Abstract
Background
At lower concentrations copper (Cu), zinc (Zn) and nickel (Ni) are trace metals essential for some bacterial enzymes. At higher concentrations they might alter and inhibit microbial functioning in a bioreactor treating wastewater. We investigated the effect of incremental concentrations of Cu, Zn and Ni on the bacterial community structure and their metabolic functions by shotgun metagenomics. Metal concentrations reported in previous studies to inhibit bacterial metabolism were investigated.
Results
At 31.5 μM Cu, 112.4 μM Ni and 122.3 μM Zn, the most abundant bacteria were Achromobacter and Agrobacterium. When the metal concentration increased 2 or fivefold their abundance decreased and members of Delftia, Stenotrophomonas and Sphingomonas dominated. Although the heterotrophic metabolic functions based on the gene profile was not affected when the metal concentration increased, changes in the sulfur biogeochemical cycle were detected. Despite the large variations in the bacterial community structure when concentrations of Cu, Zn and Ni increased in the bioreactor, functional changes in carbon metabolism were small.
Conclusions
Community richness and diversity replacement indexes decreased significantly with increased metal concentration. Delftia antagonized Pseudomonas and members of Xanthomonadaceae. The relative abundance of most bacterial genes remained unchanged despite a five-fold increase in the metal concentration, but that of some EPS genes required for exopolysaccharide synthesis, and those related to the reduction of nitrite to nitrous oxide decreased which may alter the bioreactor functioning.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Background
Metal contamination of water resources comes mostly from municipal, industrial, agricultural and mining wastewater [18, 24, 33]. In water, metals can be found as free ions or complexed with organic ligands [24]. Although their toxicity has been well described, some metals, such as iron (Fe), copper (Cu), manganese, cobalt, nickel (Ni), selenium and zinc (Zn) are essential nutrients for several organisms. Because of their intrinsic chemical orbitals, Fe and Cu may participate in redox reactions as those catalyzed by oxidoreductases or Fenton catalyzers [10, 33].
Copper is found naturally in the environment (< 200,000 mg Kg−1 in soil, < 69 mg L−1 in water and 0.79 × 10–6 mg L−1 in air), but higher concentrations might be reached due to anthropogenic contamination [3]. Copper contamination is generally the result of mining or recycling, but some fungicides and herbicides also contain Cu salts. Because of its redox potential, Cu can inhibit several enzymes and produce oxidative stress in bacteria [18]. Nickel also occurs naturally in the environment (< 165 mg Kg−1 in soil, < 18.2 mg L−1 in water, < 0.18 × 10–6 mg L−1 in air) and contamination might occur through mining, refining of oil or through coal-fired power plants. Humans are exposed to Ni by tobacco and e-cigarettes, food and water or by contact with materials or soil containing Ni [4]. Only some proteins are known to use or contain Ni, with urease, the most important one, using Ni as cofactor [40]. Nickel may affect the bacterial membrane, inhibit or deactivate enzymes and produce oxidative stress [18]. Zinc is found at 71 mg Kg−1 in the upper continental crust and is the most important transition metal in humans after iron [13]. Zinc-finger motifs are involved in gene expression. This and other functions make Zn ions essential for eukaryotic growth and cell division [14]. Zinc is also found in proteins involved in redox activity, e.g. alcohol dehydrogenase and some isoenzymes of superoxide dismutase [12]. At higher concentrations, i.e., higher than those needed for metabolism, which is usually > mM level depending on the organism, Zn may negatively affect bacterial growth and biomass production [18].
Although the effect of metals on bioreactor microbiota has been described [1, 7, 23], most of the current knowledge comes from studies with axenic cultures. In bacteria, some enzymes contain or use Cu, Zn or Ni as cofactors making them essential for bacterial metabolism. Despite that, these three metals may be toxic, mutagenic or even lethal for bacteria when their concentration become too high in the environment, i.e. higher than those needed for a normal metabolic functioning [21, 24]. As mentioned before, at higher concentrations Cu and Ni may produce oxidative stress in bacteria while Zn reduces bacterial growth.
As a response, bacteria may produce biofilms or chelates, or use other mechanisms to mitigate the negative effects of metals [24]. An increase in energy metabolism by bacteria has been suggested as a response to metal stress in river sediments, which significantly reduced their richness [41]. Similarly, in wastewater bioreactors, metals induced bacteria to produce metalloproteins (like cytochrome or heme-carriers), siderophores [23], or mutations leading to antibiotic and metal resistance [21]. As metals may alter the microbiota community structure in wastewater treatment plants, they might reduce organic matter biodegradation [7, 23]. Studying the impact of these metals on the bacterial community structure may help to understand their effect at a micro-ecosystem level and in the treatment of wastewater.
In this study, two bioreactors were used with activated sludge and fed continuously with synthetic wastewater as described in Esquivel-Rios et al. [17]. One bioreactor was supplied with increased concentrations of Cu, Ni and Zn and the bacterial community was determined each time the amount of metals applied increased. An unamended bioreactor kept under the same operational conditions served as control. The aim of this study was to investigate how the increased application of Cu, Ni and Zn affected the bacterial community and its metabolic function capabilities as determined through shotgun metagenomics. It was hypothesized that the bacterial functionality (as measured by abundance of genes encoding for them) would follow any observed changes in bacterial community structure.
Methods
Bioreactors
Two stirred tank bioreactors were operated for 166 days. Details of the wastewater used to inoculate the bioreactors and how they were maintained can be found in Esquivel et al. [17]. The 6-L bioreactors were inoculated with 1 L of wastewater sludge (from Mexico city), fed for nine weeks prior to experiment with synthetic water (see Annex I), i.e., > 30 working volumes were used to acclimate the inoculum. Experimental conditions are described in Supplementary File Annex I, and these conditions were used also during experiment. One of the bioreactors was amended with metals, i.e. the synthetic water applied to the bioreactor contained the metal divalent ions at 31.5 μM Cu, 112.4 μM Ni and 122.3 μM Zn (considered the HM-1 phase). After 25 days of this first treatment, the metal concentration was doubled in the synthetic water to 62.8 μM Cu, 224.9 μM Ni and 244.6 μM Zn (considered the HM-2 phase). After an additional 45 days, the water applied was increased five-fold to 157.4 μM Cu, 562.2 μM Ni and 611.5 μM Zn (considered the HM-5 phase). The metal concentrations in the bioreactor were derived from the inorganic salts added. Their bioavailability is likely to be lower due to the high organic matter content in bioreactors. Despite that, those concentrations were previously found to respirometrically inhibit the activity of an inoculum under the same operating conditions [17]. Samples were taken just before the metal concentration was increased two fold and five-fold and at the end of the experiment, i.e. after 166 days. As such, three samples were taken from the metal amended bioreactor. The unamended bioreactor, which served as control, was applied with the same synthetic water without metals and sampled at the beginning (considered the CON-b sample) and the end of the experiment (considered the CON-e sample).
DNA extraction and sequencing
Metagenomic DNA was extracted by mechanical homogenization and purified by enzymatic/alkaline treatment. Water samples were centrifuged and the pellets obtained were treated with 1.2 mL lysis buffer I (80 μL lysozyme in 10 mM Tris–HCl pH 8). After 1 h incubation at 37 ºC, 1 mL buffer II (0.1 M NaCl, 0.5 M Tris–HCl, 12% SDS, pH 8.0) and 0.5 g sterile sand was added to the samples. Samples were vortexed for 10 min and placed on dry ice (-70 ºC) for 20 min. Microtubes were heat-shocked at 70 ºC for 20 min and centrifuged. The supernatants were transferred to new tubes and added with 115 μL 0.5 M EDTA pH 8.0 and 5 M potassium acetate pH 5 in a 1:9 volume ratio. After refrigeration at 4 ºC for 10 min, samples were centrifuged at 13,000 rpm at 4 ºC for 10 min. The supernatant was transferred to new tubes containing 400 μL chloroform-isoamyl alcohol-phenol solution (24:1:25). Samples were vortexed and centrifuged again as described above. The aqueous phase was transferred to new tubes and the organic phase was extracted. All aqueous extracts from the same sample were pooled together. The DNA was precipitated with 13% PEG (1:1 volume ratio), vortexed and incubated at -20 ºC overnight. Samples were centrifuged and pellets were washed with 500 μL chilled 70% ethanol. After centrifugation, pellets were air-dried before been resuspended with 50 μL sterile water.
The integrity of the DNA was confirmed by agarose electrophoresis and quantified on a NanoDrop™ 3300 (Thermo Scientific, Carlsbad, CA). The DNA was sequenced by Macrogen Inc. (Seoul, Korea) using a HiSeq2000 Illumina® 2 × 100 paired-end platform. The raw sequence datasets were deposited in the NCBI under BioProject ID PRJNA1009125, BioSample ID SAMN37147121 and SRA numbers: SRR26071885, SRR26071886, SRR26071887, SRR26071888, SRR26071889. The number of reads were > 20 million sequences/sample (Supplementary Table I).
Bioinformatic analysis
After trimming and adapter-elimination with trimmomatic and paired with Flash [8], contigs were assembled with SPADES [5], and annotated using Kraken2 [37] and the Genbank nr bacteria database. Relative abundances were calculated as a percentage of the total sum of final sequence counts.
Diversity replacement of the bacterial genera was calculated according to Legendre [20]:
where: b are the unique genera of sample 1 (genera found only in one sample and not in others), c are the unique genera of sample 2, and N is the total number of genera of both samples. Replacement for all samples were calculated using CON-b as the reference for comparison.
DiTing was used to determine metabolic functions with "meta" presets [39] and heatmaps were generated with MATLAB 2017a. Dissimilatory nitrate reduction was calculated as given in eqs. 2 to 4 to include the specific nitrate influx pump and the possible participation of isoenzymes in different steps of nitrate reduction as given by UniProt, the KEGG database and [26].
Assimilatory nitrate reduction was defined as a three-step process:
1. Uptake
2. Reduction of nitrate to nitrite
3. Reduction of nitrite to ammonia
Statistical analysis
Hill numbers were calculated as described by [11]. Mean values and confidence intervals were calculated after 1,000 bootstrap re-sampling. Value distribution was used to generate violin plots. Venn diagrams were drawn using the bioinformatics and evolutionary genomics page [35]. Circos plots were drawn using the circosChart function from MATLAB Central File Exchange [25] with some modification for representation, but not for data computation.
Circos plots allow easy interpretation of shared organisms classified at a certain taxonomic level, but the higher the level, the less the "unique" taxon becomes. Most phyla were detected in all samples, but at lower taxonomic levels, the percentage of shared taxa decreased. For instance, in the circos plot for genus, unique genera were barely detectable due to the large number of genera, whereas at higher taxonomic level less unique bacterial groups (e.g. class or phyla) were found. Circos plots may mask the lack of sensitivity of a method determining richness, in this case richness of microorganisms, but it can be applied with genes or other characteristics plotted within circos. Despite that, circos plots allow to get a good idea of how many items (genera in our case) are shared among samples. Contrario sensu, it is easy to observe groups in a network and clustering analysis, as well as, those genera that differ from the rest.
Partial correlation was calculated with Eq. 5 and the p value was obtained from the F value Eq. 6 described by [19]:
with: rab.c the partial correlation between factor a and b subtracting the contribution of c, rab the Pearson correlation r between factors a and b, and so on.
with: 2 arises from v = n-m-1 with m = the number of variables beside "a" (two) and n the sample data number (five in this study).
Multidimensional multivariate statistics were done with MATLAB 2017a. Chi-square distances were used for Q-mode analysis as they are a better representation than Euclidean distances when multiple zeros were found [19]. Procrustes analyses were done using chi-square distances. The goodness-of-fit criterion was the sum of squared errors. Both possible transformations (control—> treated and treated—> control) were calculated and plotted. Although the components matrices for transformation differed, goodness-of-fit gave the same results. Clustering analyses were performed using the k-means method. Prior to determine the best number of clusters, an estimation of optimum clustering was done using the silhouette value as a criterion with k ranging from 2 to 20. Centered relative abundances were used for principal component analysis (PCA). Contribution of variance by eigenvalues was given as a percentage of the total variance. Network analysis was done and graphed using Gephi 0.10.1 [6]. Sample node size was ranked according to the betweenness centrality metric. Color in the figure was assigned according to community detection (modularity) using relative abundance in each sample as a weight factor. The “Force Atlas” algorithm was used for presentation and network visualization. Bray–Curtis dissimilarities were used for non-classical multidimensional scaling (NMDS) as this statistic allow to quantify compositional dissimilarity among samples. Stress normalized by the sum of squares of the inter-point distances was used as a goodness-of-fit criterion in NMDS to two dimensions (components). The relative abundance of genes were considered for the metabolic function they encode, and it was reported as counts per million (cpm) [39].
The metal concentration factor was used as descriptor transformed into dummy columns for R-mode analysis. Euclidean distances were calculated for redundancy analysis. The contribution to variance is presented as a percentage of the total variance (canonical (Yhat) plus non-canonical axes (Yres)) for each RDA-axis as described in [19].
Results
The bacterial diversity and community structure
Most of the organisms found in both bioreactors belonged to the bacterial domain (Table 1). In all samples, the relative abundance of bacteria was > 98%. Richness of genera (q = 0) significantly decreased with metal concentration (p < 0.05, Supplementary Fig. 1, Table 2). The evenness increased (q = 1) and the diversity of the dominant species decreased (q = 2) with increasing metal concentration.
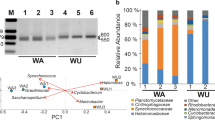
Significant differences in biodiversity were found among the samples (PERMANOVA, p < 0.01; Procrustes, p = 0.001; Supplementary Fig. 2A, dissimilarity = 0.998, PCA Supplementary Fig. 2B). Samples from the unamended bioreactor (CON-b and CON-e) were clustered together in one group and the HM-2 and HM-5 samples in a second group, whereas the HM-1 sample differed from those two groups (Fig. 1; Fig. 1B stress: 5.535 × 10–7). The network analysis confirmed this separation (Fig. 1A and C). The bacterial community in the HM-1 sample was separated clearly from the HM-2 and HM-5 samples group, and the CON-b and CON-e samples group. Although most genera were concentrated in the shared area, the relative abundance of others was clearly different. For instance, Agrobacterium and Achromobacter were grouped in the HM-1 sample, and Delftia, Stenotrophomonas and Sphingobacterium in the HM-2 and HM-5 samples. To have a better idea of the differences among the samples, genera were clustered (k-means, groups = 3, silhouette value = 0.9970), which confirmed the above mentioned groups. Agrobacterium and Achromobacter were clustered while Delftia while Stenotrophomonas and Sphingobacterium were separated from the other genera (Fig. 1C and D). The HM-1 sample had a different gradient than all other samples (Fig. 1D, redundancy analysis RDA, R2 = 0.9843).
A Network analysis, B Non-classical multidimensional scaling of the Bray–Curtis dissimilarities, C Cluster analysis, and D Redundancy analysis with the bacterial genera (black closed circles), samples (red squares) and heavy metal concentration factor (blue open circles) showing the input factor axes (blue arrows). Labels correspond to unamended bioreactor samples at the beginning (CON-b) and at the end of the experiment (CON-e), and in the metal amended bioreactor when the metal concentration was 31.5 μM Cu, 112.4 μM Ni and 122.3 μM Zn (HM-1), at a twofold (HM-2) and a fivefold (HM-5) increase in these metal concentrations
Most bacterial phyla (28 of 36) were shared among all samples, but eight of them were not found in the metal amended bioreactor, i.e. Apicomplexa, Armatimonadetes, Dictyoglomi, Ignavibacterae, Microsporidia, Nitrospirae, Synergistetes, and Thaumarchaeota (Fig. 2A and B). Their relative abundance in the unamended bioreactor was < 0.01%. Among those, the relative abundance of Pseudomonadota (formerly Proteobacteria) and Bacteroidota increased when the metal concentration increased, while Actinomycetota (formerly Actinobacteria) decreased to less than 1% (Supplementary Table 1). The most abundant classes in the control bioreactor, were Gammaproteobacteria, Alphaproteobacteria and Betaproteobacteria. In the metal amended bioreactor, the relative abundance of most bacterial classes was similar, except for that of the Betaproteobacteria, Flavobacteria and Sphingobacteriia that increased, while no members of Actinomycetia were detected (Supplementary Table 1).
A Venn diagram with the bacterial genera found in samples. B Circos plot of the amount of bacteria shared among samples at different taxonomic levels. The legends to the samples taken from the unamended and metal amended bioreactor can be found in Fig. 1
The relative abundance of some genera, e.g. Leucobacter, Microbacterium, Devosia and Acinetobacter, decreased to < 0.1% when metals were applied (Suppl. Table 1). The most abundant genera in the metal amended bioreactor were Delftia, Sphingobacterium, Stenotrophomonas and Sphingomonas, while in the control bioreactor Stenotrophomonas, Sphingobacterium, Microbacterium and Acinetobacter dominated (Supplementary Table 1). In the HM-1 phase, Achromobacter and Agrobacterium were the most abundant bacterial genera, but their relative abundance dropped when the metal concentration increased two and five-fold (HM-2 and HM-5 samples). Contrastingly, the relative abundance of others increased, e.g. Delftia, Sphingobacterium, Stenotrophomonas and Sphingomonas (Suppl. Table 1).
Most of the genera were shared among the different samples (Fig. 2A). In the unamended bioreactor, 274 unique genera of a total of 1066 genera were found at the onset the experiment (Fig. 2A). The diversity replacement index decreased from 0.51 in CON-b to 0.159 in CON-e. The diversity replacement index decreased even more when the metal concentration increased, i.e. 0.086 in HM-1, 0.059 in HM-2 and 0.045 in HM-3.
Only five bacterial genera with an abundance > 0.5% were correlated significantly with the metal concentration (Fig. 3A). Members of Delftia were the most abundant with a positive significant correlation with the metal concentrations (r = 0.986, p = 0.002) (Fig. 3A insert). The other significant correlations between the metals and the bacterial groups were negative, i.e. an unknown bacterial genus (r = -0.904, p = 0.035), other Xanthomonadaceae (r = -0.904, p = 0.035), Pseudomonas (r = -0.923, p = 0.026) and Xanthomonas (r = -0.9629, p = 0.0085) (Fig. 3B). Their partial correlations (r12.3), however, were lower when the contribution of the metal concentration was subtracted, except for the correlation between Xanthomonas and an unknown bacteria (r = 0.9854, p = 0.0146), and Delftia with unknown Xanthomonadaceae (r = -0.9812, p = 0.0188).
A Changes in the relative abundance of bacterial genera (%) with a significant variation (ANOVA p < 0.05) with the metal concentrations and B Auto-correlation matrix of the bacterial genera (%) with a significant variation (ANOVA p < 0.05) with the metal concentrations, i.e. the left panel shows the raw auto-correlation matrix as a heatmap, while the right panel a partial auto-correlation after subtracting the correlation of each genera with the metal concentration
Cluster analysis of genera gave three clusters (Fig. 1C). The first cluster included most of the bacterial genera, the second group included Agrobacterium and Achromobacter, and the third Delftia, Sphingobacterium and Stenotrophomonas (Fig. 1C). The clustering did not include input factors (Q mode), so it is not possible to relate them to an explanatory factor, such as sample or metal concentration (R mode). Network analysis gave similar results (Fig. 1A), but as samples were included in the analysis, the “group 2” Achromobacter and Agrobacterium were associated with the HM-1 sample, and Delftia, Sphingobacterium and Stenotrophomonas (3rd group in the clustering analysis) to the communities in the HM-2 and HM-5 samples (Supplementary Table 1). It seems, on the one hand, that clustering and network analysis (Fig. 1A and C) gave more information on population dynamics than circos plots. On the other hand, those analysis taken together with NMDS (Fig. 1B) and RDA (Fig. 1D) showed a difference in the bacterial community of the HM-1 sample with the HM-2 and HM-5 community.
The RDA in R-mode (explanatory variables included) allowed to understand the gradient and tendencies of the objects (samples) and explanatory variables (metal concentration factor), but information about the genera was barely noticeable. The most evident genera were Achromobacter, Agrobacterium and Delftia located in the superior quadrants (Fig. 1D). These genera represent the main differences among the HM-1, and HM2 and HM-5 communities. Although the relative abundance of Sphingobacterium and Stenotrophomonas also increased with metal concentration, the linear correlation was not significant. Indeed, Delftia was the only genus with a positive correlation between its relative abundance and the metal concentration (Fig. 3A).
Metabolic functionality analysis
The most abundant heterotrophic carbon pathways were those of anaplerotic genes (pyruvate to oxaloacetate), and fermentation to succinate (Fig. 4A). The relative abundance of autotrophic pathways, such as Calvin-Benson-Basshan (CBB) cycle and 3-hydroxypropionate bicycle, was low. The CBB cycle was found in the unamended bioreactor and in the HM-1 sample, while the 3-hydroxypropionate bicycle in all samples except in the HM-1 sample. A significant difference was found in the relative abundance of the 3-hydroxypropionate bicycle metabolic function between the control (6.353 counts per million (cpm)) and metal amended bioreactor (0.316 cpm, p = 0.005). When using the metal concentration factor as the grouping factor, only the fermentation pathway from lactate to acetate was significantly different among the samples (p = 0.049).
A Carbon bio-geocycle pathways, B Nitrogen bio-geocycle pathways, C Sulfur bio-geocycle pathways and D Miscellaneous as obtained with Diting. A black arrow indicate a significative difference with increased metal concentration, and a white arrow a significative difference with the presence or absence of the metals. The legends to the samples taken from the unamended and metal amended bioreactor can be found in Fig. 1
The most abundant genes involved in the N-cycle were those of assimilatory and dissimilatory nitrate reduction (Fig. 4B). They were not significatively different between the unamended and metal amended bioreactor. Only those related to denitrification were significatively different, i.e. reduction of nitrite to nitric oxide (nirK or nirS, p = 0.049) and from nitric oxide to nitrous oxide (norBC, p = 0.046). The nifKDH, hao, hzs and hdh genes were not detected in the samples.
The DMSP demethylation via dmdBCD were the most abundant genes involved in the S-cycle, followed by thiosulfate oxidation (SOX, doxAD and tsdA, Fig. 4C). Significant differences in sulfide oxidation to sulfur (p = 0.047), DMPS cleavage rendering DMS (p < 0.001) and DMS oxidation to DMSO (p < 0.001) were found between the unamended and metal amended bioreactor. When using the application rate of the metals (1, 2 or fivefold increase) as a grouping factor for the ANOVA, demethylation of MMPA to methanetiol (MeSH) was the only metabolic function that decreased significatively from 386.4 to 46.9 (Fig. 4C, p = 0.007).
Carbohydrate metabolism (based on the KEGG classification) was the function with the largest relative abundance, followed by amino acid metabolism (Fig. 5A). Considering the protein families, metabolism and genetic information processing were the most abundant metabolic functions with a relative abundance ranging from 3.8 × 104 to 4.61 × 104 cpm and from 4.12 × 104 to 4.9 × 104 cpm. Dissimilatory arsenic reduction was the most abundant genes in the HM-1 and HM-2 samples, but only the relative abundance of pathways related to bacterial chemotaxis (p = 0.020) and Cytochrome bc1 complex also known as complex III of respiratory chain (p = 0.026), significatively decreased in the metal amended bioreactor compared to the unamended bioreactor (Fig. 4D).
A Heatmap with the relative abundance of functions (%) as determined with the KEGG level 2 classification, B Procrustes analysis of the functional profile (R2 = 0.8886), C Non-classical multidimensional scaling of the chi-square distance of the samples, and D Abundance of genes encoding for synthesis of extracellular polymeric substances (* p < 0.05). The legends to the samples taken from the unamended and metal amended bioreactor can be found in Fig. 1
The distribution of functions changed between the unamended and metal amended bioreactor (procrustes dissimilarity = 0.889, Fig. 5B). The coordinate analysis (NMDS stress = 9.514 × 10–7) showed three different groups, i.e. samples of the unamended bioreactor were grouped, the HM-1 and HM-2 samples, and the HM-5 sample. Although the NMDS analysis of structural diversity (Fig. 2C, stress = 9.524 × 10–7) separated the HM-1 sample from the other samples, the HM-5 sample differed from the rest of the samples, i.e. the latter was found in the second quadrant in the NMDS plot, control-bioreactor samples in the third quadrant, and the HM-1 and HM-3, in the first quadrant (Fig. 5C).
The cysE, exoA, gumDI, pgaC, icaA, wcaEFL and wecA genes (involved in external polymeric substance (EPS) production) were found in both bioreactors (Fig. 5D). The genes of the exo operon (exoMOUW) were not detected when the metal concentration increased, whereas the relative abundance of wecA gene increased significantly.
Discussion
Bacterial community diversity and structure
In this study, the presence of the metals reduced the microbial diversity in the bioreactor as has been reported before [24]. In this study, the richness and diversity replacement decreased as the metal concentration increased, but not for Hill numbers at q = 1 and q = 2. In other words, dominant genera were replaced by others when the metal concentration increased. As such, the presence of the genera or species defined the dominance or evenness.
Achromobacter and Agrobacterium were the most dominant genera in the HM-1 sample, while Delftia, Stenotrophomonas and Sphingobacterium in the HM-2 and HM-5 samples. This might be explained by the increased metal concentration, or the differential impairment of metabolic functions by the metals. Members of these genera have strategies to resist metals [7, 23, 27]. Bacteria of the genera [22], Delftia [2], Stenotrophomonas [28] and Agrobacterium [38] have been found to adhere to surfaces via external polymeric substances [15]. This might lead to the formation of sediment microparticles that might immobilize metals reducing their bioavailability and toxicity [34]. This phenomenon has been observed for other genera, such as Pseudomonas, Accumulibacter, Competibacter, Mesorhizobium, Xanthomonas and Pseudoxanthomonas [15, 22, 23, 34, 36]. Some of these genera, such as Pseudomonas (relative abundance 0.3%), Mesorhizobium (0.06%), Xanthomonas (0.3%) and Pseudoxanthomonas (0.2%) were found in the metal amended bioreactor, but their relative abundance was low. The relative abundances of Pseudomonas, Xanthomonas and an unknown Xanthomonadaceae were correlated and the partial correlation between them decreased (r2) when the metal concentration was deleted from the correlation coefficients, suggesting that the apparent correlations between the genera was due mainly to the metals.
Metabolic functionality
It was hypothesized that changes in bacterial richness and diversity would also result in changes in the metabolic functions in the bioreactor samples, but that was mostly not the case. The relative abundance of most metabolic functions did not change despite the increase in metal concentration, i.e. the bacterial metabolic functionality was more stable than the bacterial community structure despite the variations in metal concentration.
Heterotrophic metabolism was the main function, and carbohydrate and amino acid metabolism the most abundant. This may suggest that despite the increases in metal concentration, organic material decomposition was not affected. This agrees with our previous report using putative/predictive functions from 16S metagenomics [1]. The reduction of nitrite to nitric oxide (nirK, nirS) and from nitrite to nitrous oxide (nifK/nirS, norBC) and DMSP demethylation, however, all decreased significantly with increased metal concentrations. In all these metabolic functions, oxido-reduction biochemical reactions are involved. The narB, nasA, nifK, nirS, nasA, and norB genes involved in the N cycle encode for proteins binding iron, while narB and nifK may bind molybdenum ions, whereas norB is from the cytochrome family. The decrease in their relative abundance might reflect the lethality of these metals to bacteria that possess those genes. The DMSP demethylation occurs thanks to proteins encoded by the dmd gene [16]. Proteins encoded by the dmdD gene bind magnesium ions (Uniprot) and products of dmdBCD are still being investigated. Dey [16] reported that dmdBCD products are involved in carbon and sulfur fixation by plankton and that a decrease in their protein expression might result in a higher production of dimethylsulfide (DMS), an air contaminant in acid rain.
The relative abundance of the metabolic functions showed only small changes. This would suggest that most of the functions are either required for the microbial community, e.g. autotrophy, or they were present and became detectable as the substrate became available, such as heterotrophy, in both bioreactors. In other words, the forces driving the occurrence of specific metabolic functions may include randomness, necessity and/or substrate availability. Apart from the different metal concentrations, conditions were similar in both bioreactors. It can thus be assumed that, despite biodiversity, the range of metabolic functions were similar as evidenced by our results. Contrario sensu, different conditions might be driving forces for changes in metabolic functions. For instance, in this study, the metals in the HM bioreactor might lead an increase in metal resistance mechanisms. Among them, the transformation and transport of metals, and EPS production are the most well-described mechanisms. The formers, transformation (redox) and transport mechanisms have been described in Lysinibacillus, Agrobacterium and some genera of the Xanthomonadaceae, such as Stenotrophomonas and Xanthomonas [28,29,30, 32, 38]. The relative abundance of Agrobacterium and Achromobacter decreased in the HM-2 and HM-5 samples, suggesting that the minimum inhibitory concentration of the metals was between that found in the HM-1 and HM-2 samples, which might also affect the metabolic functions within the bacterial community.
One of the main metabolic-related mechanisms is the production of EPS from carbohydrates and proteins. The main pathways for EPS production comprise curdlan, xanthan, colanic and alginate synthesis, which involve crd, gum, wca and alg operons [34]. Succinoglycan (exo operon), the vibrio-like polysaccharide (cysE and vps operons), the enterobacterial antigen (wec operon) and Poly-N-acetyl-glucosamine (ica and pga operons) are other, but less frequent genes encoding for EPS. The crd, gum, wca and alg operons have been detected in Agrobacterium, Xanthomonas, Enterobacteriaceae and Pseudomonas [34]. The crd operon has been reported in Agrobacterium [34], but it does not appear yet in the RefSeq gene database or KEGG. In this study, the relative abundance of the wecA gene increased with increased metal concentration. This gene has been reported in members of the Enterobacteriaceae family, such as Escherichia, Yersinia and Klebsiella [9]. The relative abundance of these genera was low in the bioreactor (≤ 0.04%) and no correlation was found between them (or the Enterobacteriaceae family) and the wecA gene. As such, further research into the prevalence of the wecA gene in other bacterial groups might be needed as the EPS production genes may also help other genera to adhere to surfaces and survive.
Although EPS production by Delftia has not yet been reported, its ability to autoaggregate and coaggregate in biofilms with other bacteria has been reported [2], which might explain its ability to colonize surfaces [15]. This coaggregation seems to be sugar-specific with Citrobacter and Enterobacter, but not with Pseudomonas, whose interaction appears to be protein–protein mediated [2]. In this study, Pseudomonas seemed to antagonize Delftia within the biofilm. It has been reported that a strain of Delftia was able to inhibit Pseudomonas growth, motility and biofilm production [31]. In this study, Delftia was also antagonistic against Xanthomonas and an unknown Xanthomonadaceae. This antagonism prevailed even after eliminating the metals from the correlation analysis, suggesting that it was independent of the presence of the metals.
Also, Achromobacter, Delftia, Pseudomonas and Stenotrophomonas have been found to produce biofilms and adhere to them [15]. A strain of Achromobacter has been reported to denitrify and be able to immobilize 10 mg L−1 (89 μM approximately) cadmium ions (Cd) in wastewater [22]. Although this immobilization of Cd has been hypothesized to occur via precipitation with carbonate, its precipitation is larger with a composite sediment containing EPS. Achromobacter and Stenotrophomonas strains carrying copLAB genes that have been isolated from an agricultural field are able to resist 0.8 mM Cu. This ability of Achromobacter together with Agrobacterium to produce EPS [34] might explain their higher abundance in the HM-2 sample. As mentioned above, the EPS may be a scaffold for Delftia to grow better when the metal concentration increased. This might have helped Delftia to adhere to the biofilm and inhibit the growth and adhesion of other bacteria, as has been described previously [9, 31]. This might explain why the relative abundance of Delftia was highest in the HM-5 sample by antagonizing Pseudomonas and other Xanthomonadaceae, and also why the abundance of Achromobacter decreased. These observations agreed with a previous study of [1] based on 16S metagenomics, where an increase in the relative abundance of Stenotrophomonas and Sphingomonas at high metal concentrations (HM-2 and HM-5) and the increase of Agrobacterium at low metal concentrations (HM-1) were reported. Although 16S metagenomics gave a good insight in the behavior of the bacterial community, shotgun metagenomics allow a deeper knowledge in microbiota diversity and gene/function prevalence, such as the EPS production genes.
Conclusion
Low concentrations of copper, zinc and nickel (31.5 μM copper, 112.4 μM nickel, 122.3 μM zinc) altered the bacterial community structure in a bioreactor seeded with wastewater that changed even more with higher concentrations of these metals. Achromobacter and Agrobacterium were enriched at low concentration of the metals, while Delftia, Stenotrophomonas and Sphingomonas at higher concentrations. Delftia was antagonistic towards Pseudomonas and other Xanthomonadaceae. Assimilatory reduction of nitrate, and assimilation of methylmercaptopropionate to dimethylsulfide was negatively affected by the metal concentration, which may impair the removal of nitrate and some organo-sulfur compounds in metal contaminated water. Functional metagenomics produced by shotgun sequencing help to improve our understanding on what mechanisms and population dynamics (e.g., antagonisms) occur in the microbial community.
Availability of data and materials
The raw sequence datasets were deposited in the NCBI under BioProject ID PRJNA1009125, BioSample ID SAMN37147121 and SRA numbers: SRR26071885, SRR26071886, SRR26071887, SRR26071888, SRR26071889.
References
Aceves-Soriano CE, Montoya-Ciriaco N, Hernández-Guzmán M, Hernández-Martínez GR, Navarro-Noya YE, Thalasso F, Dendooven L. Changes in the bacterial and microeukaryotic communities in the bioreactor upon increasing heavy metal concentrations. Front Environ Chem. 2023. https://doi.org/10.3389/fenvc.2023.1221820.
Afonso AC, Gomes IB, Saavedra MJ, Simoes LC, Simoes M. Drinking-water isolated Delftia acidovorans selectively coaggregates with partner bacteria and facilitates multispecies biofilm development. Sci Total Environ. 2023. https://doi.org/10.1016/j.scitotenv.2023.162646.
ATSDR Copper ToxFAQs. 2022. https://www.atsdr.cdc.gov/toxguides/toxguide-132.pdf Accessed 8 Sept 2023
ATSDR Nickel ToxFAQs. 2023. https://www.atsdr.cdc.gov/toxfaqs/tfacts15.pdf. Accessed 15 Sept 2023
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–77.
Bastian M, Heymann S, Jacomy M. Gephi: an open source software for exploring and manipulating networks. In: Proceedings of the third international conference on weblogs and social media, ICWSM, USA; 2009;3(1), p. 361–2.
Bhat SA, Cui G, Li W, Wei Y, Li F. Effect of heavy metals on the performance and bacterial profiles of activated sludge in a semi-continuous reactor. Chemosphere. 2020;241:125035.
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics. 2014;40(15):2114–20.
Cafalette CA, Kuklewicz J, Spellmon N, Zimmer J. Biosynthesis and Export of Bacterial Glycolipids. Annu Rev Biochem. 2020;89:741–68. https://doi.org/10.1146/annurev-biochem-011520-104707.
Čapek J, Vecerek B. Why is manganese so valuable to bacterial pathogens. Front Cell Infect Microbiol. 2023;13:943390.
Chao A, Gotelli NJ, Hsieh TC, Sander EL, MA KH, Colwell RK, Ellison AM. Rarefaction and extrapolation with Hill numbers: a framework for sampling and estimation in species diversity studies. Ecol Monographs. 2014;84(1):45–67.
ChEBI, Zinc CHEBI:29105 https://www.ebi.ac.uk/chebi/pathway.do?chebiId=CHEBI:29105. Accessed 15 Sept 2023.
Cloquet C. Carignan J, Lehmann MF, Vanhaecke F, Variation in the isotopoic composition of zinc in the natural environment and the use of zinc isotopes in biogeosciences: a review. Anal Bioanal Chem. 2008;390:451–63. https://doi.org/10.1007/s00216-007-1635-y.
da Silva LF, da Silva GCE, Ambrósio FR. A review of the role fo zinc finger proteins on hematopoiesis. J Trace Elem Medic Biol. 2023;80:127290. https://doi.org/10.1016/j.jtemb.2023.127290
Dantam J, Subbaraman LN, Jones L. Adhesion of Pseudomonas aeruginosa, Achromobacter xylosoxidans, Delftia acidovorans, Stenotrophomonas maltophilia to contact lenses under the influence of an artifical tear solution. Biofouling. 2020;36(1):32–43.
Dey M. Enzymology of microbial dimethylsulfoniopropionate catabolism. In advances in protein chemistry and structural biology, vol. 109. The Netherlands: Elsevier; 2017. p 197–224.
Esquivel-Rios I, Gonzázlez I, Thalasso F. Microrespirometric characterization of activated sludge inhibition by copper and zinc. Biodegradation. 2014;25(6):867–79.
Joshi S, Gangola S, Bhandari G, Bhandari NS, Nainwal D, Rani A, Malik S, Slama P. Rhizospheric bacteria: the key to sustainable heavy metal detoxification strategies. Frontiers Microbiol. 2023;14:1229828. https://doi.org/10.3389/fmicb.2023.1229828.
Legendre P, Legendre L. Numerical Ecology. 3rd ed. The Netherlands: Elsevier; 2012.
Legendre P. Interpreting the replacement and richness difference components of beta diversity. Global Ecol Biogeogr. 2014;23:1324–34.
Li X, Gu AZ, Zhang Y, Xie B, Li D, Chen J. Sub-lethal concentrations of heavy metals induce antibiotic resistance via mutagenesis. J Haz Mat. 2019;369:9–16. 10.1016/j.jhazmat.2019.02.006. https://doi.org/10.1016/j.jhazmat.2019.02.006
Liang DH, Hu Y. Application of a heavy metal-resistant Achromobacter sp. for the simultaneous immobilization of cadmium and degradation of sulfamethoxazole from wastewater. J Haz Mat. 2021;402:124032 https://doi.org/10.1016/j.jhazmat.2020.124032. https://doi.org/10.1016/j.jhazmat.2020.124032
Lin Y, Wang L, Xu K, Li K, Ren H. Revealing taxon-specific heavy metal resistance mechanisms in denitrifying phosphorus removal sludge using genome centric metaproteomics. Microbiome. 2021;9:67. https://doi.org/10.1186/s40168-021-01016-x.
Lors C, Mamindy-Pajany Y. Bioremediation of Heavy Metals in Sediments. In: Velázquez-Fernández JB, Muñiz-Hernández S, editors. Bioremediation: Processes, Challenges and Future Prospects. Nova Science Publishers: Inc; 2014. p. 3–18.
Mathworks [Internet]. Zhaoxu Liu, 2022 [cited 2023 May 30]. Available from: www.mathworks.com/matlabcentral/fileexchange/118655-circos-plot. Accessed 20 May 2023.
Moreno-Vivián C, Cabello P, Martínez-Luque M, Blasco R, Castillo F. Prokaryotic Nitrate Reduction: Molecular Properties and Functional Distinction among Bacterial Nitrate Reductases. J Bacteriol. 1999;181(21):573–84.
Mpongwana N, Rathilal S, Tetteh EK. Recovery strategies for heavy metal-inhibited biological nitrogen removal from wastewater treatment plants: a review. microorganisms. 2022;20:1834. https://doi.org/10.3390/microorganisms10091834.
Pages D, Rose J, Conrod S, Cuine S, Carrier P, Heulin T, Achouak W. Heavy Metal Tolerance in Stenotrophomonas maltophilia. PLoS ONE. 2008;2:e1539.
Rana R, Jaiswal G, Bansal K, Patil PB. Comparative genomics reveals the emergence of copper resistance in a non-pigmented Xanthomonas pathogen of grapevine. Environ Microbiol. 2023;15:716–26.
Ranmarine SDB, Jayaranman J, Ramsubhag. copLAB gene prevalence and diversity among Trinidadian Xanthomonas spp. black-rot lesion isolates with variable copper resistance profiles. PeerJ. 2023;11:e15657. https://doi.org/10.7717/peerj.15657.
Singh VK, Mishra A, Jha B. Anti-quorum sensing and anti-biofilm activity of Delftia tsuruhatensis extract y attenuating the quorum sensing-controlled virulence factor production in Pseudomonas aeruginosa. Front Cell Infec Microbiol. 2017;7:337. https://doi.org/10.3389/fcimb.2017.00337.
Sun S, Zhang K, Wu Y, Zhu N, Wang Y, Chen J, Leng F. Transporter drives the biosorption of heavy metals by Stenotrophomonas rhizophila JC1. Environ Sci Pollution Res. 2022;29:45380–95. https://doi.org/10.1007/s11356-022-18900-0.
Tchounwou PB, Yedjou CG, Patlolla AK, Sutton DJ. Heavy metals toxicity and the environment. EXS. 2012;101:133–64.
Vandana, Priyadarshanee M, Das S. Bacterial extracellular polymeric substance: Biosynthesis and interaction with environmental pollutants. Chemosphere. 2023;332:138876. https://doi.org/10.1016/j.chemosphere.2023.138876.
VIB-UGent Center for Plant Systems Biology, Ghent University [Internet]: Bioformatics & Evolutionary Genomics; c2023. http://bioinformatics.psb.ugent.be/webtools/Venn/. Accessed 20 May 2023.
Wang H, Zhang S, Zhang J. The copper resistance mechanism in a newly isolated Pseudoxanthomonas spadix ZSY-33.Environ Microbiol. 2023;15:484–496.
Wood DE, Lu J, Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20(1):257.
Wu Q, Li Q Zhang Y, Wan R, Peng S. Cr(VI) reduction by Agrobacterium sp. Cr-1 and Lysinibacillus sp. Cr-2, novel Cr(VI)-reducing strains isolated from chromium plant soil. Environ Sci Pollution Res. 2023;30:109724–37. https://doi.org/10.1007/s11356-023-30181-9.
Xue CX, Lin H, Zhu XY, Liu J, Zhang Y, Rowley G, Todd JD, Li M, Zhang XH. DiTing: A pipeline to infer and compare biogeochemical pathways from metagenomic and metatranscriptomic data. Front Microbiol. 2021;12:698286. https://doi.org/10.3389/fmicb.2021.698286
Zhang Y, Ying H, Xu Y. Comparative genomics and metagenomics of the metallomes. Metallomics. 2019;11:1026–43.
Zhang X, Tang S, Wang M, Sun W, Xie Y, Peng H, Zhong A, Liu H, Zhang X, Yu H, Giesy JP, Hecker M. Acid mine drainage affects the diversity and metal resistance gene profile of sediment bacterial community along a river. Chemosphere. 2019;217:790–9. https://doi.org/10.1016/j.chemosphere.2018.10.210.
Funding
Experiments were funded by CINVESTAV.
Author information
Authors and Affiliations
Contributions
J.B. V.-F. analyzed the results and wrote the manuscript, C.E. A.-S. analyzed results, F. T. maintained the bioreactors and applied the experimental design, N. M.-C. extracted the DNA, L. D. conceived the study, and helped with funding, writing and editing the manuscript. Part of the writing and editing was done while L. D. was on sabbatical leave from Cinvestav at the “Universidad Autónoma de Tlaxcala”. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Velázquez-Fernández, J.B., Aceves Suriano, C.E., Thalasso, F. et al. Structural and functional bacterial biodiversity in a copper, zinc and nickel amended bioreactor: shotgun metagenomic study. BMC Microbiol 24, 313 (2024). https://doi.org/10.1186/s12866-024-03437-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12866-024-03437-8