Abstract
Background
Gut microbial communities can contribute positively and negatively to host health. So far, eight core bacterial taxonomic clusters have been reported in honey bees. These bacteria are involved in host metabolism and defenses. Nosema ceranae is a gut intracellular parasite of honey bees which destroys epithelial cells and gut tissue integrity. Studies have shown protective impacts of honey bee gut microbiota towards N. ceranae infection. However, the impacts of N. ceranae on the relative abundance of honey bee gut microbiota remains unclear, and has been confounded during prior infection assays which resulted in the co-inoculation of bacteria during Nosema challenges. We used a novel method, the suppression of N. ceranae with specific siRNAs, to measure the impacts of Nosema on the gut microbiome.
Results
Suppressing N. ceranae led to significant positive effects on microbial abundance. Nevertheless, 15 bacterial taxa, including three core taxa, were negatively correlated with N. ceranae levels. In particular, one co-regulated group of 7 bacteria was significantly negatively correlated with N. ceranae levels.
Conclusions
N. ceranae are negatively correlated with the abundance of 15 identified bacteria. Our results provide insights into interactions between gut microbes and N. ceranae during infection.
Similar content being viewed by others
Background
Animals evolve with their associated microorganisms as a unit and symbiotic microbes can facilitate the survival of hosts toward diverse stresses. When the host encounters pathogens, the microbial community responds and the dynamics can change dramatically [1]. As microbes can have varied effects on host challenges, the context of microbial function and host interactions is essential [2, 3]. In honey bees, eight core bacterial taxa have been identified. Collectively, these microbes have been shown to impact honey bee metabolism and immune responses towards infections, altering disease susceptibility [4,5,6,7,8].
Nosema ceranae is a unicellular fungal parasite which infects honey bee mid-gut epithelial cells. Infection starts from ingestion of N. ceranae spores. The proliferation cycle of N. ceranae is approximately 4 days, at which point a large number of offspring spores are released from infected cells [9]. N. ceranae infection negatively affects honey bee physiology, behavior and immune responses [10]. N. ceranae has spread globally and this agent is one of the factors implicated in honey bee colony collapses [11, 12]. A few gene silencing efforts have shown promise in reducing N. ceranae proliferation [13,14,15].
During N. ceranae proliferation, this parasite inevitably encounters the gut microbial community. Within the honey bee gut, bacteria are dominated by eight species/clusters. Three bacteria are within the Gram-negative phylum Proteobacteria, including Gilliamella apicola, Frischella perrara and Snodgrassella alvi. Two bacteria are within the Gram-positive phylum Firmicutes, including Lactobacillus mellis, Lactobacillus kunkeei and Lactobacillus Firm5 (including L. helsingborgensis, L. melliventris and L. kimbladii). One Bifidobacterium cluster within the phylum Actinomycetes, including B. asteroids, B. actinocoloniiforme and B. bohemicum. Two other species clusters are within phylum Alphaproteobacteria of Bartonellaceae and Acetobacteraceae [16]. Some of these taxa are likely to be involved in honey and pollen digestion, along with many low-frequency opportunistic microbes [2, 7, 17]. Gut microbes show diverse responses towards N. ceranae infection [18]. Consequently, it might be possible to control N. ceranae infection by regulating gut microbes, providing a long-lasting strategy to improve overall bee health. Quantifying the responses of microbes to N. ceranae infection is a critical step in understanding these interactions. This is challenging, because inoculating honey bees with N. ceranae spores generally leads to co-inoculation with associated gut bacteria. Even after several rounds of Percoll purification, a fraction of high-throughput sequence reads from purified spores can still be aligned to bacteria [19, 20]. So, predicting responses of honey bee gut microbes towards N. ceranae proliferation using bees challenged with N. ceranae and control bees can lead to artefacts.
The gene Dicer is a key component of the RNA induced silencing complex, which is involved in developmental and physiological gene regulation [21, 22]. We previously found that targeting the gene Dicer using siRNA interrupted N. ceranae proliferation [23, 24]. We used this insight to more accurately determine associations between N. ceranae infection and gut bacteria loads.
Results
As expected, sequence reads which aligned to the bee genome decreased while reads aligned to microbes increased gradually over the experimental period (Fig. 1) (File S1). The gene expression level of the N. ceranae RNA polymerase II small subunit (RPB1, KJ373285.1) was lower in the siRNA-Dicer group compared with the infection and siRNA-scramble groups at 3 dpi (Fig. 1). In total, 15 bacterial species/strains were found during the experimental period, including three core bacteria, Snodgrassella alvi, Gilliamella apicola and Frischella perrara. Two co-regulating bacterial groups were negatively correlated with N. ceranae over the entire experimental period. The marker genes of Erwinia rhapontici, Enterobacteriaceae bacterium, Enterobacter sp. JN969314.1, Citrobacter freundii, Enterobacter aerogenes, Klebsiella pneumonia and Frischella perrara were co-expressed (co-regulation group 1 of 7 bacteria) and significantly negatively correlated with N. ceranae RPB1 (corresponding correlation − 0.36, P = 0.03). Additionally, marker genes for Serratia sp. G5_1_1BCO2, Serratia marcescens, Serratia nematodiphila and Snodgrassella alvi were co-expressed (co-regulation group 2 of 4 bacteria), which was also negatively correlated with N. ceranae levels, even though this correlation was not statistically significant (corresponding correlation − 0.29, P = 0.08). Bartonella apis, Propionibacterium sp. B4, Lactobacillus apis and Gilliamella apicola did not cluster into any co-regulation group. None of these studied 15 microbes were statistically significantly differentially expressed at each day post-infection (T-test, P > 0.05). However, the co-regulated groups (P < 0.0001) and siRNA treatment (P < 0.0001) showed significant effects on specific microbe counts during the entire experimental period within the generalized linear model. Additionally, 6 bacteria in co-regulation group 1 (E. rhapontici, E. bacterium, E. sp. JN969314.1, C. freundii, E. aerogenes, K. pneumonia), 3 bacteria in co-regulation group 2 (S. sp. G5_1_1BCO2, S. marcescens, S. nematodiphila) and G. apicola were significantly enhanced towards siRNA treatment during the entire experimental period (P < 0.001).
Read counts of microbes during the experimental period. Reads aligned to the bee genome decreased over time and the reads aligned to microbes increased. Overall, the microbes were negatively correlated with N. ceranae proliferation. X axis represents days post infection. Y axis represents the relative abundance of the studied microbes
Discussion
When a parasite infects a host, infection success depends on host immune defenses, microbial community responses, and even competition from other parasites [25]. In turn, parasites spatially alter the relative abundance of gut microbes and other symbionts [26]. Honey bee gut microbes are important for food digestion and bee immune responses [17]. In honey bees, at least 406 bacterial species and variants have been found (based on the Holobee Bar database), including eight bacterial clusters that form the core gut microbial community [16, 17]. Within this core set of bacteria, the symbionts Gilliamella apicola and Snodgrassella alvi support food digestion and pathogen defenses, even though sequence variance may lead to functional variation [27, 28]. In contrast, the bacterium Frischella perrara triggers the honey bee melanization response [29]. Honey bees and their gut bacteria both respond to stressors and infection.
As two co-regulation groups were identified, the relative abundance of bacteria may not be independent of each other in the mid-gut tissue. Additionally, all of the studied bacteria were negatively correlated with N. ceranae proliferation, suggesting N. ceranae reduces bacterial abundance, at least for the studied 15 bacteria. In the gut microbiome, diet shapes bacterial diversity and responses towards N. ceranae infection, where a negative correlation between S. alvi and N. ceranae was found, which is consistent with our data [30]. When this microbial community was disturbed by antibiotics, honey bees were more susceptible towards N. ceranae and other opportunistic pathogens [8, 31]. Administration of Lactobacillus strains as food additives showed suppressive effects on N. ceranae spores [32].
Competition for limited food resources may lead to the decrease/exclusion of other competitors, as suggested by the competitive exclusion principle [33]. Therefore, a nutritional context is essential when interpreting any responses of the bacterial community toward parasite challenges. In our data, parasites negatively affected the relative abundance of all 15 studied gut bacteria, leading to interrupted food digestion and nutrient absorption. It remains unclear whether gut bacteria can actively defend the integrity of the gut tissue in bees. However, microbes can be regulated by diet to protect the gut integrity and support pathogen resistance in other organisms [34]. In prior work, honey bee pathogens have shown negative correlations with S. alvi [35, 36]. From our data, the levels of F. perrara and S. alvi were significantly enhanced with the reduction of N. ceranae. It remains unclear why N. ceranae negatively affected gut bacteria. Even though sugar was the only food resource in our studied bees, the chance is low that N. ceranae directly competes for this food resource with bacteria, as N. ceranae proliferates within epithelia cells and bacteria live in the lumen. Tissue destruction caused by infection might lead to the deterioration of the gut lumen, thus impacting bacteria.
Although microbial treatments provide a promising chemical-free strategy to control pathogen infection [37], caution is required, as improper probiotic supplements can lead to dysbiosis of gut microbes and susceptibility toward pathogens [38]. In our study, the relationship between N. ceranae and the studied microbes was correlative. However, the siRNA Dicer treatment and the reduced N. ceranae level was causative. We provided novel insights into interactions between gut parasites and other microbes over the entire life cycle of the parasite. Our study is limited to mid-gut tissues of newly-emerged workers. It will be interesting to screen N. ceranae and bacterial interactions in older forager bees, and in bees collected during different seasons.
Conclusions
Levels of N. ceranae are negatively correlated with levels of gut bacteria. Gut bacteria were not independent with each other and were co-regulated. The relative abundance between the gut bacteria and N. ceranae was negatively correlated.
Methods
Ethics statement
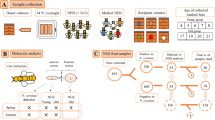
Three apiaries (with 10, 10, and 20 colonies in each apiary, respectively) for bee sample collection were established at the USDA-ARS Bee Research Laboratory, Beltsville, Maryland, USA. No specific permits were required for the described studies. Studies involved the European honey bee (Apis mellifera mellifera), which is neither an endangered nor protected species.
Infection and sample collection
N. ceranae spores were isolated from the mid-guts of one hundred heavily-infected honey bee workers from two colonies in the apiary. The abdomens of honey bee workers were homogenized using pestles in 500 mL distilled water, filtered through filtering mesh cloth (65 um pore size) and centrifuged 5 min at 3000 rpm. Spores were further purified using a Percoll gradient procedure [19]. Sealed brood frames were collected from multiple colonies and the brood frames were kept in an incubator to collect freshly emerged honey bee workers. Eighty newly-emerged workers were fed individually with 2 μL of sucrose solution as follows: (1) with 105 N. ceranae spores without siRNA treatment, as the infection group; (2) with 105 N. ceranae spores and 1.5 μg siRNA (targeting N. ceranae gene Dicer) as the siRNA-Dicer group; (3) with 105 N. ceranae spores and 1.5 μg of a non-matching (scrambled) siRNA, as the siRNA-scramble group. The sequences of the designed siRNA were provided in a previous study [23]. Forty honey bees were kept in each rearing cup. In total, six rearing cups were constructed for three treatments, with two replicates per treatment. Five bees were sampled daily from 1 to 6 days post-infection (dpi) from each cup and ultimately pooled for RNA sequencing. After being anesthetized with CO2, the mid-gut tissue was dissected and the RNA was immediately extracted with TRizol [39]. Sequencing libraries were prepared and sequenced using the Illumina Hiseq 2000 platform. In total, 36 PCR-free Illumina RNA paired-end (151 nucleotide per read) libraries of mid-gut samples were sequenced (two replicates per treatment per day), at the University of Maryland Institute for Genomic Studies.
Bioinformatics and microbial quantification
On average, 58 million reads (114 nucleotides for each paired-read member) were generated from each library, after trimming and quality control using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and SEQTK (https://github.com/lh3/seqtk). Sequencing reads were aligned to the honey bee genome assembly (Amel_HAv3.1) with the hisat2 package under default settings [40]. Reads that matched the honey bee genome were removed. On average, 14,258,564 remaining reads per library were then aligned to the Holobee ‘Bar’ database and 7,260,989 reads per library were mapped (>138X coverage of the database) including N. ceranae and other bee-associated microbes. The Holobee Bar database is a curated resource for microbes associated with honey bees (https://data.nal.usda.gov/dataset/holobee-database-v20161). In order to remove false-positive microbial classifications and restrict reads to the more prevalent bacteria responding towards the parasite infection, microbes found in fewer than 22 libraries of the sequencing libraries were removed. The counts were normalized using the weighted trimmed mean of M-values (TMM). Two replicates of each treatment for each post-infection day were used to calculate the variance in order to identity significantly regulated bacteria with edgeR [41]. The P values were corrected for multiple comparisons using false discovery rate (FDR). Significantly differentially expressed taxa met three criteria: (1) the relative abundance of the bacterium is significantly different between the infection group and the siRNA-Dicer group; (2) the relative abundance of the bacterium is significantly different between the siRNA-Dicer group and the siRNA-scramble group; and (3) the relative abundance of the bacterium is not significantly different between the siRNA-scramble group and the infection group.
Statistical analysis
Significantly differentially expressed taxa were tested using t-tests at each time point and the P value was adjusted for multiple comparisons with FDR using R (version 3.5.1). In order to analyze bacterial patterns over the infection period, co-regulation between N. ceranae and bacteria clusters was analyzed using the WGCNA package [42]. The normalized read counts of bacteria were treated as the genotype and counts of N. ceranae were treated as phenotype. To further test the effect of siRNA treatment on the microbe enrichment, a generalized linear model was performed using R (version 3.5.1). Within the linear model (counts ~ treatment + day + cage + group + microbe), counts indicate normalized microbe counts. Treatment indicates siRNA-Dicer group, siRNA-scramble group and infection group. Day indicates number of days post infection. Cage indicates each of the two cage replicates. Group indicates co-expression groups obtained from WGCNA analysis. Microbe indicates each of 15 microbe species. The code is provided in supplementary file S2.
Availability of data and materials
The raw sequencing reads are deposited in NCBI bio-project PRJNA399493.
Abbreviations
- siRNA:
-
small interfering RNA
- PRB1:
-
RNA polymerase II small subunit
- WGCNA:
-
Weight Correlation Network Analysis
References
Schwarz RS, Huang Q, Evans JD. Hologenome theory and the honey bee pathosphere. Curr Opin Insect Sci. 2015;10:1–7.
Powell JE, Martinson VG, Urban-Mead K, Moran NA. Routes of Acquisition of the gut Microbiota of the honey bee Apis mellifera. Appl Environ Microbiol. 2014;80:7378–87.
Maes PW, Rodrigues PAP, Oliver R, Mott BM, Anderson KE. Diet-related gut bacterial dysbiosis correlates with impaired development, increased mortality and Nosema disease in the honeybee (Apis mellifera). Mol Ecol. 2016;25:5439–50.
Schwarz RS, Moran NA, Evans JD. Early gut colonizers shape parasite susceptibility and microbiota composition in honey bee workers. Proc Natl Acad Sci. 2016;113:9345–50.
Kwong WK, Mancenido AL, Moran NA. Immune system stimulation by the native gut microbiota of honey bees. R Soc Open Sci. 2017;4:170003.
Zheng H, Powell JE, Steele MI, Dietrich C, Moran NA. Honeybee gut microbiota promotes host weight gain via bacterial metabolism and hormonal signaling. Proc Natl Acad Sci. 2017;114:4775–80.
Raymann K, Moran NA. The role of the gut microbiome in health and disease of adult honey bee workers. Curr Opin Insect Sci. 2018;26:97–104.
Li JH, Evans JD, Li WF, Zhao YZ, DeGrandi-Hoffman G, Huang SK, et al. New evidence showing that the destruction of gut bacteria by antibiotic treatment could increase the honey bee’s vulnerability to Nosema infection. PLoS One. 2017;12:e0187505.
Gisder S, Mockel N, Linde A, Genersch E. A cell culture model for Nosema ceranae and Nosema apis allows new insights into the life cycle of these important honey bee-pathogenic microsporidia. Environ Microbiol. 2011;13:404–13.
Paris L, El Alaoui H, Delbac F, Diogon M. Effects of the gut parasite Nosema ceranae on honey bee physiology and behavior. Curr Opin Insect Sci. 2018;26:149–54.
Higes M, Martín-Hernandez R, Garrido-Bailón E, Gonzales-Porto A, García-Palencia P, Meana A, et al. Honeybee colony collapse due to Nosema ceranae in professional apiaries. Env Microbiol Reports. 2009;1. https://doi.org/10.1111/j.1758-2229.2009.00014.x.
Higes M, Martín-Hernández R, Botías C, Bailón EG, González-Porto AV, Barrios L, et al. How natural infection by Nosema ceranae causes honeybee colony collapse. Environ Microbiol. 2008;10:2659–69.
Paldi N, Glick E, Oliva M, Zilberberg Y, Aubin L, Pettis J, et al. Effective gene silencing in a microsporidian parasite associated with honeybee (Apis mellifera) Colony declines. Appl Environ Microbiol. 2010;76:5960–4.
Li W, Evans JD, Huang Q, Rodríguez-García C, Liu J, Hamilton M, et al. Silencing the honey bee (Apis mellifera) naked cuticle gene (nkd) improves host immune function and reduces Nosema ceranae infections. Appl Environ Microbiol. 2016;82:6779–87.
Rodriguez-Garcia C, Evans JD, Li W, Branchiccela B, Li JH, Heerman MC, et al. Nosemosis control in European honey bees Apis mellifera by silencing the gene encoding Nosema ceranae polar tube protein 3. J Exp Biol. 2018;221:jeb184606. https://doi.org/10.1242/jeb.184606.
Moran NA. Genomics of the honey bee microbiome. Curr Opin Insect Sci. 2015;10:22–8.
Kwong WK, Moran NA. Gut microbial communities of social bees. Nat Rev Microbiol. 2016;14:374–84.
Rouze R, Mone A, Delbac F, Belzunces L, Blot N. The honeybee gut microbiota is altered after chronic exposure to different families of insecticides and infection by Nosema ceranae. Microbes Environ. 2019;34:226–33.
Cornman RS, Chen YP, Schatz MC, Street C, Zhao Y, Desany B, et al. Genomic analyses of the microsporidian Nosema ceranae, an emergent pathogen of honey bees. PLoS Pathog. 2009;5. https://doi.org/10.1371/journal.ppat.1000466.
ping CY, Pettis JS, Zhao Y, Liu X, Tallon LJ, Sadzewicz LD, et al. Genome sequencing and comparative genomics of honey bee microsporidia, Nosema apis reveal novel insights into host-parasite interactions. BMC Genomics. 2013;14:451.
Ota H, Sakurai M, Gupta R, Valente L, Wulff B-E, Ariyoshi K, et al. ADAR1 forms a complex with Dicer to promote microRNA processing and RNA-induced gene silencing. Cell. 2013;153:575–89.
Redfern AD, Colley SM, Beveridge DJ, Ikeda N, Epis MR, Li X, et al. RNA-induced silencing complex (RISC) proteins PACT, TRBP, and dicer are SRA binding nuclear receptor coregulators. Proc Natl Acad Sci. 2013;110:6536–41.
Huang Q, Chen Y, Neumann P, Evans JD. Effective silencing of Dicer decreases spore load of the honey bee parasite Nosema ceranae. Fungal Genomics Biol. 2016;06:1–4.
Huang Q, Li W, Chen Y, Retschnig-Tanner G, Yanez O, Neumann P, et al. Dicer regulates Nosema ceranae proliferation in honey bees. Insect Mol Biol. 2019;28:74–85.
Berrilli F, Di Cave D, Cavallero S, D’Amelio S. Interactions between parasites and microbial communities in the human gut. Front Cell Infect Microbiol. 2012;2:141.
Hubert J, Bicianova M, Ledvinka O, Kamler M, Lester PJ, Nesvorna M, et al. Changes in the Bacteriome of honey bees associated with the parasite Varroa destructor, and pathogens Nosema and Lotmaria passim. Microb Ecol. 2017;73:685–98.
Kwong WK, Engel P, Koch H, Moran NA. Genomics and host specialization of honey bee and bumble bee gut symbionts. Proc Natl Acad Sci. 2014;111:11509–14.
Engel P, Stepanauskas R, Moran NA. Hidden diversity in honey bee gut symbionts detected by single-cell genomics. PLoS Genet. 2014;10:e1004596.
Engel P, Bartlett KD, Moran NA. The bacterium Frischella perrara causes scab formation in the gut of its honeybee host. MBio. 2015;6(3):e00193–15. https://doi.org/10.1128/mBio.00193-15.
Maside X, Gómez-Moracho T, Jara L, Martín-Hernández R, De la Rúa P, Higes M, et al. Population genetics of Nosema apis and Nosema ceranae: one host (Apis mellifera) and two different histories. PLoS One. 2016;e0145609:10.
Raymann K, Shaffer Z, Moran NA. Antibiotic exposure perturbs the gut microbiota and elevates mortality in honeybees. PLoS Biol. 2017;15:e2001861.
Arredondo D, Castelli L, Porrini MP, Garrido PM, Eguaras MJ, Zunino P, et al. Lactobacillus kunkeei strains decreased the infection by honey bee pathogens Paenibacillus larvae and Nosema ceranae. Benef Microbes. 2018;9:279–90.
Khare A, Tavazoie S. Multifactorial competition and resistance in a two-species bacterial system. PLoS Genet. 2015;11:e1005715.
Forgie AJ, Fouhse JM, Willing BP. Diet-microbe-host interactions that affect gut mucosal integrity and infection resistance. Front Immunol. 2019;10:1802.
Erban T, Ledvinka O, Kamler M, Hortova B, Nesvorna M, Tyl J, et al. Bacterial community associated with worker honeybees (Apis mellifera) affected by European foulbrood. PeerJ. 2017;5:e3816.
Guo J, Wu J, Chen Y, Evans JD, Dai R, Luo W, et al. Characterization of gut bacteria at different developmental stages of Asian honey bees, Apis cerana. J Invertebr Pathol. 2015;127:110–4.
Baffoni L, Gaggia F, Alberoni D, Cabbri R, Nanetti A, Biavati B, et al. Effect of dietary supplementation of Bifidobacterium and Lactobacillus strains in Apis mellifera L. against Nosema ceranae. Benef Microbes. 2016;7:45–51.
Ptaszynska AA, Borsuk G, Zdybicka-Barabas A, Cytrynska M, Malek W. Are commercial probiotics and prebiotics effective in the treatment and prevention of honeybee nosemosis C? Parasitol Res. 2016;115:397–406.
Fries I, Chauzat M, Chen Y, Doublet V, Genersch E, Gisder S, et al. Standard methods for nosema research. In: Dietemann V, Ellis JD, Neumann P, editors. The COLOSS BEEBOOK: Volume II: Standard methods for Apis mellifera pest and pathogen research. J Apic Res; 2013. p. 52.
Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357.
Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40.
Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559.
Acknowledgements
We appreciate SWISS Institute of Bioinformatic (Vital-IT) for bioinformatic support.
Funding
The project is supported by initiation package of Jiangxi Agricultural University 050014/923230722. The funding body had no role in the design of the study, collection, analysis, data interpretation and writing the manuscript.
Author information
Authors and Affiliations
Contributions
QH and JDE designed the study, analyzed the data and wrote and manuscript. All authors gave final approval for publication.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
Authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1 File S1
sequencing alignment statistics and normalized microbe counts.
Additional file 2 File S2
code used for the statistics.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Huang, Q., Evans, J.D. Targeting the honey bee gut parasite Nosema ceranae with siRNA positively affects gut bacteria. BMC Microbiol 20, 258 (2020). https://doi.org/10.1186/s12866-020-01939-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12866-020-01939-9