Abstract
Background
Computed tomography (CT) is widely in clinics and is affected by metal implants. Metal segmentation is crucial for metal artifact correction, and the common threshold method often fails to accurately segment metals.
Purpose
This study aims to segment metal implants in CT images using a diffusion model and further validate it with clinical artifact images and phantom images of known size.
Methods
A retrospective study was conducted on 100 patients who received radiation therapy without metal artifacts, and simulated artifact data were generated using publicly available mask data. The study utilized 11,280 slices for training and verification, and 2,820 slices for testing. Metal mask segmentation was performed using DiffSeg, a diffusion model incorporating conditional dynamic coding and a global frequency parser (GFParser). Conditional dynamic coding fuses the current segmentation mask and prior images at multiple scales, while GFParser helps eliminate high-frequency noise in the mask. Clinical artifact images and phantom images are also used for model validation.
Results
Compared with the ground truth, the accuracy of DiffSeg for metal segmentation of simulated data was 97.89% and that of DSC was 95.45%. The mask shape obtained by threshold segmentation covered the ground truth and DSCs were 82.92% and 84.19% for threshold segmentation based on 2500 HU and 3000 HU. Evaluation metrics and visualization results show that DiffSeg performs better than other classical deep learning networks, especially for clinical CT, artifact data, and phantom data.
Conclusion
DiffSeg efficiently and robustly segments metal masks in artifact data with conditional dynamic coding and GFParser. Future work will involve embedding the metal segmentation model in metal artifact reduction to improve the reduction effect.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Introduction
CT images play a vital role in clinical diagnosis and radiation therapy planning, but metal artifacts caused by implants like dental fillings, hip prostheses, and implant markers can limit their usefulness. Metal artifact reduction (MAR) techniques are crucial for improving image quality by mitigating these artifacts. Various MAR methods have been proposed [1,2,3], including iteration-based [4] and projection correction [5, 6] algorithms. Recently, deep learning (DL) methods have emerged as effective tools in MAR applications [7,8,9].
Metal segmentation is fundamental to MAR, as accurate recognition of metal shape and position is essential for correcting raw data efficiently. Precise metal mask segmentation is key to obtaining accurate corrected images, as errors in segmentation can lead to incorrect artifact correction or loss of anatomical information [10]. Geometric information from metal objects is clinically important for accurate dose calculation in radiotherapy planning [11]. In CBCT, precise delineation of implant edges aids clinicians in assessing the relationship between implants and surrounding structures, improving the standardization and clinical interpretation of bone analysis [10, 12].
Metal implants appear very bright in CT images due to their high density. Metal implants create severe artifacts in CT images, such as streaks and shadows, which can obscure their boundaries and complicate accurate segmentation. Manual delineation [13] or threshold-based methods [8] have been used to identify metal objects in medical imaging. However, manual delineation is time-consuming and operator-dependent, making it unsuitable for routine clinical use. In threshold-based methods, a window width and position are selected in the CT image, with the metal object’s boundary estimated using a threshold value, often set at 2500 HU [8, 9, 14] or 3000 HU [7], to differentiate the metal from surrounding bone tissue. However, ensuring accuracy with this method can be difficult, especially near high-density anatomical structures like bone. Additionally, different thresholds may be needed for metals with varying shapes and materials. Instead of a fixed threshold value, Yazdi et al. [15] proposed using 90% of the maximum gray value as the threshold.
Various image morphological processing methods have been proposed for segmenting CT or CBCT metal masks. Pauwels et al. [10] conducted pre-threshold processing on CBCT images using manual thresholds, applied the Sobel operator for edge enhancement, and segmented the filtered images through iteratively determined fixed thresholds, with the algorithm typically taking 10–20 s. Chen et al. [16] utilized a mutual information maximization segmentation algorithm for metal artifact and mask segmentation. Karimi et al. [17] employed regional growth for segmenting metal voxels. Bal et al. [18] utilized the k-means method to segment images into air, soft tissue, normal tissue, bone, and metal categories.
Deep learning has made important advances in domains such as image segmentation [19,20,21,22,23,24,25,26], computer-aided diagnostic [27,28,29], biomedical signal processing [30, 31] and drug discovery [32,33,34] in recent years. In the realm of deep learning applied to CT metal segmentation, Hegazy et al. [35] employed U-Net for segmenting metal regions in the two-dimensional projection domain of dental CT, achieving Dice similarity indices of 0.98, 0.97, 0.93, and 0.95 for the four tested patients. Zhu et al. [36] introduced an attention-based U-Net framework for metal segmentation in the sinogram domain, utilized in one of the MAR steps. The focus of these studies on metal segmentation was primarily for subsequent MAR applications, with limited emphasis on providing quantitative segmentation results.
The diffusion probabilistic model (DPM) [37, 38] has become a popular choice for image segmentation [39] in recent years due to its random sampling process [40,41,42]. When dealing with metal segmentation, the affected area extends beyond the metal itself, posing a challenge. DiffSeg [43], a segmentation network based on the diffusion model, has shown promising results in medical segmentation and is applied to segment metal in artifact CT images. By setting image prior conditions and integrating segmentation masks at each step, DiffSeg dynamically enhances conditional features to learn the segmentation map in a multi-scale approach. The global frequency parser (GFParser) is utilized to filter high-frequency noise in the mask, and multi-scale integration is performed on skip connection paths. Various segmentation models such as U-Net [44], Attention U-Net [45], R2U-Net [46], and DeepLabV3+ [47] were compared with DiffSeg for segmenting metals from both simulated and clinical data.
The main contributions of the proposed fusion method in this paper are as follows.
(1)The study introduces DiffSeg, a novel approach for precise metal segmentation in CT images, addressing the limitations of traditional threshold methods commonly used for this task.
(2) Conditional dynamic coding and GFParser were designed. Conditional dynamic coding fuses the current segmentation mask and prior images at multiple scales, while GFParser helps eliminate high-frequency noise in the mask.
(3) DiffSeg achieves outstanding accuracy (95.81%) and Dice similarity coefficient (85.33%) compared to ground truth. Evaluation across various datasets, including clinical artifact data and phantom images, consistently demonstrates its superiority.
The rest of the paper is organized as follows. Section 2 introduces the dataset, preprocessing, segmentation network, evaluation metrics, and experimental settings. Section 3 displays the qualitative and quantitative results and ablation studies. Sections 4 and 5 elaborate discussion and the conclusion.
Method
Clinical data
A retrospective study was conducted on 100 patients without metal artifacts who underwent radiotherapy at our hospital between January 2021 and December 2023. The study included 20 cases of head and neck, 40 cases of chest, and 40 cases of abdomen. 80% of the data for each type was used for training and validation (11,280 slices), while the remaining 20% was used for testing (2,820 slices). The patient cohort consisted of 58 women and 42 men, with a mean age of 48 ± 11 years. Approval for this retrospective study was obtained from the Medical Ethics Association of Nanjing Medical University ([2020]KY154-01). Patient image data was collected using a Philips Brilliance Big Bore scanner (Philips Medical Systems, Cleveland, OH, USA) with an image matrix size of 512 × 512 and a pixel size of 0.975 mm. The scanning layer thickness was 0.25 cm. To assess metal segmentation, CT images of 5 patients with metal implants such as vertebral steel nails and femoral head implants were included. Furthermore, a case from the CTPelvic1K dataset [48] was randomly selected to evaluate segmentation performance. This dataset primarily consisted of postoperative images with metal artifacts. For quantitative analysis of segmentation performance, CT data from an ArcCheck phantom (Sun Nuclear Corporation, Melbourne, FL) containing two 2 cm titanium rods and a 002H9K phantom (CIRS Inc., Norfolk, VA) containing oval stainless steel rods were utilized. The 002H9K images were stored in 16-bit format, with a metal CT value of 11,080 HU. Data values were normalized to a range of [0,1] based on the minimum and maximum values.
Metal artifact generation
This study was conducted using simulated data sets due to the challenges in obtaining both metal artifact data sets and corresponding artifact-free data sets in clinical practice. Metal implants were inserted into clean CT images to create CT images with metal artifacts, simulating beam hardening and Poisson noise based on the simulation method by Yu et al. [49]. The CT images were generated using a fan-beam geometry with 640 uniformly sampled projection angles between 0 and 360 degrees and 793 detector bins per projection angle. The distance from the X-ray source to the rotation center was set at 107.5 cm. To simulate Poisson noise, a polychromatic X-ray source was utilized with an incident beam X-ray of 2 × 107 photons, considering the partial volume effect and scattering effect. The sinogram size of the artifact CT was 793 × 640.
The mask for metal mask simulation should be carefully designed to fit the clinical scene accurately. In this study, metal masks obtained from Zhang et al. [50] containing 100 manually segmented metal implants, such as dental fillings, spinal fixation screws, hip prostheses, coils, and wires, were utilized. However, applying the mask directly to clinical CT images presents challenges: (1) the fixed position of the mask and (2) the mask extending beyond the body outline. To address this, Matlab 2019a (The Mathworks, Natick, MA, USA) was employed to extract the metal boundary matrix from the mask. The matrix size was reduced to 250 pixels if it exceeded this size. Subsequently, a 512 × 512 matrix was generated with the mask placed at the center (horizontal directions 150 to 350, vertical directions 180 to 330) to create a new mask matrix.
The body mask is segmented using a threshold value to ensure that the metal mask does not appear partially outside the body mask due to random values. By multiplying the body mask with the metal mask, a new mask is obtained that is only within the body, which is then used to generate simulated artifact CT data. In contrast to the method by Wang et al. [8], where a layer of CT was paired with 90 masks, this study utilized a random metal mask for each CT scan. Subsequently, the artifact CT was generated and the CT values were truncated to [-1000, 3071] HU to match actual CT values [51].
Network model
DiffSeg and training
DiffSeg is based on a diffusion model, which includes two phases of forward and reverse diffusion. In the forward process, Gaussian noise is gradually added to the segment label \(\:{x}_{0}\) through a series of steps T. In the reverse process, through the reverse noise process, the neural network is trained to recover the original data, expressed as:
Where \(\:\theta\:\) is the reverse diffusion parameter. Starting from Gaussian noise,\(\:\:{p}_{\theta\:}\left({x}_{T}\right)=\mathcal{N}({x}_{T};0,I)\), where I is the identity matrix, and the reverse process converts the latent variable distribution \(\:{p}_{\theta\:}\left({x}_{T}\right)\) to the data distribution \(\:{p}_{\theta\:}\left({x}_{0}\right)\).
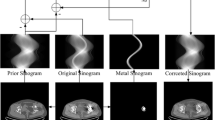
The ResU-Net network is adopted as the DPM learning network, as shown in Fig. 1. To achieve segmentation, the step size estimation function, that is, the noise function, is trained by the original image prior, which can be expressed as:
Where \(\:{E}_{t}^{I}\) is the conditional feature embed, in this case, the original image embed, and \(\:{E}_{t}^{x}\) is the segmentation map feature embedded in the current step. The two components are added and sent to the ResU-Net decoder D for reconstruction. The step-index \(\:t\) is integrated into ResU-Net by embedding and decoder features.
Specifically, the modified ResU-Net consists of a ResNet encoder following a U-Net decoder. The ResNet-34 down-sampling section includes a 7 × 7 convolutional layer with 64 filters, followed by a max pooling layer and repeated residual blocks. Each residual block comprises two 3 × 3 convolutional layers with batch normalization and identity shortcut connections. The decoder blocks use a 2 × 2 transposed convolution with a stride of 2, concatenated with a 1 × 1 convolution of the corresponding encoder feature maps. The concatenated tensor undergoes batch normalization before progressing to the subsequent decoder block. The final layer is a transposed convolution. Besides, the residual block receives time embeddings through a linear layer, SiLU activation, and another linear layer. Both I and \(\:{x}_{i}\) are encoded using distinct encoder. The results are combined by GFParser and dynamic condition coding and forwarded to the final encoding stage.
DiffSeg is trained following DPM’s standard process [37], with the loss expressed as
In each iteration, a pair of original images and segmentation labels are randomly selected for training. The iteration number t samples from a uniform distribution and \(\epsilon\) samples from a Gaussian distribution.
DPM-Solver [52] was utilized as the default sampling method during inference with a sampling step of 100 to speed up sampling.
Dynamic condition coding
Metal segmentation can be challenging due to artifacts, especially when only a static image Iraw is provided at each step in most conditional DPM. To solve this problem, dynamic conditional coding is introduced. Initially, the conditional feature map \(\:{m}_{I}^{k}\) is fused with the \(\:{x}_{t}\) encoding feature \(\:{m}_{x}^{k}\), k is the current layer index. Two feature maps are applied layer normalization and multiplied together to get a feature map. Then the feature map is multiplied with the conditional encoded feature to enhance the attentive region. The fusion mechanism \(\:\mathcal{F}\) can be expressed as:
Where \(\:\otimes\:\) represents element-by-element multiplication and LN represents layer normalization. This strategy facilitates DiffSeg dynamic positioning and calibration segmentation, while integrated embedding generates additional high-frequency noise. To mitigate this, the GFParser is proposed to constrain the high-frequency components of features.
GFParser
To mitigate high-frequency noise and improve segment details, GFParser is integrated into DiffSeg [53]. DiffSeg connects GFParser in the process of integrating features, utilizing a parameterized weight map in the Fourier space features and focusing on controlling noise-related information within the feature. As illustrated in Fig. 2, when given a decoder feature map, the initial step involves conducting a two-dimensional FFT (fast Fourier transform) along the spatial dimension, represented as \(\:M=\mathcal{F}\mathcal{F}\mathcal{T}\left[m\right]\in\:{\mathbb{C}}^{H\times\:W\times\:C}\), Where \(\:\mathcal{F}\mathcal{F}\mathcal{T}[\bullet\:]\) is two-dimensional FFT. Then, the spectrum of M is adjusted by multiplying it with a parameterized attention map A, which can be formulated as: \(\:{M}^{{\prime\:}}=A\otimes\:M\). Finally, the spatial domain is obtained by applying the inverse FFT ad \(\:{m}^{{\prime\:}}=\mathcal{i}\mathcal{F}\mathcal{F}\mathcal{T}\left[{M}^{{\prime\:}}\right]\).
GFParser serves as a trainable frequency filter that enables global modifications to components of a specific frequency, allowing it to learn how to regulate high-frequency components effectively.
Implementation details
DiffSeg employs linear noise time and noise prediction with a diffusion step T of 1000. All experiments were carried out on the PyTorch platform using 2 NVIDIA RTX 3090 GPUs. The network was trained using the AdamW optimizer with an initial learning rate of 1 × 10− 4, for 100 epochs with a batch size of 4.
Additionally, we utilized the same dataset to train and test various deep learning models such as U-Net [44], Attention U-Net [45], R2U-Net [46], and DeepLabV3+ [47] to provide a comparative evaluation of DiffSeg performance.
Verification indicators
The segmentation results were evaluated using four performance indicators: the Dice similarity coefficient (DSC), sensitivity (SE), specificity (SP), and accuracy (ACC). DSC quantifies the overlap between true and predicted values, SE measures the ability to correctly detect true positives, SP gauges the ability to correctly detect true negatives, and ACC assesses the proportion of all correct predictions. The formulas for these indicators are as follows:
Here, TP, FP, TN, and FN represent the number of true positive, false positive, true negative, and false negative pixels respectively.
Results
Segmentation results in simulated CT artifact data
Qualitative and quantitative comparisons were conducted on the segmentation results of the test set to assess the model’s accuracy. The segmentation results for DiffSeg and other models are illustrated in Fig. 3. The findings indicate that both DiffSeg and other models can effectively segment the metal mask in simulated data, closely resembling the ground truth in terms of mask shape and size. Specifically, U-Net and Attention U-Net exhibit less prominent masks compared to the ground truth in Fig. 3(a). In Fig. 3(b), the masks generated by R2U-Net and DeepLabv3 + appear more rounded than the actual mask, with slight alterations and some loss of details. The metal shape produced by DeepLabV3 + is slightly smaller than the ground truth. However, in Fig. 3(c)(d), the discrepancies between the segmentation results of various models and the actual masks are minimal.
Table 1 shows the mean values of DSC, SE, SP, and ACC for different models. DiffSeg reached the highest value among all evaluation indicators, with DSC at 95.45% and ACC at 97.89%. DSC serves as a comprehensive metric for assessing segmentation performance, and a Wilcoxon signed-rank test was conducted to compare DSC between DiffSeg and the other models. Except for DeeplabV3+, all p-values were less than 0.05, indicating significant differences in DSC between DiffSeg and the other models.
Segmentation results in clinical CT artifact data
Clinical CT images with metal artifacts were utilized to validate the efficacy of DiffSeg using real clinical data. Figure 4 illustrates the outcomes of metal segmentation in clinical CT images. Due to the lack of a corresponding ground truth, the display range of [2000,3000] HU was used to display artifact CT images (i.e. adjusted images) to better evaluate segmentation performance. It should be noted that adjusted images are not binary masks and are further checked by a senior physician. Adjusted images have more metal details than the threshold segmentation results. The traditional model for clinical CT image segmentation was found to be suboptimal when compared to simulated CT data. In Fig. 4(a), the Attention U-Net segmentation results appear smaller; in Fig. 4(b), both U-Net and Attention U-Net results lack prominence, while DeepLabV3 + fails to capture a hole in the mask. In Fig. 4(c), U-Net, Attention U-Net, and DeepLabV3 + exhibit missing parts of the screw handle. Figure 4(d) showcases data from the CLINIC metal dataset, where the Attention U-Net segmentation results are incomplete in shape, and the third-party masks from U-Net and DeepLabV3 + are fragmented.
The segmentation outcomes of different methods on phantom data are visually compared in Fig. 5 to facilitate a more direct assessment. In Fig. 5(a), it is evident that the partition boundaries produced by U-Net, Attention U-Net, R2U-Net, and DeepLabV3 + appear somewhat jagged. While DiffSeg can accurately segment local metal boundaries, it exhibits minor imperfections at edges and corners. Notably, DiffSeg successfully segments the ionization chamber between titanium rods, although it only captures a portion of the circular arrangement. Among traditional methods, R2U-Net stands out for effectively segmenting the surrounding ionization chamber. In a similar vein, for phantom image B in Fig. 5(b), the segmentation results from DiffSeg align more closely with the ground truth and demonstrate superior generalization compared to other models.
Comparison of results of DiffSeg and threshold segmentation
For comparison with commonly used threshold segmentation methods, Fig. 6 presents the results of DiffSeg and threshold segmentation in simulated and phantom data. T2500 and T3000 refer to threshold methods based on 2500 HU and 3000 HU, respectively. The results in Fig. 6(a) and (b) show that T2500 and T3000 can outline the metal, although the resulting shape is slightly larger than the ground truth. In contrast, the shape of the DiffSeg segmentation results closely resembles the ground truth. Regarding the titanium rod in Fig. 6(c), the shape obtained by T2500 is somewhat prominent and larger than the ideal result. The left titanium rod segmented by T3000 is nearly square and close to the ground truth, while the upper side of the right titanium rod appears somewhat prominent. Furthermore, the threshold method successfully segments the ionization chamber located between the titanium rods.
Table 2 presents the quantitative outcomes of the threshold segmentation technique applied to simulated data. The SE metric for threshold segmentation is 100%, signifying that the segmentation outcomes completely contain the ground truth, as demonstrated in Fig. 6. Nonetheless, the DSC values for the segmentation outcomes using thresholds T2500 and T3000 were 82.92% and 84.19%, respectively, which are lower than the 95.45% achieved by DiffSeg.
Ablation experiment
Ablation experiments were conducted using simulation data and clinical data to validate the effectiveness of DiffSeg’s dynamic conditional coding and GFParser. The visualization results are presented in Fig. 7, where DiffSeg_1 indicates the lack of dynamic conditional coding and GFParser, and DiffSeg_2 indicates the absence of dynamic conditional coding. In Fig. 7(a), the segmentation outcomes of DiffSeg_1 and DiffSeg_2 closely resemble those of DiffSeg, except that DiffSeg_2 lacks boundary protrusion. Figure 7(b) shows that DiffSeg_1 can segment predominantly one side of the steel nail, while the right side of DiffSeg_2 appears discontinuous, and DiffSeg performs well in segmenting the metal. Lastly, in Fig. 7(c), DiffSeg_1 exhibits some burrs along its boundary, whereas DiffSeg_2 fails to identify the intermediate ionization chamber.
Table 3 presents the quantitative outcomes of the ablation experiment conducted on simulated data. The results indicate that employing dynamic conditional coding proves to be a successful approach for DPM, leading to a 0.79% enhancement in DSC. GFParser, which utilizes dynamic conditional coding, successfully mitigates high-frequency noise, thereby enhancing the segmentation results and contributing to a 1.1% improvement in DSC for DiffSeg.
Visualization results of dynamic conditional encoding and GFParser ablation study in DiffSeg. (a), (b) and (c) are simulation data, clinical data, and phantom data, with a display range of [-500,1500] HU. DiffSeg_1 indicates the absence of dynamic conditional coding and GFParser, and DiffSeg_2 indicates the absence of dynamic conditional coding
Influence of segmentation results on metal artifact correction
The study further examined the impact of segmentation results on normalized metal artifact reduction (NMAR) [3] outcomes for artifact CT and phantom CT, as illustrated in Fig. 8. The first two rows present segmentation and NMAR outcomes for clinical artifact CT, while the last two rows depict results from a titanium rod phantom. NMAR_DiffSeg, NMAR_T2500, and NMAR_T3000 represent the NMAR outcomes corresponding to each segmentation method. In clinical artifact CT, there is a notable disparity between DiffSeg and threshold segmentation methods, with DiffSeg yielding a smaller segmentation mask. Notably, NMAR_DiffSeg retained some bone information, highlighted by a red arrow, which was less discernible in NMAR_T2500 and NMAR_T3000. Additionally, testing on titanium rods CT (excluding the surrounding ionization chambers) revealed that NMAR_DiffSeg exhibited a clearer demarcation line with square bars, indicated by a yellow arrow, attributed to its smaller partition result and reduced impact on the partial reconstruction of normal tissue.
NMAR results from DiffSeg and threshold segmentation, with clinical artifact CT results for the first two rows and phantom CT results for the last two rows. DiffSeg, T2500, and T3000 present DiffSeg results and threshold results based on 2500 HU and 3000 HU. NMAR_DiffSeg, NMAR_T2500, and NMAR_T3000 are the corresponding NMAR results, and the display range is [-500,1500] HU
Discussion
CT images are commonly used in clinical diagnosis due to their ease of acquisition. However, metal artifacts present a challenge to image quality and treatment planning. While existing MAR algorithms [54,55,56] can effectively remove metal artifacts, accurate metal segmentation is crucial. Most current metal segmentation methods rely on simple threshold segmentation in uncorrected CT images or specific image processing techniques, which may result in inaccurate metal segmentation or hinder clinical applications [36]. Therefore, this study introduces DiffSeg, a diffusion model-based segmentation network, for metal segmentation. The standard encoder-decoder architecture of U-Net, as a classical segmentation network, has the advantages of simple structure and good segmentation effect by integrating the characteristics of the encoding stage in the decoding stage. However, the inherent property of convolution is easy to cause the boundary ambiguity of segmentation results. The diffusion model is a data generation technique that simulates diffusion processes in nature to synthesize new data. It starts with a simple, noisy signal, gradually adds details and patterns, and eventually generates complex new data [57,58,59]. DiffSeg incorporates dynamic conditional coding fusion to combine the current segmentation mask and prior images at multiple scales, enhancing feature extraction and image detail recovery. Additionally, GFParser is utilized to reduce high-frequency noise in the mask, further improving segmentation accuracy and achieving precise metal segmentation.
By comparing the qualitative and quantitative results of the traditional deep learning model and DiffSeg for metal segmentation in artifact CT, this study demonstrates the feasibility and effectiveness of the diffusion model for metal segmentation. In simulated data, both traditional methods and DiffSeg effectively segment the entire metal masks, with DiffSeg showing superior identification of fine protruding boundaries. However, when applied to clinical artifact data, the traditional method’s segmentation performance significantly decreases compared to simulated data, resulting in a greater deviation of partial metal boundaries. In contrast, DiffSeg produces results closer to quasi-ground truths, exhibiting a more regular shape. Furthermore, when analyzing titanium rod simulation data, the traditional method’s boundary differs from the actual square shape, while DiffSeg excels in restoring the complete shape of the metal.
This study analyzes the impact of different threshold values in medical imaging. A smaller threshold may lead to normal tissue being mistaken for metal, reducing image detail. Conversely, larger thresholds may misidentify metal implants as tissue, leaving metal artifacts in the final image. Commonly used thresholds like 2500 HU or 3000 HU provide an approximate shape of the metal but may result in artifacts extending beyond the metal itself. Segmentation of titanium rod data demonstrates that the threshold method can accurately identify the ionization chamber metal within the rod, a task that the traditional method struggles with. Additionally, the segmentation results from DiffSeg are smaller than those from threshold methods, potentially better preserving normal tissue information in techniques like NMAR for metal artifact reduction.
Limitations in this study include: (1) This exploratory study focuses on metal segmentation, with plans to incorporate DiffSeg into MAR in the future. (2) The effectiveness of the metal segmentation algorithm is influenced by factors such as type, quantity, and size of the metal. To enhance model robustness, future work will involve gathering a more diverse range of metal shapes and training them in a semi-supervised manner. (3) Each batch’s inference time in the DPM inference stage is approximately 3 s. Future efforts will aim to reduce this time further by leveraging advancements in the diffusion model and parallel computation [60,61,62].
Conclusions
DiffSeg, a diffusion model network utilizing dynamic condition coding and frequency-domain feature parsing, enables precise metal segmentation in CT images. The dynamic condition coding merges the segmentation mask with the image’s prior information effectively, while the global frequency parser aids in high-frequency noise reduction within the mask. Comparative results demonstrate that DiffSeg achieved 95.45% and 97.89% in terms of DSC and accuracy, allowing for finer metal boundary segmentation. DiffSeg demonstrated better robustness relative to other traditional models in metal segments from clinical CT, artifact data, and phantom data.
Data availability
The datasets in the study are available from the corresponding author on reasonable request.
Abbreviations
- MAR:
-
Metal artifact reduction
- DL:
-
Deep learning
- DPM:
-
Diffusion probabilistic model
- GFParser:
-
Global frequency parser
- DSC :
-
Dice similarity coefficient
- SE:
-
Sensitivity
- SP:
-
Specificity
- ALT:
-
ACC accuracy
- T2500:
-
Threshold methods based on 2500 HU
References
Chang Z, Ye DH, Srivastava S, Thibault J-B, Sauer K, Bouman C. Prior-guided metal artifact reduction for iterative X-ray computed tomography[J]. IEEE Trans Med Imaging. 2018;38(6):1532–42.
Mehranian A, Ay MR, Rahmim A, Zaidi H. X-ray CT metal artifact reduction using wavelet domain L0 sparse regularization[J]. IEEE Trans Med Imaging. 2013;32(9):1707–22.
Meyer E, Raupach R, Lell M, Schmidt B, Kachelrieß M. Normalized metal artifact reduction (NMAR) in computed tomography[J]. Med Phys. 2010;37(10):5482–93.
Zhang X, Wang J, Xing L. Metal artifact reduction in x-ray computed tomography (CT) by constrained optimization[J]. Med Phys. 2011;38(2):701–11.
Jeong KY, Ra JB. Metal artifact reduction based on sinogram correction in CT[C]. 2009 IEEE Nuclear Science Symposium Conference Record (NSS/MIC). 2009:3480–3483.
Prell D, Kyriakou Y, Struffert T, Dörfler A, Kalender W. Metal artifact reduction for clipping and coiling in interventional C-arm CT[J]. Am J Neuroradiol. 2010;31(4):634–9.
Lyu Y, Lin W-A, Lu J, Zhou SK. Dudonet++: encoding mask projection to reduce ct metal artifacts[J]. arXiv preprint arXiv:200100340, 2020.
Wang H, Li Y, Zhang H, Meng D, Zheng Y, InDuDoNet+. A deep unfolding dual domain network for metal artifact reduction in CT images[J]. Med Image Anal. 2023;85:102729.
Li Z, Gao Q, Wu Y, Niu C, Zhang J, Wang M, Wang G, Shan H. Quad-Net: quad-domain network for CT metal artifact reduction[J]. IEEE Transactions on Medical Imaging; 2024.
Pauwels R, Jacobs R, Bosmans H, Pittayapat P, Kosalagood P, Silkosessak O, Panmekiate S. Automated implant segmentation in cone-beam CT using edge detection and particle counting[J]. Int J Comput Assist Radiol Surg. 2014;9:733–43.
Wang J, Xing L. A binary image reconstruction technique for accurate determination of the shape and location of metal objects in x-ray computed tomography[J]. J X-Ray Sci Technol. 2010;18(4):403–14.
Lee S, Woo S, Yu J, Seo J, Lee J, Lee C. Automated CNN-based tooth segmentation in cone-beam CT for dental implant planning[J]. IEEE Access. 2020;8:50507–18.
Zhang Y, Zhang L, Zhu XR, Lee AK, Chambers M, Dong L. Reducing metal artifacts in cone-beam CT images by preprocessing projection data[J]. Int J Radiation Oncology* Biology* Phys. 2007;67(3):924–32.
Wang H, Li Y, Meng D, Zheng Y. Adaptive convolutional dictionary network for CT metal artifact reduction[J]. arXiv preprint arXiv:220507471, 2022.
Yazdi M, Lari MA, Bernier G, Beaulieu L. An opposite view data replacement approach for reducing artifacts due to metallic dental objects[J]. Med Phys. 2011;38(4):2275–81.
Chen Y, Li Y, Guo H, Hu Y, Luo L, Yin X, Gu J, Toumoulin C. CT metal artifact reduction method based on improved image segmentation and sinogram in-painting[J]. Mathematical Problems in Engineering, 2012; 2012.
Karimi S, Cosman P, Wald C, Martz H. Segmentation of artifacts and anatomy in CT metal artifact reduction[J]. Med Phys. 2012;39(10):5857–68.
Bal M, Spies L. Metal artifact reduction in CT using tissue-class modeling and adaptive prefiltering[J]. Med Phys. 2006;33(8):2852–9.
Ansari MY, Abdalla A, Ansari MY, Ansari MI, Malluhi B, Mohanty S, Mishra S, Singh SS, Abinahed J, Al-Ansari A. Practical utility of liver segmentation methods in clinical surgeries and interventions[J]. BMC Med Imaging. 2022;22(1):97.
Ansari MY, Mangalote IAC, Masri D, Dakua SP. Neural network-based fast liver ultrasound image segmentation[C]. 2023 international joint conference on neural networks (IJCNN). 2023:1–8.
Ansari MY, Mangalote IAC, Meher PK, Aboumarzouk O, Al-Ansari A, Halabi O, Dakua SP. Advancements in deep learning for B-mode ultrasound segmentation: a comprehensive review[J]. IEEE Trans Emerg Top Comput Intell. 2024:1–24.
Ansari MY, Mohanty S, Mathew SJ, Mishra S, Singh SS, Abinahed J, Al-Ansari A, Dakua SP. Towards developing a lightweight neural network for liver CT segmentation[C]. International Conference on Medical Imaging and Computer-Aided Diagnosis. 2022:27–35.
Han Z, Jian M, Wang G-G, ConvUNeXt. An efficient convolution neural network for medical image segmentation[J]. Knowl Based Syst. 2022;253:109512.
Jafari M, Auer D, Francis S, Garibaldi J, Chen X. DRU-Net: an efficient deep convolutional neural network for medical image segmentation[C]. 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI). 2020:1144–1148.
Xie Y, Zhang J, Shen C, Xia Y. CoTr: efficiently bridging CNN and transformer for 3d medical image segmentation[C]. Medical Image Computing and Computer Assisted Intervention (MICCAI). 2021:171–80.
Bakkouri I, Bakkouri S. 2MGAS-Net: multi-level multi-scale gated attentional squeezed network for polyp segmentation[J]. SIViP, 2024:1–10.
Akhtar Y, Dakua SP, Abdalla A, Aboumarzouk OM, Ansari MY, Abinahed J, Elakkad MSM, Al-Ansari A. Risk assessment of computer-aided diagnostic software for hepatic resection[J]. IEEE Trans Radiation Plasma Med Sci. 2021;6(6):667–77.
Rai P, Ansari MY, Warfa M, Al-Hamar H, Abinahed J, Barah A, Dakua SP, Balakrishnan S. Efficacy of fusion imaging for immediate post‐ablation assessment of malignant liver neoplasms: a systematic review[J]. Cancer Med. 2023;12(13):14225–51.
Bakkouri I, Afdel K. Convolutional neural-adaptive networks for melanoma recognition[C]. Image and Signal Processing. 2018:453–60.
Ansari MY, Qaraqe M, Righetti R, Serpedin E, Qaraqe K. Enhancing ECG-based heart age: impact of acquisition parameters and generalization strategies for varying signal morphologies and corruptions[J]. Front Cardiovasc Med. 2024;11:1424585.
Ansari MY, Qaraqe M, Charafeddine F, Serpedin E, Righetti R, Qaraqe K. Estimating age and gender from electrocardiogram signals: a comprehensive review of the past decade[J]. Artif Intell Med. 2023;146:102690.
Chandrasekar V, Ansari MY, Singh AV, Uddin S, Prabhu KS, Dash S, Al Khodor S, Terranegra A, Avella M, Dakua SP. Investigating the use of machine learning models to understand the drugs permeability across placenta[J]. IEEE Access. 2023;11:52726–39.
Ansari MY, Qaraqe M, Mefood. A large-scale representative benchmark of quotidian foods for the middle east[J]. IEEE Access. 2023;11:4589–601.
Ansari MY, Chandrasekar V, Singh AV, Dakua SP. Re-routing drugs to blood brain barrier: a comprehensive analysis of machine learning approaches with fingerprint amalgamation and data balancing[J]. IEEE Access. 2022;11:9890–906.
Hegazy MA, Cho MH, Cho MH, Lee SY. U-net based metal segmentation on projection domain for metal artifact reduction in dental CT[J]. Biomed Eng Lett. 2019;9:375–85.
Zhu Y, Zhao H, Wang T, Deng L, Yang Y, Jiang Y, Li N, Chan Y, Dai J, Zhang C. Sinogram domain metal artifact correction of CT via deep learning[J]. Comput Biol Med. 2023;155:106710.
Ho J, Jain A, Abbeel P. Denoising diffusion probabilistic models[J]. Adv Neural Inf Process Syst. 2020;33:6840–51.
Rombach R, Blattmann A, Lorenz D, Esser P, Ommer B. High-resolution image synthesis with latent diffusion models[C]. Proceedings of the IEEE/CVF conference on computer vision and pattern recognition. 2022:10684–10695.
Rahman A, Valanarasu JMJ, Hacihaliloglu I, Patel VM. Ambiguous medical image segmentation using diffusion models[C]. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. 2023:11536–11546.
Wolleb J, Sandkühler R, Bieder F, Valmaggia P, Cattin PC. Diffusion models for implicit image segmentation ensembles[C]. International Conference on Medical Imaging with Deep Learning. 2022:1336–1348.
Kim B, Oh Y, Ye JC. Diffusion adversarial representation learning for self-supervised vessel segmentation[J]. arXiv preprint arXiv:220914566, 2022.
Guo X, Yang Y, Ye C, Lu S, Peng B, Huang H, Xiang Y, Ma T. Accelerating diffusion models via pre-segmentation diffusion sampling for medical image segmentation[C]. 2023 IEEE 20th International Symposium on Biomedical Imaging (ISBI). 2023:1–5.
Wu J, Fu R, Fang H, Zhang Y, Yang Y, Xiong H, Liu H, Xu Y. MedSegDiff: medical image segmentation with diffusion probabilistic model[C]. Medical Imaging with Deep Learning. 2024:1623–39.
Ronneberger O, Fischer P, Brox T. U-net: convolutional networks for biomedical image segmentation[C]. Medical Image Computing and Computer-assisted Intervention (MICCAI). 2015:234–41.
Oktay O, Schlemper J, Folgoc LL, Lee M, Heinrich M, Misawa K, Mori K, McDonagh S, Hammerla NY, Kainz B. Attention u-net: learning where to look for the pancreas[J]. arXiv preprint arXiv:180403999, 2018.
Alom MZ, Hasan M, Yakopcic C, Taha TM, Asari VK. Recurrent residual convolutional neural network based on u-net (r2u-net) for medical image segmentation[J]. arXiv preprint arXiv:180206955, 2018.
Chen L-C, Papandreou G, Kokkinos I, Murphy K, Yuille AL, Deeplab. Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs[J]. IEEE Trans Pattern Anal Mach Intell. 2017;40(4):834–48.
Liu P, Han H, Du Y, Zhu H, Li Y, Gu F, Xiao H, Li J, Zhao C, Xiao L. Deep learning to segment pelvic bones: large-scale CT datasets and baseline models[J]. Int J Comput Assist Radiol Surg. 2021;16:749–56.
Yu L, Zhang Z, Li X, Xing L. Deep sinogram completion with image prior for metal artifact reduction in CT images[J]. IEEE Trans Med Imaging. 2020;40(1):228–38.
Zhang Y, Yu H. Convolutional neural network based metal artifact reduction in x-ray computed tomography[J]. IEEE Trans Med Imaging. 2018;37(6):1370–81.
Wang T, Xia W, Huang Y, Sun H, Liu Y, Chen H, Zhou J, Zhang Y. DAN-Net: dual-domain adaptive-scaling non-local network for CT metal artifact reduction[J]. Phys Med Biol. 2021;66(15):155009.
Lu C, Zhou Y, Bao F, Chen J, Li C, Zhu J. Dpm-solver: a fast ODE solver for diffusion probabilistic model sampling in around 10 steps[J]. Adv Neural Inf Process Syst. 2022;35:5775–87.
Cui J, Zeng P, Zeng X, Wang P, Wu X, Zhou J, Wang Y, Shen D. TriDo-Former: a triple-domain transformer for direct PET reconstruction from low-dose sinograms[C]. International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI). 2023:184–94.
Agrawal H, Hietanen A, Särkkä S. Deep learning based projection domain metal segmentation for metal artifact reduction in cone beam computed tomography[J]. IEEE Access. 2023;11:100371–82.
Arabi H, Zaidi H. Deep learning–based metal artefact reduction in PET/CT imaging[J]. Eur Radiol. 2021;31:6384–96.
Wang Z, Vandersteen C, Demarcy T, Gnansia D, Raffaelli C, Guevara N, Delingette H. Deep learning based metal artifacts reduction in post-operative cochlear implant CT imaging[C]. Medical Image Computing and Computer Assisted Intervention (MICCAI). 2019:121–9.
Dakua SP. LV segmentation using stochastic resonance and evolutionary cellular automata[J]. Int J Pattern Recognit Artif Intell. 2015;29(03):1557002.
Dakua SP, Abinahed J, Zakaria A, Balakrishnan S, Younes G, Navkar N, Al-Ansari A, Zhai X, Bensaali F, Amira A. Moving object tracking in clinical scenarios: application to cardiac surgery and cerebral aneurysm clipping[J]. Int J Comput Assist Radiol Surg. 2019;14:2165–76.
Mohanty S, Dakua SP. Toward computing cross-modality symmetric non-rigid medical image registration[J]. IEEE Access. 2022;10:24528–39.
Esfahani SS, Zhai X, Chen M, Amira A, Bensaali F, AbiNahed J, Dakua S, Younes G, Baobeid A, Richardson RA. Lattice-boltzmann interactive blood flow simulation pipeline[J]. Int J Comput Assist Radiol Surg. 2020;15:629–39.
Zhai X, Chen M, Esfahani SS, Amira A, Bensaali F, Abinahed J, Dakua S, Richardson RA, Coveney PV. Heterogeneous system-on-chip-based Lattice-Boltzmann visual simulation system[J]. IEEE Syst J. 2019;14(2):1592–601.
Zhai X, Amira A, Bensaali F, Al-Shibani A, Al‐Nassr A, El‐Sayed A, Eslami M, Dakua SP, Abinahed J. Zynq SoC based acceleration of the lattice boltzmann method[J]. Concurrency Computation: Pract Experience. 2019;31(17):e5184.
Acknowledgements
Not applicable.
Funding
This work is supported by the National Natural Science Foundation of China (No. 62371243), Changzhou Social Development Program (No. CE20235063), General Program of Jiangsu Provincial Health Commission (No. M2020006), and Jiangsu Provincial Key Research and Development Program Social Development Project (No. BE2022720), Jiangsu Provincial Medical Key Discipline Cultivation Unit of Oncology Therapeutics (Radiotherapy) (No. JSDW202237), the National Natural Science Foundation of Jiangsu (No. BK20231190).
Author information
Authors and Affiliations
Contributions
K.X.,L.G.G., Y.T.Z and H.Z. conceived the study. K.X., L.G.G.,J.W.S. and X.Y.N designed the study. L.G.G, T.L. and J.F.S. implemented the methods and performed the data analysis. K.X. and X.Y.N performed the image and statistical analysis. K.X.,L.G.G., Y.T.Z and X.Y.N. drafted the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The experimental protocol was established, according to the ethical guidelines of the Helsinki Declaration and was approved by the ethics committee of the Affiliated Changzhou No.2 People’s Hospital of Nanjing Medical University (approval number: [2020]KY154-01) and waived the requirement for written informed consent from patients.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Xie, K., Gao, L., Zhang, Y. et al. Metal implant segmentation in CT images based on diffusion model. BMC Med Imaging 24, 204 (2024). https://doi.org/10.1186/s12880-024-01379-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12880-024-01379-1