Abstract
Background
Tobacco bacterial wilt (TBW) caused by Ralstonia solanacearum is the most serious soil-borne disease of tobacco that significantly reduces crop yield. However, the limited availability of resistance in tobacco hinders breeding efforts for this disease.
Results
In this study, we conducted hydroponic experiments for the root expression profiles of D101 (resistant) and Honghuadajinyuan (susceptible) cultivars in response to BW infection at 0 h, 6 h, 1 d, 3 d, and 7d to explore the defense mechanisms of BW resistance in tobacco. As a result, 20,711 and 16,663 (total: 23,568) differentially expressed genes (DEGs) were identified in the resistant and susceptible cultivars, respectively. In brief, at 6 h, 1 d, 3 d, and 7 d, the resistant cultivar showed upregulation of 1553, 1124, 2583, and 7512 genes, while the susceptible cultivar showed downregulation of 1213, 1295, 813, and 7735 genes. Similarly, across these time points, the resistant cultivar had downregulation of 1034, 749, 1686, and 11,086 genes, whereas the susceptible cultivar had upregulation of 1953, 1790, 2334, and 6380 genes. The resistant cultivar had more up-regulated genes at 3 d and 7 d than the susceptible cultivar, indicating that the resistant cultivar has a more robust defense response against the pathogen. The GO and KEGG enrichment analysis showed that these genes are involved in responses to oxidative stress, plant–pathogen interactions, cell walls, glutathione and phenylalanine metabolism, and plant hormone signal transduction. Among the DEGs, 239 potential candidate genes were detected, including 49 phenylpropane/flavonoids pathway-associated, 45 glutathione metabolic pathway-associated, 47 WRKY, 48 ERFs, eight ARFs, 26 pathogenesis-related genes (PRs), and 14 short-chain dehydrogenase/reductase genes. In addition, two highly expressed novel genes (MSTRG.61386-R1B-17 and MSTRG.61568) encoding nucleotide-binding site leucine-rich repeat (NBS-LRR) proteins were identified in both cultivars at 7 d.
Conclusions
This study revealed significant enrichment of DEGs in GO and KEGG terms linked to glutathione, flavonoids, and phenylpropane pathways, indicating the potential role of glutathione and flavonoids in early BW resistance in tobacco roots. These findings offer fundamental insight for further exploration of the genetic architecture and molecular mechanisms of BW resistance in tobacco and solanaceous plants at the molecular level.
Similar content being viewed by others
Background
Nicotiana tabacum (2n = 48) is an economically major non-edible cash crop, with its dried leaves used for industrial products, including cigarettes, cigars, and shisha tobacco, and stems used for biofuel production [1,2,3,4]. However, bacterial wilt (BW), a soil-borne disease caused by R. solanacearum that seriously damages leaf quality and causes yield losses in tobacco, is distributed in all tobacco production areas, especially warm-temperate or tropical and subtropical areas [5,6,7]. The disease also attacks other Solanaceae crops such as potato, eggplant, tomato, pepper, and sweet potato [6,7,8,9,10]. Production losses due to BW varies from 10–20% in peanut [6], 10–15% in potato [11], 20–50% in chili [12, 13], and 20–30% in ginger [14]. Tobacco bacterial wilt (TBW) disease incidence significantly reduces tobacco production by 10–30% worldwide [7, 15,16,17], reaching 15–35% [6] in the major growing areas of Sichuan, Guangdong, Hunan, Guizhou, and Hubei provinces in China [18]. However, TBW control is a global problem, with traditional methods (chemical control, tobacco-rice rotation, and soil fumigation) inadequate for reducing yield and economic losses [16, 19]. Therefore, studies are needed to investigate BW's defense mechanism or genetic basis and identify genes responsive to the infection to develop BW-resistant tobacco cultivars.
During evolution, Solanaceae plants developed complex defense mechanisms against pathogens [20], such as inhibitory substances, metabolites (alkaloids, phenols, etc.), and hormones [21]. Several quantitative trait loci (QTLs) have been reported for BW resistance in tobacco [7, 16, 22]; for example, Lan et al. [16] identified eight QTLs,qBWR17a/17b, qBWR2, qBWR6, qBWR12, and qBWR24a/24b/24c. In another study, using bi-parental mapping, four QTLs (qBWR-3a/3b and qBWR-5a/5b) were detected for TBW resistance [23]. Habe et al. [24] used QTL mapping analysis in potato, detecting five QTLs (qBWR1, qBWR2, qBWR3, qBWR4, and qBWR5) in response to BW infection. Similarly, Nguyen et al. Chae et al. [12, 25], and Zai et al. [26] reported 8, 31, and 14 significant SNPs for BW resistance in tomato, pepper, and common beans, respectively, using association mapping. These QTLs/SNPs, especially those with significant effects [27], will assist marker-assisted breeding of BW-resistant cultivars [7, 28]. Thus, Few QTLs/QTNs associated with BW resistance in tobacco have been identified compared to other Solanaceae species: pepper, tomato, potato, and eggplant [6, 11,12,13,14].
Moreover, transcriptomics, metabolomics, genomics, and proteomics are practical and diverse methodologies for elucidating and comprehensively understanding complex biological mechanisms under various stressful conditions in plants [17, 29,30,31]. Of these, transcriptomics analysis with RNA-seq or microarrays has been used to unravel the molecular basis and genes associated with the specific biological processes in solanaceous crops, which could provide BW resistance for developing superior cultivars [9, 30]. In light of previous studies, transcriptomics and proteomics analysis showed that the methionine cycle (MTC), phenylpropanoid biosynthesis, glutathione metabolism, and gamma-aminobutyric acid biosynthesis pathways played essential roles in tomato BW [9, 29, 32]. [32] used resistant and susceptible tomato genotypes to demonstrate that 140 up-regulated DEGs were related to hormones, lignin, and pathogenesis in the resistant genotype, while no changes occurred in the susceptible genotype. Similarly, Li et al. [8] identified 302 DEGs associated with potato BW, of which 81 were considered for BW resistance involved in signal transduction, terpenoids, pathogen recognition, hypersensitive response, and protection [8].
For TBW, Gao et al. [9] identified 158 and 835 DEGs in resistant and susceptible accessions, respectively, in a seedling root transcriptome study, while Shi et al. [29] reported that indole-3-acetic acid (IAA) and abscisic acid (ABA) plays significant roles in BW resistance in transcriptomics and metabolomics studies. A study reported the up-regulation of 6,233 genes in tobacco-resistant cultivars in response to BW infection. These genes were enriched in cell walls, ABC transporters, endocytosis, and glutathione metabolism [33]. Similarly, Li et al. [17] and Pan et al. [33] used the tobacco stem transcriptome to detect genes, identifying the phenylpropanoid metabolic pathway for tobacco defense against BW. Furthermore, many studies have reported the root transcriptome in pepper [34], potato [35], eggplant [36], and tomato [37] as compared to tobacco. For example, root transcriptomes at 1, 3, and 5 d identified 115 genes in response to pepper BW infection [34]. Moreover, transcription factors (TFs) [38], including WRKY, ERFs, NAC, MYB, bHLH, and bZip, and gene families like P450, MAPK, and DRGs identified in Solanaceae crops are involved in pathogen resistance [17, 29, 39]. Further, recent studies have reported candidate genes associated with plant defense against BW, including StMKK1 in potato [40], CaNAC2c in pepper [41], SlNAP1 in tomato [42], and NbPDKs in tobacco [43]. Limited transcriptomic studies have been conducted to investigate the genes involved in tobacco's resistance to R. solanacearum. Therefore, exploring and analyzing defense-related genes in tobacco through transcriptomics can provide valuable insights into the interaction between tobacco and R. solanacearum.
Controlling tobacco bacterial wilt in China is difficult due to its high severity and limited availability of resistant tobacco cultivars. Recently, we identified 52 candidate genes and 38 quantitative trait nucleotides (QTNs) using association mapping of 94 tobacco accessions with 126,602 SNPs [7]. Findings from the studies mentioned above in other solanaceous species can serve as a reference for advancing tobacco breeding efforts. However, the genetic basis and molecular mechanism of BW resistance in tobacco may differ from those in other solanaceous species. Until now, few transcriptome profiling studies of BW resistance have been reported in tobacco. Therefore, in this study, we used RNA-seq to analyze and compare transcriptomes in the roots of D101 [resistant (R)] and Honghuadajinyuan [susceptible (S)] cultivars at early (6 h, 1 d) and late (3 d and 7 d) stage of seedlings after inoculation. The study aimed to understand the early and late response of gene expression to R. solanacearum infection in roots and to identify the genes conferring root resistance to R. solanacearum through gene ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), and functional enrichment analysis. We identified significant divergence between the resistant and susceptible cultivars in their pathogen responses and also identified potential genes associated with BW resistance in tobacco plants for gene cloning, molecular studies, and breeding purposes.
Results
Phenotypic divergence of tobacco cultivars under R. solanacearum infection
Firstly, we performed disease assays to determine the response of tobacco cultivars (D101 and Honghuadajinyuan) to bacterial wilt. We observed a significant difference between accessions in disease symptoms at 3 d and 7 d. Notably, the TBW symptoms (leaf wilting) started to appear in Honghuadajinyua after 3 d exposure to infection, with no symptoms in D101 (Fig. 1A). At 7 d, Honghuadajinyua stems looked black, with some plant death (Fig. 1B-C), whereas D101 seedlings had no similar or obvious symptoms (Fig. 1A). The roots of R and S cultivars showed clear disease symptoms at 7 d compared to control condition (Additional file 2: Fig. S1). These findings suggest that the D101 cultivar is highly resistant and Honghuadajinyua is susceptible to TBW infection (Fig. 1B).
Disease symptoms in D101 (resistant; R) and Honghuadajinyuan (susceptible; S) tobacco cultivars after inoculation with R. solanacearum. Seedlings of R (A) and S (B) cultivars at 0 d, 1 d, 3 d, and 7 d after infection. The leaves show clear disease symptoms at 3 and 7 days in both cultivars as compared to 1 and 3 days, but the leaves of the S cultivar at 7 d show black color and plant death. C Bacterial wilt symptoms in leaves and stem of tobacco seedlings
Overview of RNA sequencing
A total of 1,458.74 million raw reads were collected with an average of 48.62; each library had 40.81–58.89 million raw reads (Additional file 1: Table S1). A total of 1,453.41 (99.64%) million clean reads ranging from 40.66 (99.58%) to 58.67 (99.68%) were obtained after removing adopters, low-quality reads, and unknown bases (Additional file 1: Table S1). The clean reads data was high quality, with a Q30 base rate ranging from 92.78–94.94%, with an average GC content of 42.90%. Further, clean reads were aligned with the reference genome of tobacco (Nitab 4.5), with ~ 1,314.89 (90.80%) million total reads mapped to the genome (Additional file 1: Table S1), of which 1,235.21 (85.29%) million reads were uniquely mapped to transcripts, with an average 41.17 (85.29%) and range 33.62–52.03 (68.44–90.39%), indicating that the sequenced data were suitable for further analysis. Finally, 81,534 genes were identified, of which 69,500 were known, and 12,034 were new. The PC analysis showed 81.70% overall variation, of which PC1 and PC2 had 67.60% and 14.10%, respectively, variation in expression (Fig. 2A). The hierarchical clustering analysis showed similarities between the biological replicates of each R and S cultivar sample (Fig. 2B). Through PC and clustering analysis, the resistant and susceptible accessions clustered from each other, with their replicates located nearby (Fig. 2A-B). The R-7d and S-7d after infection were clustered separately from other time points. All the time points of R and S cultivars except R-7d and S-7d had significant positive correlations (r = 0.60–0.70) with other time points (Fig. 2C). The boxplots showed differences between different time points (Fig. 2D).
Overview of tobacco root transcriptomic data based on bacterial wilt resistant (R) and susceptible (S) tobacco cultivars at five time points. A Principal component analysis represents 81.70% overall variation in expression; B Cluster analysis using FPKM values of each sample with replicates indicates the similarities between the biological replicates of both cultivars; C Correlation analysis between resistant and susceptible tobacco cultivars at four time points between the samples; D log10 transformation of FPKM values at four time-points show the differences between the samples at each time points
Mining differentially expressed genes (DEGs) under bacterial wilt
A pairwise comparison of gene expression levels between inoculation and control treatments identified 23,568 (19,130 known and 4,438 novels) DEGs in R and S at four time points (Fig. 3A and Additional file 1: Table S2). In the R cultivar, 20,711 (87.88%) genes were differentially expressed, of which 2,587 (1,553 up and 1,034 down), 1,873 (1,124 up and 749 down), 4,269 (2,583 up and 1,686 down), and 18,598 (7,512 up and 11,086 down) DEGs were identified at R-6 h, R-1d, R-3d, and R-7d, respectively, and 277 DEGs were common at all stages (Fig. 3B-C and Additional file 1: Table S2). Likewise, S samples at S-6 h, S-1d, S-3d, and S-7d had 3,166 (1,953 up and 1,213 down), 3,085 (1,790 up and 1,295 down), 3,147 (2,334 up and 813 down), and 14,115 (6,380 up and 7,735 down) DEGs, respectively (Fig. 3B), with 554 commonly identified at all time-points (Fig. 3D and Additional file 1: Table S2). In addition, 157 DEGs were commonly induced at 6 h, 1 d, 3 d, and 7 d in R and S cultivars (Fig. 3B-C). The R cultivar had 1.10 and 1.17 times more up-regulated genes than the S cultivar at 3 d and 7 d, respectively, while the S cultivar had 1.26 and 1.59 times more up-regulated genes than the R cultivar at 6 h and 1 d (Additional file 1: Table S2). These results indicate that TBW infection had a more substantial regulatory effect on gene regulation at 3 d and 7 d in R cultivars and 6 h and 1 d in S cultivars.
Analysis of differentially expressed genes in resistant (R) and susceptible (S) tobacco cultivars in response to bacterial wilt infection. A Volcano plots represent significant genes in R and S cultivars at 6 h, 1 d, 3 d, and 7 d relative to CK-0 h, with log2FC values transformed into –log10 (FDR). Blue and red dots indicate up-regulated and down-regulated DEGs (log2FC ≥ ± 1, FDR ≤ 0.05); black dots represent non-significant. B The number of genes differentially expressed at different time points. The highest number of DEGs were identified at 7 d in R and S cultivars. C-D The Venn diagram shows the number of unique and common DEGs in R and S tobacco cultivars at each time point
Gene ontology (GO) enrichment analysis of DEGs
The GO enrichment analysis revealed that 1,236 and 1,121 GO terms were significantly enriched in R and S cultivars, respectively. Information on the DEGs and their corresponding GO terms is in Fig. 4, Additional file 2: Fig. S2–S5 and Additional file 1: Table S3. For the R cultivar, 1,554, 1,206, 2,712, and 12,024 DEGs at R-6 h, R-1d, R-3d, and R-7d, respectively, were involved in 235, 288, 452, and 746 GO terms (Fig. 4A-B, Additional file 2: Fig. S2, S3 and Additional file 1: Table S3), including ‘GO:0005975 carbohydrate metabolic,’ ‘GO:0071554 cell wall organization or biogenesis,’ ‘GO:0016491 oxidoreductase activity,’ ‘GO:0006979 response to oxidative stress,’ ‘GO:0005618 cell wall,’ ‘GO:0006749 glutathione metabolic process,’ ‘GO:0009698 phenylpropanoid metabolic process,’ and ‘GO:0046271 phenylpropanoid catabolic process’ (Fig. 4 and Additional file 1: Table S3). For the S cultivar, 2,012, 1,926, 2,014, and 9,096 DEGs were significantly enriched for 263, 530, 532, and 576 GO terms at S-6 h, S-1d, S-3d, and S-7d, respectively (Fig. 4, Additional file 2: Fig. S3–S5 and Additional file 1: Table S3), including ‘GO:0016740 transferase activity,’ ‘GO:0008017 microtubule binding,’ ‘GO:0048046 apoplast,’ ‘GO:0015631 tubulin binding,’ ‘GO:0005874 microtubule,’ ‘GO:0046527 glucosyltransferase,’ ‘GO:0003700 transcription factor,’ ‘GO:0004871 signal transducer activity,’ ‘GO:0006749 glutathione metabolic process,’ and ‘GO:0009812 flavonoid metabolic process,’ (Additional file 2: Fig. S3–S5 and Additional file 1: Table S3). Additionally, genes involved in the defense-related process, such as pathogen-associated molecular pattern perception, signaling, and the activation of defense-related enzymes and proteins, were enriched in the R cultivar (Fig. 4A-B and Additional file 1: Table S3). Interestingly, the S cultivar had fewer genes enriched in the above-mentioned biological processes, suggesting that these processes are less active or less efficient in the susceptible cultivar (Fig. 4C-D and Additional file 1: Table S3), which could help explain why this cultivar is more susceptible to TBW infection than the R-resistant cultivar. Finally, the top 10 potential up-regulated and downregulated DEGs, their expression pattern, and GO terms at all time points were identified, which might play an essential role in disease resistance (Table 1).
Gene ontology (GO) enrichment analysis of up- and down-regulated genes in bacterial wilt resistant (R) and susceptible (S) tobacco cultivars at 6 h and 1 d. The top significant GO terms of the three categories are shown at R-6 h (A), R-1d (B), S-6 h (C), and S-1d (D) at P ≤ 0.05. The circular diagram represents the enrichment, GO terms with the number of up- and down-regulated genes. The outer circular line indicates the GO ID of each term, the middle circular line indicates the q-value of enrichment with the total number of genes, and the inner circular lines indicate the up-regulated (purple) and down-regulated (blue) number of genes
KEGG enrichment analysis of DEGs
Compared with the control, the DEGs at 6 h, 1 d, 3 d, and 7 d were significantly assigned to 11, 13, 17, and 35 pathways in the R cultivar (Fig. 5A-B and Additional file 1: Table S4) and 10, 14, 19, and 40 pathways in the S cultivar (Fig. 5C-D and Additional file 2: Fig. S8-S9), respectively. In the R cultivar, the highest number of DEGs were enriched in phenylpropanoid biosynthesis, pyruvate metabolism, plant-pathogen interaction, phenylalanine metabolism, carbon metabolism, plant hormone signal transduction, glutathione metabolism, MAPK signaling pathway, steroid biosynthesis, peroxisome, and circadian rhythm, involved in response to R. solanacearum at all time-points (Fig. 5A-B and Additional file 1: Table S4). Similarly, biosynthesis of various antibiotics, ribosomes, biosynthesis of amino acids, glycolysis/gluconeogenesis, plant hormone signal transduction, glutathione metabolism, phenylpropanoid biosynthesis, photosynthesis, plant-pathogen interaction, and phenylalanine metabolism were involved in the response of the S cultivar to BW infection at all time-points (Additional file 2: Fig. S8-S9 and Additional file 1: Table S4). KEGG analysis revealed the enrichment of several pathways, including MAPK signaling, phenylpropanoid biosynthesis, plant hormone signal transduction, and phenylalanine and glutathione metabolism in both R and S cultivars (Fig. 5). Additionally, a substantial number of highly up-regulated genes in these cultivars were associated with phenylalanine metabolism, which serves as a precursor for various secondary metabolites in plants.
KEGG analysis of differentially expressed genes in bacterial wilt resistant (R) and susceptible (S) tobacco cultivars at 6 h and 1 d. The top KEGG pathways are listed at R-6 h (A), R-1d (B), S-6 h (C), and S-1d (D) at P ≤ 0.05. The circular diagram represents the enrichment KEGG pathways with the number of up- and down-regulated genes. The outer circular line indicates the pathway ID of each pathway, middle circular line indicates the q-value of enrichment with total number of genes, and inner circular lines indicate the up-regulated (purple) and down-regulated (blue) number of genes
Identification of potential candidate genes associated with bacterial wilt resistance
Based on GO and KEGG enrichment and functional annotation analysis, we evaluated potential candidate genes related to R. solanacearum resistance (Figs. 6 and 7 and Additional file 1: Tables S5–S7), including those involved in phenylalanine metabolism (stilbenoid, diarylheptanoid, and gingerol biosynthesis//ko00945 and flavonoid biosynthesis//ko00941) and glutathione metabolism pathways, and TFs (WRKYs, ARFs, and ERFs) and gene families (SDR, NBS-LR, PRs, and CKX). Hence, DEGs within these pathways or TFs were selected as potential candidate genes for subsequent investigations.
Heatmaps of differentially expressed genes enriched in glutathione and phenylpropane pathways involved in bacterial wilt resistance in tobacco. A Number of DEGs involved in the phenylpropane pathway; B Number of DEGs involved in glutathione pathway and showed different expression levels at each time point
Pathway-enriched genes for TBW resistance
Forty-nine DEGs involved in the phenylpropane pathway (e.g., flavonoid biosynthesis, stilbenoid, diarylheptanoid, and gingerol biosynthesis) were identified (Fig. 6A and Additional file 1: Table S5), with most up-regulated at 3 d and 7 d. For example, Nitab4.5_0017074g0010 and Nitab4.5_0000024g0150 encoding acyl-sugar acyltransferase 3-like were up-regulated at all time points. At the same time, the R cultivar had higher expression levels of three genes (Nitab4.5_0002623g0010, Nitab4.5_0002775g0010, and Nitab4.5_0000041g0360) that encode vinorine synthase-like protein than the S cultivar (Fig. 6A). Specifically, at 6 h after infection, the R and S cultivars had 8 and 4 up-regulated genes, respectively (Fig. 6A). Additionally, the phenylpropane pathway was enriched in up-regulated genes in both cultivars post-infection at different time points. In the R cultivar, 12, 18, and 35 up-regulated genes were enriched at 1 d, 3 d, and 7 d post-infection, respectively, whereas the S cultivar had 4, 16, and 27 up-regulated genes enriched at the same time-points (Fig. 6A). These genes related to cytochrome P450 (CYP), flavanone 3-hydroxylase (AN3), 4-hydroxyphenylpyruvate dioxygenase (HPD), 4-hydroxyphenylpyruvate dioxygenase (HPD), aromatic-L-amino-acid decarboxylase-like (TDC1), aspartate aminotransferase 1, partial (ASP), and phenylalanine ammonia-lyase G4-like (PAL), enhancing plant resistance against R. solanacearum (Fig. 6A and Additional file 1: Table S5). Moreover, in the glutathione metabolic pathway, 45 genes encoding 5-oxoprolinase-like (OXP), glutathione S-transferase (GST), ornithine decarboxylase-like (ODC), and L-ascorbate peroxidase 6 (APX), had higher expression at all time-points except 6 h (Fig. 6B). Specifically, the R cultivar up-regulated more genes than the S cultivar at 3d and 7 d post-infection (23 and 43, respectively) (Fig. 6B).
Transcription factor genes for TBW resistance
WRKY, ARF, and ERF transcription factors are essential regulators of plant resistance [17]. Among them, the WRKY family is the most well-known and plays a vital role in modulating the transcription of resistance-related genes and regulating various plant defense processes. In this study, 47 WRKY genes were significantly expressed in both cultivars at 6 h, 1 d, 3 d, and 7 d (Fig. 7A and Additional file 1: Table S6). Among these, 19 WRKY genes (WRKY70, WRKY75, WRKY11, WRKY15, WRKY23, WRKY40, WRKY43, WRKY51, WRKY61, WRKY26, and WRKY50 family genes) were simultaneously up-regulated in R and S cultivar at 3 and 7, and 14 WRKY genes (WRKY20, WRKY24, WRKY26, WRKY40, WRKY51, WRKY71, WRKY72, WRKY6, WRKY12, WRKY28, WRKY41, and WRKY53 family genes) were only up-regulated in R cultivar at 7d (Fig. 7A). Notably, four genes, Nitab4.5_0007605g0020 (WRKY50), Nitab4.5_0001000g0160 (WRKY51), Nitab4.5_0000048g0080 (WRKY70), and Nitab4.5_0014092g0020 (WRKY45) were up-regulated in R cultivars at all time-points except 7d (Fig. 7A and Additional file 1: Table S6). It is speculated that three genes (Nitab4.5_0004702g0020, Nitab4.5_0005781g0050, and Nitab4.5_0000005g0190) of WRKY6 TFs were identified at R-6 h and S-6 h with a high expression that could act as positive regulators of TBW resistance (Fig. 7A).
Moreover, ERFs have essential biological functions in various life activities, including plant growth, development, and environmental response. We identified 48 ERFs DEGs in both cultivars (Fig. 7B and Additional file 1: Table S6), of which 5 (ERF1), 2 (ERF2), 9 (ERF3), Nitab4.5_0002813g0010 (ERF4), Nitab4.5_0000734g0070 (ERF5), 4 (ERF10), and 5 (ERF109) family genes were up-regulated at least one-time point in R and S cultivars (Fig. 7B). Like, Nitab4.5_0002230g0030 (ERF110) and Nitab4.5_0002236g0020 (ERF1), genes were up-regulated in R cultivar at 1d, 3d, and 7d while these genes only expressed in S cultivar at 3d after infection (Fig. 7B and Additional file 1: Table S6). Moreover, one gene, Nitab4.5_0006400g0020 (ERF119), was up-regulated in both cultivars at all time points, whereas Nitab4.5_0002271g0010 (ERF2), Nitab4.5_0000734g0070 (ERF5), Nitab4.5_0002924g0050 (ERF13), and Nitab4.5_0001459g0040 (ERF016), were up-regulated only in R cultivar (Fig. 7B). In addition, eight auxin response factor (ARFs) DEGs were identified in response to BW in tobacco R and S cultivars (Fig. 7C and Additional file 1: Table S6). Three genes, Nitab4.5_0003119g0050 (ARF2), Nitab4.5_0000315g0090 (ARF6), and Nitab4.5_0002071g0010 (ARF6), were highly expressed in the early stage of infection (Fig. 7C). Two genes, Nitab4.5_0000476g0080 (ARF7) and Nitab4.5_0007373g0010 (ARF8), were up-regulated only in Resistant cultivars.
In addition, we identified TFs that were Differentially expressed at 6 h of infection in the R and S cultivars; for example, Nitab4.5_0000315g0090 (ARF6) was up-regulated at 6 h in both cultivars, while Nitab4.5_0002071g0010 (ARF6) was only up-regulated at 6 h in S cultivar. Similarly, ten WRKY genes were highly expressed at 6 h; among these, three DEGs (WRKY61, WRKY75, and WRKY28) were only identified in R cultivar, whereas seven genes, including WRKY40, WRKY50, WRKY15, WRKY45, and WRKY51 were identified in S cultivar. Seven ERFs, Nitab4.5_0002526g0070 (ERF053), Nitab4.5_0000512g0230 (ERF053), Nitab4.5_0002473g0040 (ERF053), Nitab4.5_0004682g0020 (ERF.C.3), Nitab4.5_0000333g0020 (ERF010), Nitab4.5_0000057g0300 (ERF118), and Nitab4.5_0006400g0020 (ERF119), were high up-regulated at 6 h in both cultivars, and other four (ERF106, ERF109, ERF110, and ERF.C.3), were down-regulated in S cultivar. These genes might be considered as primary responses of the plant.
Gene families involve in TBW resistance
NBS-LRR genes encode nucleotide-binding site leucine-rich repeat (NBS-LRR) proteins involved in pathogen recognition and signaling. We identified two novel genes (MSTRG.61386-R1B-17 and MSTRG.61568) up-regulated in both cultivars at 7 d (Additional file 1: Table S7). Further, 26 genes that encode pathogenesis-related (PRs) proteins were identified in both cultivars, which may be involved in various aspects of the plant defense response (Fig. 7D and Additional file 1: Table S7). Among these, Nitab4.5_0014031g0010 (PRB1), Nitab4.5_0004861g0030 (PRB1), Nitab4.5_0008835g0020 (STH-2), and Nitab4.5_0000360g0100 (TLP1) were highly differentially expressed in the R cultivar than the S cultivar at 1 d, 3 d, and 7 d (Fig. 7D). Additionally, PRB5 genes were down-regulated in both cultivars. In this study, 14 DEGs encode short-chain dehydrogenase/reductase—detected at different time points in the R and S cultivars after bacterial infection—involved in the biosynthesis of phytohormones that play a key function in defense response of plants against pathogens and BW (Fig. 7E and Additional file 1: Table S7). Nitab4.5_0011992g0020 and Nitab4.5_0000001g0060 were significantly up-regulated in both cultivars at all time points. One gene, Nitab4.5_0001703g0050 (SDR2a), was highly expressed in the R cultivar but low in the S cultivar (Fig. 7E). The remaining SDR genes showed an initial up-regulation during the early stage of infection, followed by a down-regulation after post-infection.
Validation of transcriptomic data through quantitative real-time PCR
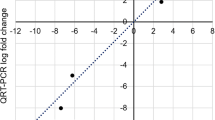
To measure the expression levels of potential resistance-related genes, 13 DEGs (11 up-regulated and 2 down-regulated) were randomly selected and analyzed by qRT-PCR at 6 h, 1 d, 3 d, and 7 d after infection. Of these, two genes (Nitab4.5_0000356g0010 and Nitab4.5_0012932g0010) associated with GST, one gene (Nitab4.5_0005779g0060) related to CKX5, and one gene (Nitab4.5_0008666g0010) related to short-chain dehydrogenase TIC32. As a result, the RT-qPCR produced similar results as the RNA-seq data at 6 h, 1 d, 3 d, and 7 d (Fig. 8A-H). The correlation analysis revealed a highly significant positive correlation between RT-qPCR and RNA-seq data at R-6 h (r = 0.95 and P = 8.5 × 10–5) and S-6 h (r = 0.96 and P = 9.8 × 10–5), indicating methodological reliability and supports the consistency of the RNA-sequencing data (Fig. 8A-H).
Validation of 13 DEGs in response to bacterial wilt resistance via RT-qPCR. The RT-qPCR and RNA-seq log2FC values were compared using correlation analysis at R-6 h (A), R-1d (B), R-3d (C), R-7d (D), S-6 h (E), S-1d (F), S-3d (G), and S-7d (H). R and P indicate the correlation coefficient and corresponding P-value. The correlation showed a significant positive relationship between the RT-qPCR and RNA-seq at each time point in R and S cultivars
Discussion
Tobacco bacterial wilt caused by R. solanacearum is a major soil-borne disease that adversely affects the quality and yield of tobacco crops. The survival of R. solanacearum in non-host or soil, and its ability to cause severe damage once the host appears makes TBW a significant economic threat [7, 9, 17, 44, 45]. The plant's transcriptional response to pathogen infection is a dynamic process involving the modulation of signals and compounds that change over time [29, 33, 46]. This study identified 239 potential candidate genes associated with the phenylpropane pathway, glutathione metabolism, WRKY, ERFs, ARFs, PRs, and NBS-LRR using root transcriptomic sequencing data of R and S cultivars at 6 h, 1 d, 3 d, and 7 d (Fig. 6–7 and Additional file 1: Tables S5–S7).
The Illumina sequencing data was reliable and accurately verified using RT-qPCR expression (Fig. 8) with a high Q30 base rate ranging from 92.78–94.94%. A total of 81,534 genes were identified, with 69,500 known and 12,034 novel genes, in line with published studies [33, 46]. The results indicate that BW infection significantly changes gene expression in R and S cultivars, with 20,711 DEGs in the R cultivar and 16,663 DEGs in the S cultivar (Fig. 3A-B and Additional file 1: Table S2). Among these, 554 were common between all samples (Fig. 3B-C), suggesting that the response to BW infection is cultivar-specific and occurs at different time-points in the two cultivars, similar to studies on tobacco [9], tomato [37], pepper [34], and Arabidopsis [47]. For example, Ishihara et al. [32] identified 140 DEGs in a resistant tomato variety and none in a susceptible variety.
The identified DEGs were involved in 1,236 and 1,121 GO terms in the R and S cultivars, respectively (Fig. 4 and Additional file 1: Table S3). The R cultivar had a stronger defense response to R. solanacearum infection than the S cultivar, as evidenced by the up-regulation of genes involved in transferase activity, cell wall organization, hormone signal transduction, abiotic stress response, and oxidative stress response. In contrast, the S cultivar had a weaker defense response, as evidenced by the up-regulation of genes involved in tetrapyrrole binding, oxidoreductase activity, and carbohydrate metabolic processes (Fig. 4 and Additional file 1: Table S3). Our results are consistent with previous studies on BW resistance in solanaceous crops, linking resistance to increased expression of genes related to defense mechanisms, cell wall organization, and oxidative stress response [29, 32,33,34, 48]. For example, Pan et al. [33] found that 3,967 DEGs were associated with endocytosis, cell wall, signal transduction, and starch/sucrose metabolism in response to BW infection.
The KEGG analysis showed that the DEGs at 6 h, 1 d, 3 d, and 7 d were significantly assigned to different pathways in the R and S cultivars (Fig. 5 and Additional file 1: Table S4). The highest up-regulated genes in the R cultivar (Additional file 2: Fig. S6-S7 and Additional file 1: Table S4) were enriched in plant hormone signal transduction, glutathione metabolism, phenylpropanoid biosynthesis, plant–pathogen interactions, and MAPK signaling pathway, consistent with studies investigating the molecular mechanisms of BW resistance in tomato [37], eggplant [36], tobacco [17, 30, 33], and potato [8, 35]. Furthermore, other studies reported the involvement of the enriched pathways in tobacco defense against R. solanacearum [26, 29, 46]. These results also align with studies that reported the activation of multiple defense mechanisms in response to TBW infection in resistant tomato and eggplant cultivars, including the activation of glutathione metabolism and phenylpropanoid biosynthesis [32, 34, 49].
The present study identified 49 potential candidate genes in the phenylpropanoid pathway (Fig. 6A). Activation of the phenylpropanoid pathway has been associated with increased resistance to BW in tobacco [9, 35, 46]. Six PAL genes (Nitab4.5_0002130g0010, Nitab4.5_0000754g0260, Nitab4.5_0012382g0010, Nitab4.5_0018680g0010, Nitab4.5_0000582g0180, and Nitab4.5_0000071g0170) were up-regulated at 3 d and 7 d post-infection (Fig. 6A). Several studies have investigated the role of the phenylpropanoid pathway in defending against BW [17, 35, 37]. For example, Park et al. [50] reported that the up-regulation of PAL genes plays a significant role in the plant defense response to BW, while the OsPAL2 mutant gene in rice contributed to the control of pathogen disease resistance [51]. This study revealed two genes (CYP98A2 and CYP98A3) of cytochrome CYP450 (Fig. 6A) that play an essential role in BW resistance in many crops [52,53,54,55]. Moreover, we identified 46 candidate genes involved in glutathione metabolism, including APX, GST, ODC, OXP, and PRP (Fig. 6B), which play a crucial role in the detoxification of ROS and the regulation of cellular signaling pathways that lead to the activation of defense mechanisms [6, 30, 37, 56, 57]. Our results showed that the R cultivar up-regulated more genes involved in glutathione metabolism than the S cultivar at 3 d and 7 d after TBW inoculation (Fig. 6B), consistent with other studies [7, 36, 58]. Overexpression of GST and GPX genes increased expression activity twofold in wild-type tobacco seedlings, contributing to chilling or salt stress tolerance [59]. Li et al. [17] reported up-regulated DEGs involved in the phenylpropanoid pathway and glutathione metabolism for TBW resistance. Therefore, the glutathione metabolism and phenylpropanoid pathway are essential defense mechanisms in tobacco plants against BW caused by R. solanacearum. Further research is needed to understand how these pathways contribute to disease resistance in tobacco.
TFs are crucial plant regulators [38, 60], with WRKY TFs playing essential roles in plant immunity against various stresses. WRKY40, WRKY6, WRKY27, and WRKY22 have been identified as positive regulators of Solanaceae crop resistance to BW in several [38, 60,61,62]. In this study, WRKY6 (Nitab4.5_0004702g0020, Nitab4.5_0005781g0050, and Nitab4.5_0000005g0190) and WRKY11 (Nitab4.5_0000286g0080) were found to exhibit a resistance response to TBW in the R cultivar (Fig. 7A), particularly in the later stage, consistent with the response pattern observed in pepper to BW [62]. These findings indicate that these genes may be important regulators of tobacco resistance against BW [63, 64]. Moreover, ERFs are a class of plant TFs involved in biological and abiotic stress responses [17, 65, 66]. In this study, 48 ERFs genes were significantly expressed in R and S cultivars at all time points (Fig. 7B). The expression pattern of ERF5 (Nitab4.5_0000734g0070) in the R cultivar was distinct from that in the S cultivar (Fig. 7B), especially at 3 d. The eight ARF genes identified in the present study were up-regulated at 3 d and 7 d after infection (Fig. C); these genes play a critical role in regulating plant growth and development to improve plant defense mechanisms against pathogens [17, 33, 39]. Pathogenesis-related proteins (PRs) are essential for BW resistance [67, 68], with the present study identifying 26 PR genes (PR-1, PR-4, and PR-5) in both cultivars at all time points (Fig. 7D). Two PR-1 genes (Nitab4.5_0004861g0040 and Nitab4.5_0005400g0020) were up-regulated at 3 d and 7 d in both cultivars (Fig. 7D), similar to previous studies [68, 69]. Rivière et al. [69] found that PR-1b was up-regulated in tobacco plants resistant to BW and that silencing PR-1b increased disease susceptibility. Similarly, Alamillo et al. [70] and Pruss et al. [71]reported up-regulated expression of PR-2 in tobacco infected with R. solanacearum, while silencing PR-2 increased disease susceptibility [72]. Further, we identified 14 SDR genes involved in various metabolic pathways, including detoxification and hormone biosynthesis (Fig. 7E). In tobacco, SDR genes play a role in the defense response against bacterial pathogens: Yu et al. [73] reported that SDR genes are involved in the biosynthesis of jasmonic acid, which is critical for defense response against pathogens. Overall, the present study identified 239 potential candidate genes associated with pathways, TFs, and gene families (Figs. 6 and 7 and Additional file 1: Tables S5–S7), which will help understand the defense response mechanism and could be potential targets for improving resistance to BW disease. In summary, our results revealed that DEGs associated with phenylpropane/flavonoids pathway, glutathione metabolic pathway, WRKY, ERFs, ARFs, pathogenesis-related genes (PRs), and short-chain dehydrogenase/reductase genes essential to control the resistance and programmed cell death. Of these, most of the defense-related genes were up-regulated in the R cultivar compared to the S cultivar at an early stage of infection. Therefore, our hypothesized model indicates the involvement of defense-related and susceptible-related genes networking of R. solanacearum in the tobacco system (Fig. 9).
A model pathway involved in resistance against tobacco bacterial wilt. The tobacco reaction to R. solanacearum infection and the systematic pathway involved in resistance. The signals from the pathogens were recognized through related receptors (R genes) and transcription factors, which triggered the plant defense mechanism. After that, defense-related molecules/genes, including PR-proteins, metabolites, glutathione and phenylpropane genes, were activated to cause resistance to pathogens through high expression levels
Conclusion
This study used high throughput sequencing to identify DEGs and related pathways in resistant and susceptible cultivars under R. solanacearum infection at early and late seedling stages. In early plant response after 6 h, 2 ARFs, 10 WRKY, and 11 ERFs genes were differentially expressed, which might be the plants' primary response. The R and S cultivars had similar response levels at the early stage, but the R cultivar-related DEGs had specific expression patterns in response to infection after 7 d. Finally, 239 potential candidate genes were identified using GO, KEGG, functional enrichment analysis, and literature search. These DEGs in the resistant cultivar are related to glutathione metabolism and phenylpropanoid biosynthesis, TFs, and PRs, and are likely the main substances conferring root resistance against BW infection in tobacco. Additionally, two novel genes (MSTRG.61386-R1B-17 and MSTRG.61568) were up-regulated in both cultivars at 7 d, which can be used for functional studies to find the exact role in BW resistance. The finding of this study shows the molecular mechanisms involved in tobacco resistance to bacterial wilt and provides an important source for controlling and breeding tobacco plants resistant to TBW.
Methods
Plant material and growth conditions
Two tobacco cultivars, D101 (R) and Honghuadajinyuan (S), were received from Nanxiong Scientific Research Institute of Guangdong Research Institute of Tobacco Science, Shaoguan, China. The experiments were conducted in the greenhouse of Guangzhou University, China. Seeds of both cultivars were surface-sterilized with distilled water and spread on a black plastic tray with white and pit seedling sponges. The tray was covered with a layer of fresh-keeping film and placed in the growth chamber to germinate at 28℃, 60% relative humidity, and 16 h/8 h light/dark photoperiod. After germination, seedlings in the crosswise stage were transferred into hydroponic boxes (41 × 24 × 14 cm; length × width × height). Hoagland’s nutrient solution was added to the aerated hydroponic containers, comprising 945 mg/L Ca(NO3)2·4H2O, 607 mg/L KNO3, 115 mg/L (NH4)2HPO4, 493 mg/L MgSO4·7H2O, 30 mg/L [-CH2N(CH2COONa)CH2COO]2Fe, 2.86 mg/L H3BO3, 2.13 mg/L MnSO4.4H2O, 0.22 mg/L ZnSO4.7H2O, 0.08 mg/L CuSO4.5H2O, and 0.02 mg/L (NH4)2MoO4.
Inoculation treatment and sample collection
The highly pathogenic R. solanacearum strain HBLC5 was isolated from tobacco plants obtained from Hukou experimental station in Nanxiong City as described in our previous studies [7, 15]. For inoculation preparation, the R. solanacearum strain HBLC5 was cultured on BG medium and reproduced in Liquid LB medium by shaking at 30℃ and 180 rpm for two days. Five-leaf stage plants were infected with 300 mL cell suspension (adjusted to 1 × 108 CFU/mL and OD600nm = 0.1) by irrigating roots in one pot. The control was treated with sterile water instead of the bacterial solution. Each tobacco variety was grown in three pots, each with 60 seedlings. Tobacco roots from three individual seedlings taken from one pot were considered one biological replicate. Therefore, three biological replicates of each variety were harvested from three pots at every time point. After inoculation, the roots from R and S cultivars were collected at R/S-0 h, R/S-6 h, R/S-1d, R/S-3d, and R/S-7d, and the samples were stored at -80 °C after freezing in liquid nitrogen until RNA extraction.
RNA extraction, library preparation, and Illumina sequencing
The total RNA of tobacco seedlings roots at five-six leaf stage (~ 30 days) was extracted from three biological replicates at each time-point using the TRIzol Reagent Kit (Invitrogen, Carlsbad, CA, USA) with the manufacturer’s instructions. In short, RNA was extracted from the frozen samples, and DNase I (TakaraBio, Japan) was used to remove DNA contamination. The RNA quality and integrity were assessed through 1% agarose gel electrophoresis and a microplate spectrophotometer (BioTek Company, USA). To develop the cDNA libraries, mRNA with polyA tails was purified using Oligo(dT) magnetic beads and fragmented. The first and second strands of cDNA were synthesized with random hexamer primers and DNA polymerase I, respectively. Adapters were ligated to double-stranded cDNAs, which were then exposed to PCR amplification and purification. The quality of the cDNA libraries was measured using an Agilent 2100 Bioanalyzer, and sequencing was performed on the Illumina Novaseq 6000 platform by Genedenovo Biotechnology Co., Ltd in Guangzhou, China.
Sequencing data analysis and genomic annotation
The sequenced raw reads were processed using fastp v0.18.0 software [74] to determine the reliability and quality of sequenced data and remove the reads that contained adaptors, ≤ 10% unknown bases, and low-quality reads at Q ≤ 20%. Clean reads were used to determine Q20, Q30, and GC contents and for all further downstream analyses. The Nitab4.5 reference genome of N. tabacum [75] and gene annotation files were downloaded from the solgenomics database (https://solgenomics.net/organism/Nicotiana_tabacum/genome). The first step was to align the clean reads (i.e., reads filtered for quality and adapter sequences) to a reference genome. In this case, the clean reads were aligned to Nitab4.5 using HISAT2 v2.1.0 [76] and Bowtie2 v2.2.5 [77]. After alignment, the sequences of all assembled genes were aligned to different databases for functional annotation, including the Nitab4.5 reference genome, UniProt, NT, KEGG, GO, and COG using Trinotate (http://trinotate.github.io/) software with default parameters except E ≤ 1e-5. Then, transcripts were reconstructed using StringTie v2.2.0 [78] based on the HISAT2 alignment. Transcript abundance was measured as FPKM values using RSEM software [79]. Finally, the principal component analysis (PCA) and Pearson’s correlation coefficients were calculated using the R4.1.0 environment (http://www.r-project.org/) based on the FPKM expression data.
Identification of differentially expressed genes
DESeq2 R package [80] was used to perform differential expression analysis by pairwise comparison of the control and treatments at 6 h, 1 d, 3 d, and 7 d. The P-value was adjusted using the Benjamini–Hochberg method with an FDR threshold of 0.05, and genes were identified as up- or down-regulated based on the |log2FC|> 1 criteria. The hierarchical K-mean clustering method was used to analyze the DEGs using the hclust function in R version 4.1.0 (http://www.r-project.org/), and DEGs were then visualized using a heatmap as log2FC input.
GO and KEGG enrichment analysis
GO enrichment analysis of DEGs was performed using the GO database (http://www.geneontology.org/) with a significance threshold of corrected P ≤ 0.05. KEGG enrichment analysis was conducted using the online KEGG database (https://www.genome.jp/kegg/kegg1.html) at P ≤ 0.05 as a significance threshold to identify significant biological functions and metabolic pathways [81]. The clusterProfiler v3.4.4 software was used to visualize the top GO terms and KEGG pathways [82].
Gene expression analysis through RT-qPCR
Thirteen genes were randomly selected with different expression profiles to validate sequencing data by RT-qPCR using specific primers presented in Additional file 1: Table S8. Reverse transcription was completed using the HiScript® III RT SuperMix for PCR (+ gDNA wiper) Kit (Vazyme Biotech Co., Ltd). The qRT-PCR was performed using 1 µL cDNA template mixed with 10 µL 2 × ChamQ SYBR qPCR Master Mix (Vazyme Biotech Co., Ltd) and 0.4 µL each of the forward and reverse primers in a final volume of 20 µL. Amplification consisted of 40 cycles of 95℃ for 10 s, 60℃ for 30 s, and 95℃ for 15 s, followed by a denaturation step to generate the melting curves. The Ntubc2 (accession number: AB026056) was selected as an internal reference gene, and gene expression profiles were measured using the 2-∆∆CT method. Three technical and biological replicates were used for each sample of cultivars at all time points.
Availability of data and materials
The study data sets can be accessed at NCBI database under accession PRJNA935858 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA935858).
Abbreviations
- DEGs :
-
Differentially expressed genes
- R:
-
Resistant
- S:
-
Suceptible
- TF:
-
Transcriptional factors
References
Berbeć AK, Matyka M. Biomass characteristics and energy yields of tobacco (Nicotiana tabacum l.) cultivated in eastern Poland. Agric. 2020;10:1–12.
Li H, Ikram M, Xia Y, Li R, Yuan Q, Zhao W, et al. Genome-wide identification and development of InDel markers in tobacco (Nicotiana tabacum L.) using RAD-seq. Physiol Mol Biol Plants. 2022;28:1077–89.
Sierro N, Battey JND, Ouadi S, Bakaher N, Bovet L, Willig A, et al. The tobacco genome sequence and its comparison with those of tomato and potato. Nat Commun. 2014;5:3833.
Tong Z, Xiu Z, Ming Y, Fang D, Chen X, Hu Y, et al. Quantitative trait locus mapping and genomic selection of tobacco (Nicotiana tabacum L.) based on high-density genetic map. Plant Biotechnol Rep. 2021;15:845–54.
Hayward AC. Biology and epidemiology of bacterial wilt caused by Pseudomonas Solanacearum. Annu Rev Phytopathol. 1991;29:65–87.
Jiang G, Wei Z, Xu J, Chen H, Zhang Y, She X, et al. Bacterial wilt in China: History, current status, and future perspectives. Front Plant Sci. 2017;8:1549.
Lai R, Ikram M, Li R, Xia Y, Yuan Q, Zhao W, et al. Identification of novel quantitative trait nucleotides and candidate genes for bacterial wilt resistance in tobacco (Nicotiana tabacum L.) using genotyping-by-sequencing and multi-locus genome-wide association studies. Front Plant Sci. 2021;12:744175.
Li GC, Jin LP, Wang XW, Xie KY, Yang Y, van der Vossen EAG, et al. Gene transcription analysis during interaction between potato and Ralstonia solanacearum1. Russ J Plant Physiol. 2010;57:685–95.
Gao W, Chen R, Pan M, Tang W, Lan T, Huang L, et al. Early transcriptional response of seedling roots to Ralstonia solanacearum in tobacco (Nicotiana tabacum L.). Eur J Plant Pathol. 2019;155:527–36.
Basit A, Farhan M, Di MW, Ding HX, Ikram M, Farooq T, et al. Enhancement of resistance by poultry manure and plant hormones (salicylic acid & citric acid) against tobacco mosaic virus. Saudi J Biol Sci. 2021;28:3526–33.
Delgado R, Morillo E, Buitrón J, Bustamante A, Sotomayor I. First report of Moko disease caused by Ralstonia solanacearum race 2 in plantain ( Musa AAB) in Ecuador. New Dis Reports. 2014;30:23–23.
Chae S-Y, Lee K, Do J-W, Hong S-C, Lee K-H, Cho M-C, et al. QTL mapping of resistance to bacterial wilt in pepper plants (Capsicum annuum) using genotyping-by-sequencing (GBS). Horticulturae. 2022;8:115.
Du H, Chen B, Zhang X, Zhang F, Miller SA, Rajashekara G, et al. Evaluation of Ralstonia solanacearum infection dynamics in resistant and susceptible pepper lines using bioluminescence imaging. Plant Dis. 2017;101:272–8.
Guji MJ, Yetayew HT, Kidanu ED. Yield loss of ginger (Zingiber officinale) due to bacterial wilt (Ralstonia solanacearum) in different wilt management systems in Ethiopia. Agric Food Secur. 2019;8.
Ikram M, Xiao J, Li R, Xia Y, Zhao W, Yuan Q, et al. Identification of superior haplotypes and candidate genes for yield-related traits in tobacco (Nicotiana tabacum L.) using association mapping. Ind Crops Prod. 2022;189 October:115886.
Lan T, Zheng S, Yang L, Wu S, Wang B, Zhang S, et al. Mapping of quantitative trait loci conferring resistance to bacterial wilt in tobacco (Nicotiana tabacum L.). Plant Breed. 2014;133:672–7.
Li YY, Wang L, Sun GW, Li XH, Chen ZG, Feng J, et al. Digital gene expression analysis of the response to Ralstonia solanacearum between resistant and susceptible tobacco varieties. Sci Rep. 2021;11:1–16.
Liu Y, Wu D, Liu Q, Zhang S, Tang Y, Jiang G, et al. The sequevar distribution of Ralstonia solanacearum in tobacco-growing zones of China is structured by elevation. Eur J Plant Pathol. 2017;147:541–51.
Nishi T, Tajima T, Noguchi S, Ajisaka H, Negishi H. Identification of DNA markers of tobacco linked to bacterial wilt resistance. Theor Appl Genet. 2003;106:765–70.
Dodds PN, Rathjen JP. Plant immunity: Towards an integrated view of plantĝ€" pathogen interactions. Nat Rev Genet. 2010;11:539–48.
Dermastia M. Plant Hormones in Phytoplasma Infected Plants. Front Plant Sci. 2019;10:447.
Lai R, Li R, Xia Y, Guo P, Yuan Q, Zhao W. SSR marker-based genetic diversity analysis of tobacco germplasm and association analysis with resistance to bacterial wilt. Acta Tabacaria Sin. 2018;24:67–81.
Qian Y liang, Wang X sheng, Wang D zhou, Zhang L na, Zu C long, Gao Z liang, et al. The detection of QTLs controlling bacterial wilt resistance in tobacco (N. tabacum L.). Euphytica. 2013;192:259–66.
Habe I, Miyatake K, Nunome T, Yamasaki M, Hayashi T. Qtl analysis of resistance to bacterial wilt caused by ralstonia solanacearum in potato. Breed Sci. 2019;69:592–600.
Nguyen TT, Le NT, Sim S-C. Genome-wide association study and marker development for bacterial wilt resistance in tomato (Solanum lycopersicum L.). Sci Hortic (Amsterdam). 2021;289:110418.
Zia B, Shi A, Olaoye D, Xiong H, Ravelombola W, Gepts P, et al. Genome-wide association study and genomic prediction for bacterial wilt resistance in common bean (Phaseolus vulgaris) core collection. Front Genet. 2022;13 May:1–13.
Ikram M, Han X, Zuo JF, Song J, Han CY, Zhang YW, et al. Identification of QTNs and their candidate genes for 100-seed weight in soybean (Glycine max L.) using multi-locus genome-wide association studies. Genes (Basel). 2020;11:1–22.
Ikram M, Lai R, Xia Y, Li R, Zhao W, Siddique KHM, et al. Genetic dissection of tobacco (Nicotiana tabacum L.) plant height using single-locus and multi-locus genome-wide association studies. Agronomy. 2022;12:1047.
Shi H, Xu P, Yu W, Cheng Y, Ding A, Wang W, et al. Metabolomic and transcriptomic analysis of roots of tobacco varieties resistant and susceptible to bacterial wilt. Genomics. 2022;114:110471.
Yang L, Wei Z, Valls M, Ding W. Metabolic profiling of resistant and susceptible tobaccos response incited by Ralstonia pseudosolanacearum causing bacterial wilt. Front Plant Sci. 2022;12 January.
Ikram M, Chen J, Xia Y, Li R, Siddique KHM, Guo P. Comprehensive transcriptome analysis reveals heat-responsive genes in flowering Chinese cabbage (Brassica campestris L. ssp. chinensis) using RNA sequencing. Front Plant Sci. 2022;13:1077920.
Ishihara T, Mitsuhara I, Takahashi H, Nakaho K. Transcriptome analysis of quantitative resistance-specific response upon Ralstonia solanacearum infection in tomato. PLoS ONE. 2012;7:e46763.
Pan X, Chen J, Yang A, Yuan Q, Zhao W, Xu T, et al. Comparative transcriptome profiling reveals defense-related genes against Ralstonia solanacearum infection in tobacco. Front Plant Sci. 2021;12.
Hwang J, Choi Y, Kang J, Kim S, Cho M, Mihalte L, et al. Microarray analysis of the transcriptome for bacterial wilt resistance in pepper (Capsicum annuum L.). Not Bot Horti Agrobot Cluj-Napoca. 2011;39:49–57.
Zuluaga AP, Solé M, Lu H, Góngora-Castillo E, Vaillancourt B, Coll N, et al. Transcriptome responses to Ralstonia solanacearum infection in the roots of the wild potato Solanum commersonii. BMC Genomics. 2015;16:246.
Peng J, Wang P, Fang H, Zheng J, Zhong C, Yang Y, et al. Weighted gene co-expression analysis network-based analysis on the candidate pathways and hub genes in eggplant bacterial wilt-resistance: A plant research study. Int J Mol Sci. 2021;22:13279.
French E, Kim BS, Rivera-Zuluaga K, Iyer-Pascuzzi AS. Whole root transcriptomic analysis suggests a role for auxin pathways in resistance to Ralstonia solanacearum in tomato. Mol Plant-Microbe Interact. 2018;31:432–44.
Dang F, Wang Y, She J, Lei Y, Liu Z, Eulgem T, et al. Overexpression of CaWRKY27, a subgroup IIe WRKY transcription factor of Capsicum annuum, positively regulates tobacco resistance to Ralstonia solanacearum infection. Physiol Plant. 2014;150:397–411.
Gutterson N, Reuber TL. Regulation of disease resistance pathways by AP2/ERF transcription factors. Curr Opin Plant Biol. 2004;7:465–71.
Chen X, Wang W, Cai P, Wang Z, Li T, Du Y. The role of the MAP kinase−kinase protein StMKK1 in potato immunity to different pathogens. Hortic Res. 2021;8:117.
Cai W, Yang S, Wu R, Cao J, Shen L, Guan D, et al. Pepper NAC-type transcription factor NAC2c balances the trade-off between growth and defense responses. Plant Physiol. 2021;186:2169–89.
Wang J, Zheng C, Shao X, Hu Z, Li J, Wang P, et al. Transcriptomic and genetic approaches reveal an essential role of the NAC transcription factor SlNAP1 in the growth and defense response of tomato. Hortic Res. 2020;7:209.
Kiba A, Fukui K, Mitani M, Galis I, Hojo Y, Shinya T, et al. Silencing of phosphoinositide dependent protein kinase orthologs reduces hypersensitive cell death in nicotiana benthamiana. Plant Biotechnol. 2020;37:363–7.
Drake-Stowe K, Bakaher N, Goepfert S, Philippon B, Mark R, Peterson P, et al. Multiple disease resistance loci affect soil-borne disease resistance in tobacco (Nicotiana tabacum). Phytopathology. 2017;107:1055–61.
Abbas A, Khan SU, Khan WU, Saleh TA, Khan MHU, Ullah S, et al. Antagonist effects of strains of Bacillus spp. against Rhizoctonia solani for their protection against several plant diseases: Alternatives to chemical pesticides. C R Biol. 2019;342:124–35.
Pan X, Chen J, Yang A, Yuan Q, Zhao W, Xu T, et al. Comparative transcriptome profiling reveals defense-related genes against Ralstonia solanacearum Infection in Tobacco. Front Plant Sci. 2021;12:757–75.
Deslandes L, Olivier J, Theulières F, Hirsch J, Feng DX, Bittner-Eddy P, et al. Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. Proc Natl Acad Sci U S A. 2002;99:2404–9.
Mehmood S, Ahmed W, Ikram M, Imtiaz M, Mahmood S, Tu S, et al. Chitosan modified biochar increases soybean (Glycine max l.) resistance to salt-stress by augmenting root morphology, antioxidant defense mechanisms and the expression of stress-responsive genes. Plants. 2020;9:1–25.
Jiang N, Fan X, Lin W, Wang G, Cai K. Transcriptome analysis reveals new insights into the bacterial wilt resistance mechanism mediated by silicon in tomato. Int J Mol Sci. 2019;20:761.
Park N Il, Xu H, Li X, Kim YS, Lee MY, Park SU. Overexpression of phenylalanine ammonia-lyase improves flavones production in transgenic hairy root cultures of Scutellaria baicalensis. Process Biochem. 2012;47:2575–80.
Tonnessen BW, Manosalva P, Lang JM, Baraoidan M, Bordeos A, Mauleon R, et al. Rice phenylalanine ammonia-lyase gene OsPAL4 is associated with broad spectrum disease resistance. Plant Mol Biol. 2015;87:273–86.
Boachon B, Junker RR, Miesch L, Bassard JE, Höfer R, Caillieaudeaux R, et al. CYP76C1 (Cytochrome P450)-mediated linalool metabolism and the formation of volatile and soluble linalool oxides in arabidopsis flowers: A strategy for defense against floral antagonists. Plant Cell. 2015;27:2972–90.
Han JY, Hwang HS, Choi SW, Kim HJ, Choi YE. Cytochrome P450 CYP716A53v2 catalyzes the formation of protopanaxatriol from protopanaxadiol during ginsenoside biosynthesis in panax ginseng. Plant Cell Physiol. 2012;53:1535–45.
Han JY, Kim HJ, Kwon YS, Choi YE. The Cyt P450 enzyme CYP716A47 catalyzes the formation of protopanaxadiol from dammarenediol-II during ginsenoside biosynthesis in Panax ginseng. Plant Cell Physiol. 2011;52:2062–73.
Li H, Pinot F, Sauveplane V, Werck-Reichhart D, Diehl P, Schreiber L, et al. Cytochrome P450 family member CYP704B2 catalyzes the v-hydroxylation of fatty acids and is required for anther cutin biosynthesis and pollen exine formation in rice. Plant Cell. 2010;22:173–90.
Foyer CH, Noctor G. Oxidant and antioxidant signalling in plants: A re-evaluation of the concept of oxidative stress in a physiological context. Plant, Cell Environ. 2005;28:1056–71.
Safdar L Bin, Almas F, Sarfraz S, Ejaz M, Ali Z, Mahmood Z, et al. Genome‐wide association study identifies five new cadmium uptake loci in wheat. Plant Genome. 2020;13.
Gullner G, Komives T, Király L, Schröder P. Glutathione S-Transferase Enzymes in Plant-Pathogen Interactions. Front Plant Sci. 2018;9.
Roxas VP, Smith RK, Smith RK, Allen RD. Overexpression of glutathione s-transferase/glutathione peroxidase enhances the growth of transgenic tobacco seedlings during stress. Nat Biotechnol. 1997;15:988–91.
Hussain A, Li X, Weng Y, Liu Z, Ashraf MF, Noman A, et al. CaWRKY22 acts as a positive regulator in pepper response to ralstonia solanacearum by constituting networks with caWRKY6, caWRKY27, caWRKY40, and caWRKY58. Int J Mol Sci. 2018;19:1426.
Ashraf MF, Yang S, Wu R, Wang Y, Hussain A, Noman A, et al. Capsicum annuum hsfb2a positively regulates the response to ralstonia solanacearum infection or high temperature and high humidity forming transcriptional cascade with cawrky6 and cawrky40. Plant Cell Physiol. 2018;59:2608–23.
Cai H, Yang S, Yan Y, Xiao Z, Cheng J, Wu J, et al. CaWRKY6 transcriptionally activates CaWRKY40, regulates Ralstonia solanacearum resistance, and confers hightemperature and high-humidity tolerance in pepper. J Exp Bot. 2015;66:3163–74.
Ali MA, Azeem F, Nawaz MA, Acet T, Abbas A, Imran QM, et al. Transcription factors WRKY11 and WRKY17 are involved in abiotic stress responses in Arabidopsis. J Plant Physiol. 2018;226:12–21.
Ali MA, Wieczorek K, Kreil DP, Bohlmann H. The beet cyst nematode heterodera schachtii modulates the expression of WRKY transcription factors in syncytia to favour its development in arabidopsis roots. PLoS ONE. 2014;9:e102360.
Xu ZS, Chen M, Li LC, Ma YZ. Functions and Application of the AP2/ERF transcription factor family in crop improvement. J Integr Plant Biol. 2011;53:570–85.
Thirunavukkarasu N, Hossain F, Mohan S, Shiriga K, Mittal S, Sharma R, et al. Genome-wide expression of transcriptomes and their co-expression pattern in subtropical maize (Zea mays L.) under waterlogging stress. PLoS One. 2013;8:e70433.
Gorjanović S. A review: Biological and technological functions of barley seed pathogenesis- related proteins (PRS). J Inst Brew. 2009;115:334–60.
van Loon LC, Rep M, Pieterse CMJ. Significance of Inducible Defense-related Proteins in Infected Plants. Annu Rev Phytopathol. 2006;44:135–62.
Rivière MP, Marais A, Ponchet M, Willats W, Galiana E. Silencing of acidic pathogenesis-related PR-1 genes increases extracellular β-(1→3)-glucanase activity at the onset of tobacco defence reactions. J Exp Bot. 2008;59:1225–39.
Alamillo JM, Saénz P, García JA. Salicylic acid-mediated and RNA-silencing defense mechanisms cooperate in the restriction of systemic spread of plum pox virus in tobacco. Plant J. 2006;48:217–27.
Pruss GJ, Lawrence CB, Bass T, Li QQ, Bowman LH, Vance V. The potyviral suppressor of RNA silencing confers enhanced resistance to multiple pathogens. Virology. 2004;320:107–20.
Graham TL, Graham MY, Subramanian S, Yu O. RNAi silencing of genes for elicitation or biosynthesis of 5-deoxyisoflavonoids suppresses race-specific resistance and hypersensitive cell death in Phytophthora sojae infected tissues. Plant Physiol. 2007;144:728–40.
Yu S, Sun Q, Wu J, Zhao P, Sun Y, Guo Z. Genome-wide identification and characterization of short-chain dehydrogenase/reductase (SDR) gene family in Medicago truncatula. Int J Mol Sci. 2021;22:9498.
Chen S, Zhou Y, Chen Y, Gu J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. In: Bioinformatics. 2018. p. i884–90.
Edwards KD, Fernandez-Pozo N, Drake-Stowe K, Humphry M, Evans AD, Bombarely A, et al. A reference genome for Nicotiana tabacum enables map-based cloning of homeologous loci implicated in nitrogen utilization efficiency. BMC Genomics. 2017;18:448.
Kim D, Langmead B, Salzberg SL. HISAT: A fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–60.
Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9.
Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33:290–5.
Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550.
Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30.
Yu G, Wang LG, Han Y, He QY. ClusterProfiler: An R package for comparing biological themes among gene clusters. Omi A J Integr Biol. 2012;16:284–7.
Acknowledgements
The authors would like to thank the Nanxiong Scientific Research Institute of Guangdong Research Institute of Tobacco Science, Shaoguan, China, for providing the tobacco accessions.
Funding
This work was funded by the Scientific and Technological Projects of Guangdong Tobacco Corporation (201403, 201702). The funder was not involved in the study design, data collection, analysis, interpretation of data, the writing of this article, or the decision to submit it for publication.
Author information
Authors and Affiliations
Contributions
PG and QY designed the study, HZ, RL, YX, MI, and WZ performed the experiments and analyzed the data, YX, MI, RL, QY, and WZ provided materials and analysis tools, MI, HZ, RL, YX, KHMS, and PG wrote the manuscript, and all authors have read and approved the final version.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
No specific permissions were required for the collection of plant material that was conducted in this study. The collection of leaf samples and use in the study comply with relevant institutional, national, and international guidelines and legislation. Tobacco accessions collected from the United States of America (USA) and China were kindly provided by Dr. Weicai Zhao (Guangdong Research Institute of Tobacco Science, Shaoguan 512029, China).
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Zhang, H., Ikram, M., Li, R. et al. Uncovering the transcriptional responses of tobacco (Nicotiana tabacum L.) roots to Ralstonia solanacearum infection: a comparative study of resistant and susceptible cultivars. BMC Plant Biol 23, 620 (2023). https://doi.org/10.1186/s12870-023-04633-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-023-04633-w