Abstract
Background
Antimicrobial resistant bacteria among hospitalized patients are becoming a major public health threat worldwide, mainly in developing countries. Infections by these multidrug resistant pathogens cause high rate of mortality, prolong hospital stays, and affect individual and country economies in greater amounts. Thus, this study aimed to assess the bacterial profile, antimicrobial susceptibility status, and associated factors of isolates from hospitalized patients at the Dessie Comprehensive Specialized Hospital.
Methodology
This hospital-based cross-sectional study was conducted between February and April 2021. Consecutive sampling was used to select the study participants. All bacterial isolates were identified using standard bacteriological techniques. Antibiotic susceptibility testing was performed using disk diffusion technique. The data was analyzed using SPSS version 25. Descriptive statistics and logistic regression were used. A P-value of less than 0.05 was considered statistically significant.
Results
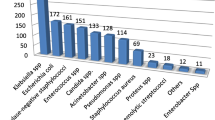
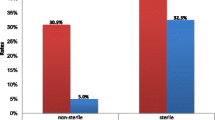
Of 384 clinical samples (blood, urine, stool, wound, vaginal discharge, and ear discharge) processed 180 (46.9%) were culture positive. Overall, Escherichia coli was the predominant isolate (41; 22.8%), followed by Staphylococcus aureus (36; 20%). Most of the isolates were from blood (70; 38.9%). The level of overall drug resistance of the gram-negative bacteria isolates for ampicillin, tetracycline, and cotrimoxazole was (104; 88.1%), (79; 75.9%), and (78; 75.0%), respectively. The overall multidrug rate of isolates was 143 (79.4%). Variables such as history of invasive procedures, chronic underlying diseases, history of hospitalization, and habit of eating raw animal products were statistically significant for the acquisition of bacterial infection.
Conclusions and recommendation
E. Coli and S. aureus were the most common isolates. Most of the isolates were resistant to commonly prescribed antibiotics. And also, consumption of raw animal products, chronic underlying disease, previous hospitalization, history of invasive procedures, and educational status were associated with the acquisition of bacterial infections. Therefore, routine antimicrobial susceptibility testing, proper patient management, wise use of antibiotics in clinical settings and health education are recommended.
Similar content being viewed by others
Introduction
The global distribution and growing number of pathogens resistant to antimicrobial drugs are potentially one of the greatest threats to global health, especially in sub-Saharan Africa, where the burden of infectious diseases is high. According to recent estimates, antimicrobial resistant (AMR) infections cause approximately 700,000 deaths per year, with the number expected to rise to 10 million by 2050 if current trends continue [1, 2].
Blood stream, urinary tract, wound, and lower respiratory tract infections are the most common illnesses caused by both gram-positive and gram-negative bacteria [3,4,5,6,7,8]. The most common pathogens associated with bacterial infections in both health care and community settings are Escherichia coli (E. coli), Staphylococcus aureus (S. aureus), Klebsiella pneumonia (K. pneumonia), Pseudomonas aeruginosa (P. aeruginosa), Streptococcus pneumoniae (S. pneumonia), and Salmonella spp [5, 9].
Microbial diseases cause 25% of the world’s 60 million yearly fatalities. Despite great improvements in infection control methods, infections caused by drug-resistant organisms remain a major source of morbidity and mortality in hospitalized patients and the community, especially in sub-Saharan African countries [5, 6]. Populations with low immunity, such as pediatrics, pregnant women, and patients with chronic illnesses who frequently visit health facilities, have a high incidence of bacterial infections, which increase mortality and morbidity [6, 10].
A recent report estimated that 10 million deaths will be attributed to AMR by 2050, and 100 trillion dollars of the world’s economic output will be lost if substantive efforts are not made to address this threat [2, 11]. Sub-Saharan Africa, including Ethiopia, has few comprehensive antimicrobial surveillance strategies and scarce infection prevention and control programs [1, 5, 6]. Unwise antimicrobial use and poor guidelines for over-the-counter sales of antibiotics are increasing in these countries, increasing the emergence and spread of multidrug resistant (MDR) pathogens [1, 5, 11].
Even if much data is required for the intervention of AMR at the national level, there is insufficient information showing the country-wide prevalence of the bacteria, antimicrobial susceptibility results, and MDR profiles. Furthermore, due to insufficient human resources, laboratory facilities, and diagnostic kits for antimicrobial susceptibility testing, data are scarce in hospitals. Therefore, this study aimed to identify the bacterial profiles, antimicrobial susceptibility patterns, and associated factors among hospital admitted patients at Dessie Comprehensive Specialized Hospital, Northeast Ethiopia.
Methods
Study area
The study was conducted at the Dessie Comprehensive Specialized Hospital. Dessie Comprehensive Specialized Hospital is a referral hospital that provides healthcare services for more than four million people in the area. It has more than 700 client loads per day (source: Dessie Comprehensive Specialized Hospital Report, 2020). The hospital consists of six major wards: emergency, surgical, medical, obstetrics and gynecology, pediatric, neonatal intensive care unit (NICU), and adult intensive care.
Study design, period, and population
A hospital-based cross-sectional study was conducted among hospitalized patients at the Dessie Comprehensive Specialized Hospital from February to April 2021. Patients admitted to the medical, surgical, obstetrics, gynecology, pediatric, and intensive care unit (ICU) wards for less than 48 h and who had clinical evidence of bacterial infection were considered the study population.
Sample size and sampling technique
A single population proportion formula was used to calculate the sample size, assuming a 95% confidence level, 5% margin of error, and taking 50% prevalence of community acquired bacterial infections in hospitalized patients. The final sample size was 384. The study participants were consecutively included until the required sample size was achieved.
Operational definition
Hospitalized patients: Patients admitted to the medical, surgical, obstetrics, gynecology, pediatric, and intensive care unit (ICU) wards for less than 48 h and who had clinical evidence of bacterial infection.
Data collection
Information on demographic variables and clinical data was collected from each participant through face-to-face interviews using a structured questionnaire and by reviewing patients’ medical records in consultation with their respective physicians. All the data were collected and recorded using a trained data collector.
Sample collection and processing
Clinical samples, including blood, urine, stool, wound, vaginal discharge, and ear discharge, were collected based on the type of disease which progressed in the participants. Samples were collected during the study period, transported, and processed according to standard procedures [12].
Blood sample
Venous blood (10, 2, and 1 mL from adults, children, and neonates, respectively) was aseptically collected. Immediately after collection, blood samples were added to tryptic soy broth (TSB) (Oxoid, Ltd.) in two bottles for each patient at approximately 1 h intervals [13] and incubated at 37 oC. If growth was observed, subculturing was performed on MacConkey agar (MAC) (HiMedia, India) and blood agar (BA) (HiMedia, India) plates. For chocolate agar (HiMedia, India) it was incubated in a 5–10% CO2 atmosphere using a candle jar at 37 °C for 24 to 48 h. When no visible growth was detected, the TSB tubes were incubated for 7 days before being reported as negative.
Urine sample
Ten milliliters of freshly voided midstream urine were collected using a sterile, wide-mouthed, and leak-proof container. For patients with a urinary catheter, the aperture was cleaned in a circular motion from the aperture opening outward using a cleansing solution. The open end of the catheter was placed in the specimen container, and 5–10 mL of urine was dripped into the container. Using a calibrated wire loop, 1 µL (0.001 mL) of a well-mixed urine sample was inoculated into a cysteine lactose electrolyte-deficient medium (CLED) (HiMedia, India) and incubated for 24 h. Colonies were counted to check for the presence of significant growth, and those yielding bacterial growth of ≥ 105 CFU/mL were considered significant bacteriuria [12]. The colonies from CLED agar were subcultured in MAC (Oxoid, Ltd.) and BA (Oxoid, Ltd.) and incubated at 37 °C for 24 h.
Wound sample
Wound swab/pus aspirate samples from each participant were collected aseptically from the depth of the wound with cotton swabs dipped in normal saline using the Levine technique. The sample was immersed in a container of brain-heart infusion transport medium. The brain heart infusion culture was incubated for 24 h, sub-cultured on MAC agar and BA plates, and re-incubated for 24 h at 37 oC.
Sputum sample
Two milliliters of sputum samples were collected from patients suspected of having a bacterial lower respiratory tract infection using sterile, clean, wide-necked, leak-proof containers and transported within 2 h [13]. Gram staining was performed on purulent portions of each sputum sample. Sputum samples were examined to determine the appropriateness of the culture. Samples with fewer than 10 polymorphonuclear neutrophils per epithelial cell were considered contaminated by saliva and rejected. Those sputum specimens of good quality were inoculated with MAC, BA, and chocolate (5–10% CO2) and incubated for 24–48 h at 35–37 °C [13].
Stools sample
Two grams of fecal specimen were collected, and rectal swabs were collected from children that could not provide a stool. The sample was transported using Cary-Blair transport medium (Oxoid, Basingstoke, UK), inoculated onto MAC, and incubated at 35–37 °C for 24 h. The sample was cultured on xylose lysine deoxycholate (XLD) agar from patients suspected of salmonella and Shigella spp. infections.
Cerebrospinal fluid sample (CSF)
Two to five milliliters of CSF were collected aseptically using two tubes by an experienced professional between the fourth and fifth lumbar vertebrae of the arachnoid space and were transported immediately. One tube was centrifuged for Gram staining, and the other tube was used for culturing [13]. For cloudy specimens, Gram staining was performed before centrifugation; if not cloudy, CSF was centrifuged for Gram staining. After centrifugation, the sediment was cultured on MAC, BA, and chocolate (5–10% CO2) and incubated for 24–48 h at 35–37 °C [12].
Vaginal and ear discharges sample
Vaginal and ear discharges were collected aseptically using sterile cotton swabs and transported using tryptic soya broth media. Vaginal and ear discharge was inoculated into MAC, BA, and chocolate (5–10% CO2) and incubated for 24–48 h at 35–37 °C [12].
All isolates were preliminarily screened by colony morphology, pigment production (pink to colorless flat or mucoid colonies), and Gram staining techniques. Further identification of the isolates was performed based on relevant biochemical tests [12]. Gram-negative bacteria were identified based on indole production, H2S production on KIA agar, citrate utilization, urease tests, carbohydrate fermentation on KIA, motility tests, and LDC tests. Gram-positive bacteria were identified using the catalase, coagulase, novobiocin, and bacitracin sensitivity tests.
Antimicrobial susceptibility testing
The antimicrobial susceptibilities of the isolates were determined according to the criteria of the Clinical and Laboratory Standard Institute 2020 (CLSI) using the Kirby-Bauer disk diffusion method on Mueller-Hinton Agar [14]. Antimicrobial disks used for susceptibility testing were cefoxitin (30 µg), clindamycin (2 µg), erythromycin (15 µg), and penicillin G (10 IU). Ampicillin (10 µg), cefotaxime (30 µg), ciprofloxacin (5 µg), chloramphenicol (30 µg), trimethoprim-sulfamethoxazole (1.25/23.25 µg), tetracycline (30 µg), and gentamicin (10 µg) were used for both gram-negative and gram-positive bacteria, whereas ceftriaxone (30 µg) and amoxicillin-clavulanic acid (20/10 µg) were used for gram-negative bacteria. The inoculated plates with antimicrobial disks were incubated at 35–37 0C for 16–18 h, and the diameters of the zones of inhibition were measured using a ruler and interpreted according to CLSI standards [15]. Bacterial isolates that are resistant to three or more antibiotics from different classes are considered multi-drug resistant [16].
Data and laboratory quality control
The questionnaires were checked for completeness during and after data collection. All laboratory assays were performed while maintaining quality control procedures. The sterility of the medium was assessed by incubating 5% of the batch at 35–37 0C overnight. Antimicrobial sensitivity testing disks were checked using standardized reference strains of S. aureus ATCC25923, E. coli ATCC25922, and P. aeruginosa ATCC27853 [15].
Data analysis
Data were analyzed using the Statistical Package for Social Sciences (SPSS) version 25. Descriptive statistics, such as frequency and percentage, were calculated and are presented in the tables. Binary logistic regression was used to assess the association between individual explanatory variables and dependent variables. Odds ratios were calculated using the respective confidence intervals. Furthermore, variables with P ≤ 0.2 in the bivariable analysis were entered into multivariable logistic regression. A P-value less than 0.05 was considered to be statistically significant.
Result
Socio-demographic and clinical characteristics
Among the 384 study participants enrolled in the study, 230 (59.9%) were male. The age of the study participants ranged from 4 days to 85 years, with a median age of 36 years. The majority of the participants were rural residents (238; 62%) (Table 1).
Bacterial isolation rate
A single specimen was collected from each participant, and the overall culture positivity rate was 180 (46.9%). Among the isolates, gram negative bacteria were predominant (119/180; 66.1%) and from all bacteria E. coli (41; 22.8%) was the most prevalent, followed by S. aureus (36; 20.0%), and K. pneumoniae (21; 11.7%). The majority of the isolates were from blood samples (70; 38.9%) and urine samples (68; 37.8%) (Table 2).
Antimicrobial resistance profile of gram negative bacterial isolates
The overall resistance rates of gram negative bacteria to ampicillin, tetracycline, and trimethoprim-sulfamethoxazole were highest at 88.1%, 75.9%, and 75%, respectively. However, these isolates exhibited the lowest overall resistance to ceftriaxone (41.2%). From the isolates E. coli showed the lowest resistance to ceftriaxone (39%) but the highest for ampicillin (95.1%) and trimethoprim-sulfamethoxazole (87.8%). The level of resistance of K. pneumoniae was highest for ampicillin (81.0%) and tetracycline (76.2%) (Table 3).
Antimicrobial resistance patterns of gram positive bacterial isolates
The overall resistance rates of the gram-positive bacteria to ampicillin, tetracycline, and trimethoprim-sulfamethoxazole were 77.0%, 72.1%, and 71.7%, respectively. Staphylococcus aureus showed high resistance to ampicillin (80.6%), penicillin G, tetracycline (72.2% each), and trimethoprim–sulfamethoxazole (69.4%), whereas S. saprophyticus showed equal resistance to ampicillin and trimethoprim–sulfamethoxazole (75%) (Table 4).
Multidrug resistance profiles of isolates
Of all bacterial isolates, 95% of the isolates were resistant to at least one of the selected antibiotics. The overall multidrug resistance rate (bacteria resistant to three or more different classes of drugs) of isolated bacteria was 143 (79.4%). Among gram negative bacteria a higher MDR rate was shown to K. pneumoniae and E. coli (90.5% and 90.2%, respectively). Among the gram-positive bacteria, the MDR rates of S. aureus and S. saprophyticus were 78.8% and 75%, respectively (Table 5).
Associated factors of bacterial infection
Bivariable logistic regression analysis was performed for the sociodemographic and clinical variables. Those with P ≤ 0.2 were further analyzed using multivariable logistic regression. In the multivariate analysis, clients who couldn’t read and write had higher odds of having bacterial infection than those who had a degree (AOR = 3.84; 95% CI = 1.83–8.03). Participants who had chronic underlying disease were 1.83 times more likely to develop bacterial infection (95% CI = 1.05–3.17) as compared with those who did not. Moreover, history of having an invasive procedure had 2.09 times the odds of acquiring bacterial infection than their counterparts (95% CI = 1.06–4.49) (Table 6).
Discussion
Our study provides a comprehensive description of the epidemiology and microbiological causes of community-acquired infections in a specialized hospital in Northeast Ethiopia. In this study, the prevalence of bacterial infection was 46.9%. This was consistent with studies conducted in Debre Markos, Ethiopia, at 46.7% and 48.7% [17, 18], Bahir Dar, Ethiopia, at 49.4% [19], and Tanzania, at 42.2% [20]. However, this was higher than findings from studies conducted in Hawassa, Ethiopia, at 28% [21] and Tanzania, at 14.8% [22]. Lower findings were observed when compared with studies from Ethiopia, at 64% [23] and Pakistan, at 87.17% [24]. This variation may be due to geographic differences, seasonal variations, differences in study methods, and variations in infection control programs.
In the current study, gram-negative bacteria were the dominant isolates, accounting for 66.1%. This was supported by studies conducted in Ethiopia (67.8%) [21] and Indonesia (68.6%) [25]. The most predominant gram-negative isolate in this study was E. coli (22.8%), followed by K. pneumoniae (11.7%), which is similar to other studies conducted in Ethiopia [26, 27] and Uganda [28]. This might be due to their major abundance as a normal flora and the fact that they can easily be transported to sterile areas of the body and cause infection [29].
In contrast, 33.9% of the isolates were gram-positive bacteria. Among these, S. aureus was the leading isolate. This finding is supported by studies conducted in Gondar, Ethiopia [27], and Indonesia [25]. Overall, E. coli and S. aureus were present at higher proportions. This finding is similar to those of other studies from Hawassa and Jimma, Ethiopia [21, 23], and Zimbabwe [30].
In the current study, 171 (95%) of the isolates were resistant to at least one antimicrobial tested, which is similar to a study conducted in Bahir Dar (94.4%) [26]. However, this result was higher than that reported in Debre Markos and Gondar, Ethiopia [18, 27]. This may be due to increased overuse of antibiotics and self-medication, resulting in antibiotic resistance. Generally, amplified resistance is caused by the use of broad-spectrum agents, and the use of medications as a result of insufficient doses or incomplete treatment courses plays a significant role in the emergence of antimicrobial resistance [31].
In this study, the overall multi-drug resistance profile of isolates was 79.4%, which was comparable with findings from Debre Markos, Ethiopia (76.1%) [32]. However, this result was higher compared to studies from Indonesia (47.7%) [25] and Bahir Dar, Ethiopia (61.8%) [26]. This indicates that antibiotic resistance, especially MDR, is a growing risk in Ethiopia. This might be linked to the misuse, overuse, and inappropriate use of antibacterial agents. Hospitals receive referrals from many districts and distant rural villages. These patients received different antibiotic treatments from the referring centers or over-the-counter centers, usually with inappropriate doses, before arriving at the hospital.
Among the gram-negative isolates, the levels of MDR for K. pneumoniae and E. coli were 90.5% and 90.2%, respectively. The level of MDR for gram-positive bacteria such as S. aureus and S. saprophyticus were 78.8% and 75%. This finding also showed that gram-negative isolates had a higher rate of MDR compared to other studies, similar to the findings in Bahir Dar [33]. The difference in MDR levels between gram-negative and gram-positive bacteria may be due to different gram-negative mechanisms, including altered target sites, β-lactamase production, decreased antibiotic penetration, and efflux pumps [34].
In this study, sociodemographic and clinical factors were analyzed as independent risk factors for bacterial infection. Using multivariable logistic regression, different variables were identified as contributing factors to bacterial infection. The presence of chronic underlying disease, previous hospitalization, history of invasive procedures, and consumption of raw animal products showed a statistically significant association with the acquisition of bacterial infection.
According to our study, patients with chronic underlying disease had 1.83 times the odds of increased acquisition of bacteria than patients who did not have an underlying disease (95% CI = 1.05–3.17). This might be because participants with underlying chronic diseases may be exposed to many drugs, frequently interact with health professionals, and have a poor immune status, which can contribute to bacterial infection. This finding is supported by those of other studies conducted in Ethiopia [35, 36] and China [37].
The other independent factor associated with bacterial infection was a history of hospitalization. Having a history of previous hospitalization had 1.96 times the odds of increased acquisition of bacteria than patients with no history of previous hospitalization (95% CI = 1.09–3.54). This finding is supported by other studies conducted in Brazil and India [38, 39].
The educational status of the illiterate individuals and those who read and write was also significantly associated with the acquisition of bacteria. Participants with no formal education lacked personal hygiene knowledge. This is similar with studies in Ethiopia [40, 41].
Another factor associated with the acquisition of bacterial infections was the history of invasive procedures. In this study, participants who had a history of an invasive procedure had 2.09 times the odds of acquiring bacterial infection than those who didn’t have such a history (95% CI = 1.06–4.49). This finding is supported by other studies conducted in Ethiopia [42].
Conclusions and recommendation
The magnitude of community-acquired bacterial infection was 47%. Gram-negative pathogens were predominant that account for more than half of the cases. E. Coli and S. aureus were the most common pathogens identified. Antimicrobial resistance among common isolates was high for routinely used antibiotics and the rate of MDR was alarming. Chronic underlying disease, previous hospitalization, history of invasive procedures, consumption of raw animal products, and educational status of illiterate, and read and write had significant association were acquisition of bacterial infection. Strengthening of antimicrobial resistance surveillance system, antibiotic stewardship programs and effective antibiotic policy is required to halt the transmission of disease including drug resistant bacteria in the community.
Data availability
The findings of this study were generated from the data collected and analyzed based on the stated methods and materials. All data have already been found in the manuscript, and there are no supplementary files. The original data supporting this finding are available upon request.
Abbreviations
- AIDS:
-
Acquired Immune Deficiency Virus
- AMR:
-
Anti-Microbial Resistance
- BSI:
-
Blood Stream Infection
- CLED:
-
Cysteine Lactose Electrolyte Deficient
- CLSI:
-
Clinical and Laboratory Standard Institute
- CoNS:
-
Coagulase Negative Staphylococcus
- CSF:
-
Cerebrospinal Fluid
- HIV:
-
Human Immune Deficiency Virus
- ICU:
-
Intensive Care Unit
- LRTI:
-
Lower Respiratory Tract Infection
- MDR:
-
Multi Drug Resistant
- MRSA:
-
Methicillin-Resistant Staphylococcus Aureus
- MR-VP:
-
Methyl Red-Vogues Proskauer
- NICU:
-
Neonatal Intensive Care Unit
- TSI:
-
Triple Sugar Iron
- UTI:
-
Urinary Tract Infection
- WHO:
-
World Health Organization
- XLD:
-
Xylose Lysine Decarboxylate
References
Leopold SJ, van Leth F, Tarekegn H, Schultsz CJJAC. Antimicrobial drug resistance among clinically relevant bacterial isolates in sub-saharan Africa: a systematic review. 2014;69(9):2337–53.
O’Neil, JJARTacfth. Nations wonHW. Rev Antibiotic Resisitance. 2014:1–16.
Rai S, Yadav UN, Pant ND, Yakha JK, Tripathi PP, Poudel A et al. Bacteriological profile and antimicrobial susceptibility patterns of bacteria isolated from pus/wound swab samples from children attending a tertiary care hospital in Kathmandu, Nepal. 2017;2017.
Scott K, George AS. Ved RRJHrp, systems. Taking stock of 10 years of published research on the ASHA programme: examining India’s national community health worker programme from a health systems perspective. 2019;17:1–17.
Beyene G, Tsegaye WJE. Bacterial uropathogens in urinary tract infection and antibiotic susceptibility pattern in jimma university specialized hospital. Southwest Ethiopia. 2011;21(2):141–6.
Khan S, Priti S, Ankit SJI. Bacteria etiological agents causing lower respiratory tract infections and their resistance patterns. 2015;19(4):240.
Regha I, Sulekha BJIJMMTD. Bacteriological profile and antibiotic susceptibility patterns of lower respiratory tract infections in a tertiary care hospital. Cent Kerala. 2018;4(4):186–90.
Khurana S, Bhardwaj N, Kumari M, Malhotra R, Mathur, PJJolp. Prevalence, etiology, and antibiotic resistance profiles of bacterial bloodstream infections in a tertiary care hospital in Northern India: a 4-year study. 2018;10(04):426–31.
Tacconelli E. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development. 2017.
Carcillo JA, Dean JM, Holubkov R, Berger J, Meert KL, Anand KJ et al. Inherent risk factors for nosocomial infection in the long stay critically ill child without known baseline immunocompromise: a post–hoc analysis of the CRISIS trial. 2016;35(11):1182.
Lim C, Takahashi E, Hongsuwan M, Wuthiekanun V, Thamlikitkul V, Hinjoy S et al. Epidemiology and burden of multidrug-resistant bacterial infection in a developing country. 2016;5:e18082.
Cheesbrough M. District laboratory practice in tropical countries, part 2. Cambridge University Press; 2006.
Vandepitte J, Verhaegen J, Engbaek K, Rohner P, Piot P, Heuck C, et al. Basic laboratory procedures in clinical bacteriology. World Health Organization; 2003.
Desta M, Amha H, Anteneh Bishaw K, Adane F, Assemie MA, Kibret GD et al. Prevalence and predictors of uterine rupture among Ethiopian women: a systematic review and meta-analysis. 2020;15(11):e0240675.
Weinstein MP, Lewis JS. The Clinical and Laboratory Standards Institute Subcommittee on Antimicrobial susceptibility testing: background, Organization, functions, and processes. J Clin Microbiol. 2020;58(3).
Magiorakos A-P, Srinivasan A, Carey Rt, Carmeli Y, Falagas Mt G, Ct, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–81.
Abebe M, Tadesse S, Meseret G, Derbie A. Type of bacterial isolates and antimicrobial resistance profile from different clinical samples at a Referral Hospital, Northwest Ethiopia: five years data analysis. BMC Res Notes. 2019;12(1):1–6.
Mulu W, Abera B, Yimer M, Hailu T, Ayele H, Abate D. Bacterial agents and antibiotic resistance profiles of infections from different sites that occurred among patients at Debre Markos Referral Hospital, Ethiopia: a cross-sectional study. BMC Res Notes. 2017;10(1):1–9.
Moges F, Eshetie S, Abebe W, Mekonnen F, Dagnew M, Endale A, et al. High prevalence of extended-spectrum beta-lactamase-producing gram-negative pathogens from patients attending Felege Hiwot Comprehensive Specialized Hospital, Bahir Dar, Amhara region. PLoS ONE. 2019;14(4):e0215177.
Kumburu HH, Sonda T, Mmbaga BT, Alifrangis M, Lund O, Kibiki G, et al. Patterns of infections, aetiological agents and antimicrobial resistance at a tertiary care hospital in northern Tanzania. Tropical Med Int Health. 2017;22(4):454–64.
Amsalu A, Geto Z, Asegu D, Eshetie S. Antimicrobial resistance pattern of bacterial isolates from different clinical specimens in Southern Ethiopia: a three year retrospective study. Afr J Bacteriol Res. 2017;9(1):1–8.
Mnyambwa NP, Mahende C, Wilfred A, Sandi E, Mgina N, Lubinza C, et al. Antibiotic susceptibility patterns of bacterial isolates from routine clinical specimens from Referral hospitals in Tanzania: a prospective hospital-based observational study. Infect drug Resist. 2021;14:869.
Gashe F, Mulisa E, Mekonnen M, Zeleke G. Antimicrobial Resistance Profile of different clinical isolates against third-generation cephalosporins. J Pharm. 2018;2018:5070742.
Sabir R, Alvi SFD, Fawwad A. Antimicrobial susceptibility pattern of aerobic microbial isolates in a clinical laboratory in Karachi-Pakistan. Pakistan J Med Sci. 2013;29(3):851.
Masyeni S, Sukmawati H, Siskayani AS, Dharmayanti S, Sari K. Antimicrobial susceptibility pattern of Pathogens isolated from various specimens in Denpasar-Bali: a two years retrospective study. Biomedical Pharmacol J. 2018;11(1):493–502.
Yitayeh L, Gize A, Kassa M, Neway M, Afework A, Kibret M, et al. Antibiogram profiles of Bacteria isolated from different body site infections among patients admitted to GAMBY Teaching General Hospital, Northwest Ethiopia. Infect Drug Resist. 2021;14:2225–32.
Adane A, Belay G, Tamirat KS. Microbiological Profile and Drug-Resistance Pattern of pathogens among patients who visited the University of Gondar Comprehensive Specialized Hospital, Ethiopia. Infect Drug Resist. 2020;13:4449.
Odoki M, Almustapha Aliero A, Tibyangye J, Nyabayo Maniga J, Wampande E, Drago Kato C et al. Prevalence of bacterial urinary tract infections and associated factors among patients attending hospitals in Bushenyi district, Uganda. International journal of microbiology. 2019;2019.
Tadesse S, Kahsay T, Adhanom G, Kahsu G, Legese H, Derbie A. Prevalence, antimicrobial susceptibility profile and predictors of asymptomatic bacteriuria among pregnant women in Adigrat General Hospital, Northern Ethiopia. BMC Res Notes. 2018;11(1):1–6.
Mhondoro M, Ndlovu N, Bangure D, Juru T, Gombe NT, Shambira G, et al. Trends in antimicrobial resistance of bacterial pathogens in Harare, Zimbabwe, 2012–2017: a secondary dataset analysis. BMC Infect Dis. 2019;19(1):1–9.
Worku S, Gelaw A, Aberra Y, Muluye D, Derbie A, Biadglegne F. Bacterial etiologies, antibiotic susceptibility patterns and risk factors among patients with ear discharge at the University of Gondar Hospital, Northwest Ethiopia. Asian Pac J Trop Dis. 2017;7(1):36–42.
Negussie A, Mulugeta G, Bedru A, Ali I, Shimeles D, Lema T, et al. Bacteriological profile and antimicrobial susceptibility pattern of blood culture isolates among septicemia suspected children in selected hospitals Addis Ababa, Ethiopia. Int J Biol Med Res. 2015;6(1):4709.
Fenta A, Dagnew M, Eshetie S, Belachew T. Bacterial profile, antibiotic susceptibility pattern and associated risk factors of urinary tract infection among clinically suspected children attending at Felege-Hiwot comprehensive and specialized hospital, Northwest Ethiopia. A prospective study. BMC Infect Dis. 2020;20(1):1–10.
Peterson E, Kaur PJF. Antibiotic resistance mechanisms in bacteria: relationships between resistance determinants of antibiotic producers, environmental bacteria, and clinical pathogens. 2018;9:2928.
Afenigus A, Shbabawu A, Melese T. Surgical site infection and associated factors among adult patients admitted in west and east Gojjam Zone hospitals, Amhara region. Ethiopia Nurse Care Open Acces J. 2019;6(3):107–12.
Shimekaw M, Tigabu A, Tessema B. Bacterial Profile, Antimicrobial susceptibility pattern, and Associated Risk factors among patients with wound infections at Debre Markos Referral Hospital, Northwest, Ethiopia. Int J Low Extrem Wounds 2020:1534734620933731.
Cheng K, Li J, Kong Q, Wang C, Ye N, Xia G. Risk factors for surgical site infection in a teaching hospital: a prospective study of 1,138 patients. Patient Prefer Adherence. 2015;9:1171.
Fram D, Okuno MFP, Taminato M, Ponzio V, Manfredi SR, Grothe C, et al. Risk factors for bloodstream infection in patients at a Brazilian hemodialysis center: a case–control study. BMC Infect Dis. 2015;15(1):1–9.
Pathak A, Upadhayay R, Mathur A, Rathi S, Lundborg CS. Incidence, clinical profile, and risk factors for serious bacterial infections in children hospitalized with fever in Ujjain, India. BMC Infect Dis. 2020;20(1):1–11.
Mechal T, Hussen S, Desta M. Bacterial Profile, Antibiotic Susceptibility Pattern and Associated factors among patients attending adult OPD at Hawassa University Comprehensive Specialized Hospital, Hawassa, Ethiopia. Infect Drug Resist. 2021;14:99.
Gutema T, Weldegebreal F, Marami D, Teklemariam Z. Prevalence, antimicrobial susceptibility pattern, and associated factors of urinary tract infections among adult diabetic patients at Metu Karl Heinz Referral Hospital, Southwest Ethiopia. International journal of microbiology. 2018;2018.
Shiferaw WS, Aynalem YA, Akalu TY, Petrucka PM. Surgical site infection and its associated factors in Ethiopia: a systematic review and meta-analysis. BMC Surg. 2020;20:1–15.
Acknowledgements
This research was supported by the College of Medicine and Health Sciences at Wollo University. We would like to acknowledge the Dessie Comprehensive Specialized Hospitals for their permission to conduct laboratory work in their setting. We also acknowledge the Wollo University Microbiology Laboratory for permission to conduct the antimicrobial sensitivity tests.
Funding
This research was not funded.
Author information
Authors and Affiliations
Contributions
A.S. was involved in conceptualization, collected the data, performed the microbiological investigation, analyzed the data, and critically edited the manuscript. Ag.S. was conducted designed and supervised the entire study, critically revised and contributed to the scientific content of the manuscript. Ab.S. was involved in designed and supervised the entire study, critically revised and contributed to the scientific content of the manuscript. S.T. was involved the analysis and interpretation of the data, substantially revised and critically edited the manuscript, and contributed to the scientific content of the manuscript. W.A. was involved in the analysis and interpretation of the data, substantially revised and critically edited the manuscript, and contributed to the scientific content of the manuscript.
Corresponding author
Ethics declarations
We declare that this manuscript is original and has not been presented or published in this University or institution. And also, all methods were achieved based on appropriate guidelines and regulations.
Ethical approval and Informed consent
This study followed the ethical standards of the Declaration of Helsinki. And also, this study protocol was reviewed and approved by the Research and Ethics Review Committee of the College of Medicine and Health Sciences of Wollo University. Written permission was obtained from the Dessie Comprehensive Specialized Hospital. The purpose and procedures of the study were explained to the participants, parents, and guardians. Written informed consent from the age of 18 and above and/or assent from those less than 18 years old study participants along with written informed consent from their respective parents/ caregiver/guardians was obtained. The confidentiality of all study participants was maintained.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Sisay, A., Seid, A., Tadesse, S. et al. Assessment of bacterial profile, antimicrobial susceptibility status, and associated factors of isolates among hospitalized patients at Dessie Comprehensive Specialized Hospital, Northeast Ethiopia. BMC Microbiol 24, 116 (2024). https://doi.org/10.1186/s12866-024-03224-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12866-024-03224-5