs of homozygosity, heterozygosity-enriched regions, and population structure in cattle populations selected for different breeding goals. BMC Genomics. 2022;23.
Carrara ER, Lopes PS, Veroneze R, Pereira RJ, Zadra LEF, Peixoto MGCD. Assessment of runs of homozygosity, heterozygosity-rich regions and genomic inbreeding estimates in a subpopulation of Guzerá (Bos indicus) dual-purpose cattle. J Anim Breed Genet. 2024;141:207–19.
Howrigan D, Simonson M, genomics MK-B. 2011 undefined. Detecting autozygosity through runs of homozygosity: a comparison of three autozygosity detection algorithms. BMC Genomics. 2011;12:460.
Leroy G. Inbreeding depression in livestock species: review and meta-analysis. Anim Genet. 2014;45:618–28.
Sumreddee P, Toghiani S, Hay EH, Roberts A, Agrrey SE, Rekaya R. Inbreeding depression in line 1 Hereford cattle population using pedigree and genomic information. J Anim Sci. 2019;97:1–18.
Lozada-Soto EA, Maltecca C, Lu D, Miller S, Cole JB, Tiezzi F. Trends in genetic diversity and the effect of inbreeding in American Angus cattle under genomic selection. Genet Sel Evol. 2021;53.
Forneris NS, Garcia-Baccino CA, Cantet C, Vitezica RJ. Estimating inbreeding depression for growth and reproductive traits using pedigree and genomic methods in Argentinean Brangus cattle. J Anim Sci. 2021;99:1–10.
Beef ABIEC. Report 2023. https://www.abiec.com.br/wpcontent/uploads/BeefReport2022atualizadojun2022.pdf. Acessed 12 Jun 2024.
Vozzi AP, Marcondes RC, Magnabosco CU, Bezerra LAF, Lôbo RB. Structure and genetic variability in Nellore (Bos indicus) cattle by pedigree analysis. Genet Mol Biol. 2006;29:3.
Fernandes Júnior GA, de Oliveira HN, Carvalheiro R, Cardoso DF, Fonseca LFS, Ventura RV, et al. Whole-genome sequencing provides new insights into genetic mechanisms of tropical adaptation in Nellore (Bos primigenius indicus). Sci Rep. 2020;10:9412.
Zavarez LB, Utsunomiya YT, Carmo AS, Neves HHR, Carvalheiro R, Ferencakovic M, et al. Assessment of autozygosity in Nellore cows (Bos indicus) through high-density SNP genotypes. Front Genet. 2015;16:542.
Santana ML, Pereira RJ, Bignardi AB, Ayres DR, Menezes GRO, Silva LOC, et al. Structure and genetic diversity of Brazilian Zebu cattle breeds assessed by pedigree analysis. Livest Sci. 2016;187:6–15.
Mercadante MEZ, Packer IU, Razook AG, Cyrillo JNSG, Figueiredo LA. Direct and correlated responses to selection for yearling weight on reproductive performance of Nelore cows. J Anim Sci. 2003;81:376–84.
Cardoso DF, De Albuquerque LG, Reimer C, Qanbari S, Erbe M, Do Nascimento AV et al. Genome-wide scan reveals population stratification and footprints of recent selection in Nelore cattle. Genet Sel Evol. 2018;50.
Benfica LF, Sakamoto LS, Magalhães AFB, De Oliveira MHV, De Albuquerque LG, Cavalheiro R et al. Genetic association among feeding behavior, feed efficiency, and growth traits in growing indicine cattle. J Anim Sci. 2020;98.
Kim ES, Cole JB, Huson H, Wiggans GR, Van Tassel CP, Crooker BA et al. Effect of artificial selection on runs of homozygosity in U.S. Holstein cattle. PLoS ONE. 2013;8.
Goszczynski D, Molina A, Terán E, Morales-Durand H, Ross P, Cheng H et al. Runs of homozygosity in a selected cattle population with extremely inbred bulls: descriptive and functional analyses revealed highly variable patterns. PLoS ONE. 2018;13.
Hidalgo J, Cesarani A, Garcia A, Sumreddee P, Larios N, Mancin E, et al. Genetic background and inbreeding depression in romosinuano cattle breed in Mexico. Animals. 2021;11:1–20.
Kardos M, Taylor HR, Ellegren H, Luikart G, Allendorf FW. Genomics advances the study of inbreeding depression in the wild. Evol Appl. 2016;9:1205–18.
Browning SR, Browning BL. Identity by descent between distant relatives: detection and applications. Annu Rev Genet. 2012;46:617–33.
Mc Parland S, Kearney JF, Rath M, Berry DP. Inbreeding trends and pedigree analysis of Irish dairy and beef cattle populations. J Anim Sci. 2007;85:322–31.
Ferenčaković M, Hamzić E, Gredler B, Solberg TR, Klemetsdal G, Curik I, et al. Estimates of autozygosity derived from runs of homozygosity: empirical evidence from selected cattle populations. J Anim Breed Genet. 2013;130:286–93.
Marras G, Gaspa G, Sorbolini S, Dimauro C, Ajmone-Marsan P, Valentini A, et al. Analysis of runs of homozygosity and their relationship with inbreeding in five cattle breeds farmed in Italy. Anim Genet. 2015;46:110–21.
Gurgul A, Szmatoła T, Topolski P, Jasielczuk I, Żukowski K, Bugno-Poniewierska M. The use of runs of homozygosity for estimation of recent inbreeding in Holstein cattle. J Appl Genet. 2016;57:527–30.
Keller MC, Visscher PM, Goddard ME. Quantification of inbreeding due to distant ancestors and its detection using dense single nucleotide polymorphism data. Genetics. 2011;189:237–49.
VanRaden PM. Efficient methods to compute genomic predictions. J Dairy Sci. 2008;91:4414–23.
Hill WG, Weir BS. Variation in actual relationship as a consequence of mendelian sampling and linkage. Genet Res (Camb). 2011;93:47–64.
Gutiérrez-Reinoso MA, Aponte PM, Cabezas J, Rodriguez-Alvarez L, Garcia-Herreros M. Genomic evaluation of primiparous high-producing dairy cows: inbreeding effects on genotypic and phenotypic production–reproductive traits. Animals. 2020;10:1–18.
VanRaden PM. Symposium review: how to implement genomic selection. J Dairy Sci. 2020;103:5291–301.
Legarra A, Aguilar I, Colleau JJ. Short communication: methods to compute genomic inbreeding for ungenotyped individuals. J Dairy Sci. 2020;103:3363–7.
Gutiérrez-Reinoso MA, Aponte PM, García-Herreros M. A review of inbreeding depression in dairy cattle: current status, emerging control strategies, and future prospects. J Dairy Res. 2022;89:3–12.
Meuwissen THE, Sonesson AK, Gebregiwergis G, Woolliams JA. Management of genetic diversity in the era of Genomics. Front Genet. 2020;11.
Fisher RA. A fuller theory of junctions in inbreeding. 1954.
Howard JT, Pryce JE, Baes C, Maltecca C. Invited review: inbreeding in the genomics era: inbreeding, inbreeding depression, and management of genomic variability. J Dairy Sci. 2017;100:6009–24.
FAO. In vivo conservation of animal genetic resources. FAO animal production and heath guidelines 14. 2013. Accessed 11 Mar 2023.
Schmidek A, Mercadante MEZ, Costa MJRP, Razook AG, Figueiredo LAF. Falha na primeira mamada em bezerros Guzerá: fatores predisponentes e parâmetros genéticos. R Bras Zootec. 2008;37:998–1004.
Schmidek A, Costa MJRP, Mercadante MEZ, Toledo LM, Cyrillo JNSG, Branco RH. Genetic and non-genetic effects on calf vigor at birth and preweaning mortality in Nellore calves. R Bras Zootec. 2013;42:421–7.
Fernandes Júnior GA, Rosa GJM, Valente BD, Carvalheiro R, Baldi F, Garcia DA et al. Genomic prediction of breeding values for carcass traits in Nellore cattle. Genet Sel Evol. 2016;48.
Meyer K, Johnston DJ, Graser HU. Estimates of the complete genetic covariance matrix for traits in multi-trait genetic evaluation of Australian Hereford cattle. Aust J Agric Res. 2004;55:195–210.
Tonussi RL, Espigolan R, Gordo DGM, Magalhães AFB, Venturini GC, Baldi F, et al. Genetic association of growth traits with carcass and meat traits in Nellore cattle. Genet Mol Res. 2015;14:18713–9.
Ghoreishifar M, Milad Vahedi S, Salek Ardestani S, Khansefid M, Pryce JE. Genome-wide assessment and mapping of inbreeding depression identifies candidate genes associated with semen traits in Holstein bulls. BMC Genomics. 2023;24.
Pacheco HA, Rossoni A, Cecchinato A, Peñagaricano F. Identification of runs of homozygosity associated with male fertility in Italian Brown Swiss cattle. Front Genet. 2023;14.
Pilon B, Hinterneder K, Hay EHA, Fragomeni B. Inbreeding calculated with runs of homozygosity suggests chromosome-specific inbreeding depression regions in line 1 hereford. Animals. 2021;11.
Zhang Q, Guldbrandtsen B, Bosse M, Lund MS, Sahana G. Runs of homozygosity and distribution of functional variants in the cattle genome. BMC Genomics. 2015;16.
Purfield DC, Berry DP, McParland S, Bradley DG. Runs of homozygosity and population history in cattle. BMC Genet. 2012;13:70.
Szpiech ZA, Xu J, Pemberton TJ, Peng W, Zöllner S, Rosenberg NA, et al. Long runs of homozygosity are enriched for deleterious variation. Am J Hum Genet. 2013;93:90–102.
Pemberton TJ, Absher D, Feldman MW, Myers RM, Rosenberg NA, Li JZ. Genomic patterns of homozygosity in worldwide human populations. Am J Hum Genet. 2012;91:275–92.
García-Dorado A. Understanding and predicting the fitness decline of shrunk populations: inbreeding, purging, mutation, and standard selection. Genetics. 2012;190:1461–76.
Charlesworth B. Fundamental concepts in genetics: effective population size and patterns of molecular evolution and variation. Nat Rev Genet. 2009;10:195–205.
Martikainen K, Sironen A, Uimari P. Estimation of intrachromosomal inbreeding depression on female fertility using runs of homozygosity in Finnish Ayrshire cattle. J Dairy Sci. 2018;101:11097–107.
Doekes HP, Veerkamp RF, Bijma P, De Jong G, Hiemstra SJ, Windig JJ. Inbreeding depression due to recent and ancient inbreeding in Dutch holstein-friesian dairy cattle. Genet Sel Evol. 2019;51.
Dadousis C, Ablondi M, Cipolat-Gotet C, van Kaam JT, Finocchiaro R, Marusi M et al. Genomic inbreeding coefficients using imputed genotypes: assessing differences among SNP panels in Holstein-Friesian dairy cows. Front Vet Sci. 2023;10.
Rosen BD, Bickhart DM, Schnabel RD, Koren S, Elsik CG, Tseng E, et al. De novo assembly of the cattle reference genome with single-molecule sequencing. Gigascience. 2020;9:1–9.
Wellmann R. Optimum contribution selection for animal breeding and conservation: the R package optiSel. BMC Bioinformatics. 2019;20.
Maignel L, Boichard D, Verrier E. Genetic variability of French dairy breeds estimated from pedigree information. Interbull Bull. 1996;14:49–56.
Maccluer JW, Boyce AJ, Dyke B, Weitkamp LR, Pfennig DW, Parsons CJ. Inbreeding and pedigree structure in standardbred horses. J Hered. 1983;74:394–9.
González-Recio O, López De Maturana E, Gutiérrez JP. Inbreeding depression on female fertility and calving ease in Spanish dairy cattle. J Dairy Sci. 2007;90:5744–52.
Gutiérrez JP, Cervantes I, Goyache F. Improving the estimation of realized effective population sizes in farm animals. J Anim Breed Genet. 2009;126:327–32.
Sargolzaei M, Chesnais JP, Schenkel FS. A new approach for efficient genotype imputation using information from relatives. BMC Genomics. 2014;15:478.
Purcell S, Neale B, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75.
Misztal I, Tsuruta S, Lourenco D, Masuda Y, Aguilar I, Legarra A, Vitezica Z. Manual for BLUPF90 family of programs. University of Georgia; 2014.
Meuwissen THE, Luo Z. Computing inbreeding coefficients in large populations. Genet Sel Evol. 1992;24:305–13.
McQuillan R, Leutenegger AL, Abdel-Rahman R, Franklin CS, Pericic M, Barac-Lauc L, et al. Runs of homozygosity in European populations. Am J Hum Genet. 2008;83:359–72.
Benfica LF, Brito LF, do Bem RD, Mulim HA, Glessner J, Braga LG et al. Genome-wide association study between copy number variation and feeding behavior, feed efficiency, and growth traits in Nellore cattle. BMC Genomics. 2024;25.
Doekes HP, Veerkamp RF, Bijma P, Hiemstra SJ, Windig JJ. Trends in genome-wide and region-specific genetic diversity in the dutch-flemish holstein-friesian breeding program from 1986 to 2015. Genet Sel Evol. 2018;50.
Hillestad B, Aqua V, Klemetsdal G, Hillestad B, Woolliams JA, Meuwissen T et al. Estimating Rate of Inbreeding and Effective Population Size Using Genomic Data in Norwegian Red Cattle. 2018. Page 470 in 10th World Congress of Genetics Applied to Livestock Production. Vancouver, British Columbia, Canada.
Cyrillo JNSG, Razook AG, Figueiredo LA, Neto LMB, Mercadante MEZ, Tonhati H. Estimativas De Tendências E Parâmetros Genéticos do Peso Padronizado aos 378 Dias De Idade, Medidas Corporais E Perímetro Escrotal De Machos Nelore De Sertãozinho, SP. R Bras Zootec. 2001;30:56–65.
Koch RM, Gregory KE, Chambers D, Swiger LA. Efficiency of feed use in beef cattle. J Anim Sci. 1963;22:486–94.
Instituto de Zootecnia. https://iz.agricultura.sp.gov.br/publica.php?id=473. Accessed 11 Mar 2024.
Acknowledgements
The authors thank the Institute of Animal Science (Sertãozinho, SP, Brazil) for providing the datasets used in this research, as well as the staff and interns for their support and valuable contributions.
Funding
This study was financially supported by the National Council for Scientific and Technological Development (CNPq) - grant number 434681/2018-0, by the São Paulo Research Foundation (FAPESP, 2017/10630-2 and 2017/50339-5), and by the Coordination for the Improvement of Higher Education Personnel (CAPES, Brasília, DF, Brazil; Finance Code 001).
Author information
Authors and Affiliations
Contributions
RDB, LFB, and MEZM conceived and designed the study and conducted the data analyses. RDB, LFB, MSB, JNSGC, RCC, SFMB, and MEZM contributed to the data acquisition and organization. RDB, LFB, DAS, ERC, LFB, HAM, contributed to the interpretation of the results and discussion. RDB, LFB, ERC, LFB, HAM and MEZM wrote and edited the manuscript. All authors reviewed and contributed to the editing of the manuscript and approved its final version.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Animal Care and Use Committee approval was not obtained for this study because analyses were performed on pre-existing datasets. The animal records were obtained in strict accordance with the Guidelines on Animal Welfare and Humane Slaughter of the State of São Paulo (Law No. 11.977).
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it.The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Bem, R.D., Benfica, L.F., Silva, D.A. et al. Assessing different metrics of pedigree and genomic inbreeding and inbreeding effect on growth, fertility, and feed efficiency traits in a closed-herd Nellore cattle population. BMC Genomics 25, 738 (2024). https://doi.org/10.1186/s12864-024-10641-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-024-10641-3