Abstract
Background
Periodontitis is the sixth-most common disease worldwide. The oral microbiome composition and its association with Periodontal disease (PD) have been largely explored; however, limited studies have explored the microbial profiles of both oral and toothbrushes in patients with PD. Thus, this study aimed to ascertain the oral and toothbrushes microbial composition in high-altitude populations, hypothesizing that their correlation with periodontal health would differ from those at lower altitudes, potentially indicating links between environmental factors, microbial colonization patterns, and periodontal health in distinct geographic contexts.
Methods
In the present study, we enrolled 35 individuals including 21 healthy and 14 diagnosed with PD from the Lhasa region of Tibet, China. Saliva and toothbrush samples were collected from each participant to assess the association between toothbrush usage and oral microbiome with PD using 16 S rRNA gene-specific V3-V4 regions sequencing. To assess the oral and toothbrush microbiome composition and diversity and its possible link to PD.
Results
Significantly higher Alpha diversity (Shannon index) was observed between the PD group and PD toothbrushes (p = 0.00021) and between the PD group and Healthy toothbrushes (p = 0.00041). The predominant species were Proteobacteria, Bacteroidota, Firmicutes, Actinobacteria, and Fusobacteria, with genera Pseudomonas, Veillonella, Neisseria, Acinetobacter, and Haemophilus. In addition, PICRUST2 analysis unveiled 44 significant pathways differentiating the disease and healthy groups, along with 29 pathways showing significant differences between their respective toothbrush microbial profiles. The distinct oral and toothbrush microbial composition among high-altitude populations suggests potential adaptations to the challenges of high-altitude environments.
Conclusion
This study emphasizes the importance of tailored dental care strategies, accounting for altitude and racial factors, to effectively manage periodontal health in these communities. Further research is warranted to investigate the specific microbial mechanisms and develop targeted interventions for optimizing oral health in populations across varying altitudes.
Similar content being viewed by others
Introduction
The mouth is the second richest microbiome after the gastrointestinal tract and is an important part of the human digestive system [1]. Periodontal disease [2], dental caries [3], pulp disease [4], periapical disease [5], oral mucosal disease [6], and oral cancer [7] are all related to oral microbiota. The effects of the oral microbiome extend beyond oral health, and these microbes can also cause a variety of systemic diseases, including cardiovascular diseases (CVD) [8], digestive diseases [9], lung diseases [10], autism [11], obesity [12], obstetric complications [13], Alzheimer’s disease [14], rheumatoid arthritis [15], etc. Periodontal disease, characterized by chronic inflammation leading to progressive attachment and alveolar bone loss, is one of the most important oral diseases after caries and one of the major causes of the global burden of chronic disease [16]. Periodontal disease is multifactorial and is the result of unbalanced tissue loss and increase driven by infection, chronic inflammation, and reduced healing [17]. Severe periodontitis is widespread, affecting more than 50% of adults and 11% of adults, and is the sixth most prevalent disease worldwide [18]. Periodontitis can seriously impair an individual’s oral health-related quality of life [19], self-esteem [20], and general well-being [21]. In addition, a large number of studies have linked periodontal disease to a variety of systemic diseases, including cardiovascular diseases, diabetes, osteoporosis, obesity, neurodegenerative diseases, infertility, adverse pregnancy outcomes, etc. [22]. The cost of treating Parkinson’s disease is high [23]. Traditional risk factors such as smoking [24], alcohol consumption [25], poor diet [26], lack of exercise [27], stress [28], distress, and psychological coping resistance [29] have been extensively discussed. However, the specific interactions between the oral and toothbrush microbiota and their role in the occurrence and progression of PD are largely uncharted territory in current research.
The toothbrush acts as a reservoir for a variety of microorganisms, originating from both humans and the surrounding environment [30]. The microbial diversity found on a used toothbrush may be linked to other characteristics of the microbiome within the built environment. Specifically, the diverse array of microorganisms on a toothbrush includes Enterococcus, Porphyromonas, Parvomona, Escherichia, Pseudomonas, Lactobacillus, Staphylococcus, Klebsiella, Clostridium, and Streptococcus, with contributions from both human-related and environmental taxa. Several factors, particularly those influencing the potential source microbiome, shape the composition of this diverse microbiome. Notably, factors such as age, oral hygiene practices, and overall clinical health, including conditions like periodontal disease, dental caries, and even oral cancer, are associated with the human oral microbiome [30]. We currently lack a comprehensive understanding of the key factors that shape the microbiome of toothbrushes. Additionally, the impact of the toothbrush microbiome on the structure and function of the oral microbiome, as well as its potential contribution to the development of periodontal disease (PD), remains unclear.
To date, there have been limited studies examining the diversity of oral and toothbrush microbial communities in high-altitude environments and investigating their potential influence on human periodontal health. This has prompted our interest in understanding variations in the characteristics of oral and toothbrush microbiota among PD patients and healthy individuals from the same ethnic group residing at high altitudes. Consider paying attention to how these modifications will affect the development of PD and the control of dysbiosis in the oral microbiome, both in the present and future scenarios.
Materials and methods
Statement of ethics
This study was approved by Northwest Minzu University’s Research Ethics Committee in Gansu, China (Approval No: XBMU20230074). After being informed of the study’s objectives, each participant gave their informed consent in accordance with the Declaration of Helsinki’s tenets.
Study participants
We recruited 35 people for this study, including 21 healthy individuals and 14 people with PD. Participants were selected based on the following criteria: They were between the ages of 18 and 60 and belonged to the same ethnic group at high altitudes. The inclusion criteria of the participants were the following: age range 18 to 60 belonging to the same racial group in a high-altitude area. Inclusion criteria for the health group: Good oral health confirmed by oral examination, absence of systemic diseases like diabetes or cardiovascular conditions, and no recent major illnesses. Age and sex distribution should match the periodontal disease group for sample representation. Volunteers must fully understand the study, participate voluntarily, and sign informed consent forms. Exclusion criteria for the healthy group: History of periodontitis, gingivitis, or other oral diseases; systemic diseases affecting oral health such as diabetes or rheumatic diseases; long-term or recent use of medications affecting oral health like antibiotics or immunosuppressants; habits like long-term smoking, drinking, or chewing betel nut; and inability to comply with sample collection requests.
Inclusion criteria for patients with periodontal disease: Diagnosed with untreated chronic periodontitis, having at least 16 evaluable teeth including 4 molars, at least 2 sites with probe depth ≥ 5 mm, and loss of attachment (AL) ≥ 2 mm across different quadrants. Patients should not have had dental cleaning within the past year or periodontal treatment within the last 6 months. Even distribution in age and gender is sought for sample representation. Participants must fully understand the study and provide voluntary informed consent. Exclusion criteria for patients with periodontal disease: Presence of other oral diseases such as oral ulcers or oral cancer, systemic diseases like diabetes or cardiovascular conditions, intake of medications affecting study results (e.g., antibiotics, immunosuppressants), and inability to comply with sample collection requirements.
Saliva and toothbrush sample collection
Saliva sample collection: Before collection, the patient should rinse his mouth with water. The patient then breathes with their mouth closed for a period of time (e.g. 30 s) to reduce contamination of the sample by microorganisms and food debris in the mouth. Next, a sterile collection device (such as a straw or bottle) is used to collect the patient’s naturally flowing saliva, about 3 to 5 ml. During the collection process, it is necessary to ensure the sterility of the collection instrument to avoid contamination of the sample. After collection, the saliva sample was placed in a sterile centrifuge tube and stored at -80℃ for subsequent analysis [31].
Toothbrush sample collection: Before taking a toothbrush sample, the patient should use a special toothbrush for daily brushing and use the toothbrush for at least 1 month. After brushing, place the toothbrush in a sterile collection bag. During the collection process, it is necessary to ensure that the toothbrush is dry and clean to avoid contamination by other substances. After collection, the toothbrushes are stored in a slatted sterile sampling bag at -80 °C for subsequent analysis.
In the process of sample collection, the aseptic operation principle should be strictly observed to ensure the accuracy and reliability of samples. At the same time, the patient is fully explained and instructed to ensure that they can properly cooperate with the sample collection.
DNA extraction and amplicon profiling
Following the manufacturer’s instructions, DNA was extracted using Tiangen Biotech (Beijing) Co., Ltd.‘s TGuide S96 Magnetic Soil/Stool DNA Kit. Then, using the Qubit dsDNA HS Assay Kit and the Qubit 4.0 Fluorometer from Invitrogen, Thermo Fisher Scientific, with headquarters in Oregon, USA, the concentration of DNA in the samples was ascertained.
The V3-V4 region of the 16 S rRNA gene was amplified using genomic DNA extracted from each sample. A universal primer set, including the Forward primer ACTCCTACGGGAGGCAGCA and Reverse primer GGACTACHVGGGTWTCTAAT, was employed for this purpose. Both forward and reverse primers were extended with sample-specific Illumina index sequences to facilitate deep sequencing. The PCR reaction occurred in a 10 µl total volume, with components such as 5–50 ng DNA template, 0.3 µl *Vn F (10µM), 0.3 µl *Vn R (10µM), 5 µl KOD FX Neo Buffer, 2 µl dNTP (2 mM each), 0.2 µl KOD FX Neo, and ddH2O up to 10 µl. The selection of Vn F and Vn R was based on the amplification area. The PCR protocol consisted of an initial denaturation at 95 °C for 5 min, followed by 25 cycles of denaturation at 95 °C for 30 s, annealing at 50 °C for 30 s, and extension at 72 °C for 40 s, concluding with a final step at 72 °C for 7 min. Subsequently, PCR amplicons underwent purification using Agencourt AMPure XP Beads (Beckman Coulter, Indianapolis, IN) and quantification using the Qubit dsDNA HS Assay Kit and Qubit 4.0 Fluorometer (Invitrogen, Thermo-Fisher Scientific, Oregon, USA). After individual quantification, the amplicons were amalgamated in equal proportions for library construction, and Illumina Novaseq 6000 was employed for sequencing.
Bioinformatics and statistical interpretation
To do quality filtering on the raw data, Trimmomatic version 0.33 was used (Bolger et al., 2014). Using Cutadapt version 1.9.1, primer sequences were found and eliminated (Martin, 2011), Utilising FLASH version 1.2.11, terminal reads were eliminated (Magoč and Salzberg, 2011), Chimeric sequences were eliminated using UCHIME version 8.1 (Edgar et al., 2011), Following the aforementioned procedures, we eventually had high-quality sequences for additional study. USEARCH (version 10.0) was used to filter out sequences with lengths fewer than 100 base pairs and an error rate greater than 2. The Database Project of Ribosomes was utilized to classify sample sequences (Cole et al., 2009). After data quality was improved, operational taxonomic units (OTUs) were created by combining more than 97% of identical clusters. R Studio version 3.2 was used to compute rarefaction curves at the OTU level and perform VENN analysis (Colwell et al., 2012). Analysis of alpha diversity indices, including the Chao1 index, Shannon-Wiener Diversity Index, Simpson Diversity Index, Phylogenetic Diversity index, Abundance-based Coverage Estimator (ACE), and Coverage, was undertaken using the QIIME2 program (https://qiime2.org/). Using the R language platform (version 3.2.1), Principal Coordinate Analysis (PCoA) based on the Bray-Curtis distance matrix was carried out to measure beta diversity. Using the Bray-Curtis similarity index, Permutational Multivariate Analysis of Variance (PERMANOVA) was used to evaluate community variance. The effect size of linear discriminant analysis (LEfSe) was used to determine the biomarker species that differentiated the two groups (Segata et al., 2011). Enhancing the information link between species is Linear Discriminant Analysis (LDA), which is further supported by significant difference tests such as pairwise Wilcoxon and Kruskal-Walli tests. Moreover, the microbial communities in both saliva and toothbrushes from both groups are predicted by Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt2).
Results
Characteristics of the patients
A total of 35 individuals participated in this study, comprising 21 healthy individuals and 14 patients with PD. Saliva and toothbrush samples were collected from each participant. Table 1 provides a full description of the clinical characteristics.
OTUs distribution
A total of 5,599,311 paired-end sequencing reads were generated from 70 samples. After paired-end read quality control and assembly, 4,935,111 clean reads were obtained. Each sample yielded a minimum of 67,883 and an average of 70,502 clean reads. In total, 19,107 operational taxonomic units (OTUs) were observed. Among them, 3,010 OTUs were shared across all groups, with 2,938 unique to the PD Saliva (PDS) group, 3,458 to the PD toothbrush (PTB) group, 5,561 to the Healthy Saliva (HS) group, and 4,140 to the Healthy Individuals Toothbrush (HTB) group, as depicted in the Venn diagram (Fig. 1).
Alpha diversity
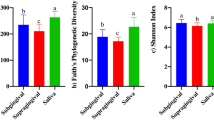
Initially, alpha diversity across all groups was assessed using Chao1, Shannon, and Simpson indices. The alpha diversity of the saliva and toothbrush microbiome did not exhibit significant differences among the four groups, as indicated by the Chao1 index (p = 0.008). Similarly, no significant differences were observed in Simpson indices (Fig. 2A and B). Notably, a significantly higher diversity, as measured by the Shannon index, was observed between the PD Saliva (PDS) and PD Toothbrush (PDT) groups (p = 0.00021) and between PDS and Healthy Individuals Toothbrush (HIT) groups (p = 0.00041) (Fig. 2C, Supplementary Table 1). The rarefaction curves in Fig. 2D demonstrated a plateau in species richness, suggesting that the sample numbers and sequencing depth covered sufficient information for subsequent analyses. A p-value less than 0.005 was considered significant.
Alpha diversity indices among 4 groups. (A). shows the Chao 1 index among all groups. (B). Represent the Simpson index among 4 groups. (C). Display the Shannon index among all groups. (D). The rarefaction curve illustrates the relationship between the number of sequence reads and the corresponding number of operational taxonomic units (OTUs) in both saliva and toothbrush samples in this study
Beta diversity
To investigate structural variations in the bacterial community between the PD Saliva (PDS) and Healthy Saliva (HS) groups, beta diversity was assessed. The overall differences were visualized through a principal coordinate analysis (PCoA) plot, with the top two principal coordinates contributing to the diversity (PC1 = 16.01% and PC2 = 4.94%). The Bray-Curtis distance-based plot (Fig. 3A) depicted a total variation of 20.95%. Furthermore, PERMANOVA results indicated a significant difference in microbial composition between the two groups (R2 = 0.149, p = 0.001), as shown in Fig. 3B. In summary, notable changes were observed in microbial taxa between saliva and toothbrush samples.
(A) shows the overall composition of the genus communities in both groups as determined by the Principal Coordinate Analysis (PCoA). Each dot represents one sample, and the distance between samples indicates the similarity or differences in the saliva and toothbrush microbiomes. The PCoA analysis was conducted using the Bray-Curtis method. (B) presents the PERMANOVA analysis, demonstrating significant differences in bacterial taxa between saliva and toothbrush samples in both groups (p < 0.005)
Comparison of microbial taxa at phylum and genus levels
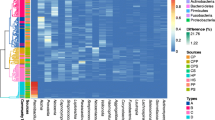
At the phylum and genus levels, the relative abundance of the top 10 operational taxonomic units (OTUs) was categorized. Proteobacteria, Bacteroidota, Firmicutes, Actinobacteria, and Fusobacteria were the main phyla that were found at the phylum level, which dominated in the PD Saliva (PDS), PD Toothbrush (PDT), Healthy Saliva (HS), and Healthy Individuals Toothbrush (HTB) groups. Specifically, Phylum Proteobacteria (70%, 28%, 27%, and 57%) showed enrichment in the PDS group compared to HS, PTB, and HTB, respectively. Other phyla, including Bacteroidota (9%, 3%, 9%, and 2%) and Firmicutes (7%, 2%, 1%, and 2%), exhibited slight variations among all groups (Fig. 4A).
Figure 4B illustrates the overall composition of saliva and toothbrush microbial communities in both groups at the genus level. Of them, Pseudomonas followed by Veillonella, Neisseria, Acinetobacter, Haemophilus, Streptococcus, Prevotella_7, Prevotella, Alloprevotella, and Brevundimonas were the dominant genera in PDS, PDT, HS, and HTB groups. Of them, the genus Pseudomonas (21%, 3%, 9%, and 1%), Veillonella (4%, 9%, 3%, and 14%) followed by Neisseria, Acinetobacter, Haemophilus, Streptococcus, Prevotella_7, Prevotella, Alloprevotella, and Brevundimonas were slightly different among PDS, PDT, HIS, and HIT groups. No significant difference was observed in saliva and toothbrush microbial taxa between PDS and Healthy groups at the genus and phylum level (p < 0.05).
Distinct bacterial taxa between both groups
Using a logarithmic LDA score cutoff of 10, LEfSe modelling was used to validate the taxonomic and statistical differences between the saliva and toothbrush microbiomes in people with periodontal disease and those who are healthy. Upon examining each participant’s taxonomic profile, LEfSe determined significant differences between the four groups at the phylum, class, order, family, genus, and genus levels, with a threshold score of LDA > 10. Using linear discriminant analysis, it was discovered that 26 taxa exhibited significant differences across all groupings. Notably, Class_Gammaproteobacteria: Orders_Pseudomonadales, and Corynebacterales: Families_Moraxellaceae, Enterobacteriaceae, and Alcaligenaceae: Genus_Emticicia, and Achromobacter in the PDS group; Families_Pasteurellaceae, Neisseriaceae, and Fusobacteriaceae: Genus_Neisseria, Alloprevotella, and Fusobacterium in PDT; Orders_Sphingomonadales: Families_Comamonadaceae, and Sphingomonadaceae: Genus_Comamonas in the HS group; and Genus_Haemophilus in HTB were found to be significantly distinct among all groups (Fig. 5A). Furthermore, Fig. 5B uses a cladogram to show how the microbiomes of the four groups differ at different evolutionary levels.
Functional analysis (KEGG annotated pathway)
Predictions of bacterial gene functions were made using the PICRUSt2 algorithm, which utilizes 16 S rRNA gene-based microbial compositions to infer information from KEGG annotated databases. Significant differences among the 44 KEGG pathways were observed between the PD Saliva (PDS) and control groups (Fig. 6A). In the PDS group, variations were noted in pathways related to aging, cancer-specific types, metabolism of other amino acids, signal transduction, neurodegenerative diseases, metabolism of terpenoids and polyketides, drug resistance (antineoplastic), amino acid metabolism, transport and catabolism, circulatory system, immune system, substance dependence, infectious diseases (viral), cardiovascular disease, cancer (overview), cell motility, infectious diseases (parasitic), nervous system, development, xenobiotics biodegradation and metabolism, and infectious diseases (bacterial). On the other hand, pathways associated with immune diseases, endocrine and metabolic diseases, signaling molecules and interaction, glycan biosynthesis and metabolism, excretory system, translation, folding, sorting and degradation, global and overview maps, sensory system, cell growth and death, metabolism of cofactors and vitamins, transcription, replication and repair, drug resistance (antimicrobial), biosynthesis of other secondary metabolites, cellular community-prokaryotes, nucleotide metabolism, and energy metabolism were down-regulated in the Healthy Saliva (HS) group compared to the PDS group.
Similarly, comparing the PTB and HTB exhibited significant differences among the 29 KEGG pathways (Fig. 6B). The functional pathways of the patient’s toothbrush showed differences in aging, cell growth, and death, immune diseases, nervous system, signaling molecules, and interaction, cancer: specific types, neurodegenerative diseases, carbohydrates metabolism, cardiovascular diseases, infectious disease: viral, drug resistance: antineoplastic, metabolism of other amino acids, drug resistance antimicrobial, biosynthesis of other secondary metabolites, signal transduction, glycan biosynthesis and metabolism, infectious disease bacterial, energy metabolism and nucleotide metabolism were down-regulated while folding, sorting and degradation, metabolism of cofactors and vitamins, cell motility, amino acids and metabolism, translation, environmental adaptation, member transport, replication and repair, cellular community-prokaryotes and circulatory system were up-regulated in HTB group. These findings suggest a significant dysregulation of functional pathways in patients, primarily exhibiting downregulation, contrasted with a notable down-regulation of functional pathways in their toothbrushes. These results underscore the potential interplay between host health and the microbial environment, highlighting the need for further investigation into the implications of these dysregulated pathways on overall oral health and disease progression.
Functional pathways between the disease and healthy groups are depicted as follows: (A) Illustrates the significant differences in saliva functional pathways between the healthy group and patients. (B) Demonstrates significant changes in toothbrush functional pathways between patients’ toothbrush samples and those of the healthy group
Functional pathways between the disease and healthy groups are depicted as follows: (A) Illustrates the significant differences in saliva functional pathways between the healthy group and patients. (B) Demonstrates significant changes in toothbrush functional pathways between patients’ toothbrush samples and those of the healthy group
RDA analysis
We assessed the influence of age, weight, hygiene, ventilation, and usage time on the composition of oral microbiota and periodontal disease using RDA analysis. Our findings showed that hygiene, Vent, and age have a great impact on periodontal disease see Fig. 7A. We further examined the correlation between clinical characteristics and microbiological taxa. Where three genera namely, Veillonella, Streptococcus, and Actinomyces showed a positive correlation with hygiene while only Streptococcus showed a positive correlation with Vent, depicted in Fig. 7B. Therefore, this implies that hygiene, Vent, and oral microbiota may significantly influence oral health and promote the development and progression of periodontal disease.
Discussion
In this study, the salivary and toothbrush microbiome of the healthy (21 subjects), and periodontal (14 subjects) groups were compared to PDS, PDT, HS, and HTB groups to ascertain the oral and toothbrush microbiome composition and their association with PD. A previous study reported significant alterations in oral microbiome profiles in patients with PDs [32]. However, the link between oral and toothbrush microbiome signatures with PD is an emerging research area. To understand the association between oral and toothbrush microbiome with PD, we compared the oral and toothbrush microbial signatures of healthy Tibetans who lived in the same geographical environment for a long time, had the same lifestyle and similar dietary habits, and Tibetans with PDs to minimize the influence of genetic background. We aimed to explore the oral and toothbrush microbiome composition and its potential link to PD.
In this study, we employed high-throughput sequencing to investigate the microbial diversity in both the salivary and toothbrush microbiomes. Notably, a reduced alpha diversity, as measured by the Shannon index, was observed in both the oral and toothbrush microbiomes when comparing PDS vs. PDT (p = 0.0002) and PDS vs. HTB (p = 0.0004). However, no significant differences were identified in the oral microbiome signatures between individuals with periodontal disease and those who were healthy. These findings are consistent with a study conducted by Blaustein, which also compared toothbrushes and human oral microbiomes. Furthermore, a different study found that toothbrush alpha diversity was greater than skin and vaginal microbiota diversity but lower than oral and gut microbiota diversity [30]. However, our results are different from these findings, therefore our findings suggest the potential adaptation to the challenges of high-altitude environments. Therefore, the study highlights the importance of developing tailored dental care strategies based on altitude and ethnic factors to effectively manage periodontal health in these communities. Additionally, beta diversity between both groups was found significantly altered. Evidence recently accumulated from the Tibetan Plateau suggests that the diversity of oral microbiota living at different elevations changes and the associated ecological mechanisms respond differently compared to lower elevations [33]. Therefore, we hypothesized that high altitude could play a crucial role in the causation of PDs. Further study is needed to specific microbial mechanisms and develop targeted interventions to optimize the oral health of populations at different altitudes.
At the phylum level, the Proteobacteria level increased in the PDS group. Proteobacteria includes a variety of human pathogens, including Brucella, Neisseria, Rickettsia, and Helicobacter pylori, which are known to be closely associated with diseases such as lung disease, inflammatory bowel disease, and metabolic disorders [34]. Notably, the toothbrush microbiome includes key members shared with the oral microbiome, establishing a link between the two. The microbial community on your toothbrush may contribute to a variety of diseases. However, there has been limited research into the microbial diversity of toothbrushes and the potential effects of these bacteria on human health. Our findings highlight the prevalence of Haemophilus, Neisseria, and Streptococcus in all groups. Because the microbial species commonly found in the oral microbiome are quite common, the toothbrush acts as an interface between the human host and the surrounding environmental microbiome [35]. A previous study had similar findings, suggesting that conserved oral-related taxa, including Haemophilus, Neisseria, and Streptococcus, play an important role in biofilm adhesion and supringival plaque formation [36]. Interestingly, our study identified two distinct genera in the PDS group, Emticicia, and Achromobacter, while Neisseria, Alloprevotella, and Fusobacterium dominated on their toothbrushes. They found that broad differences in microbial trends in toothbrushes are similar to differences in dust or on surfaces in the built environment, which act as additional “sinks” for microbes of human origin [37, 38]. Some of the less niche-specific taxa found on toothbrushes, including Enterobacteria, Kocuria, Pseudomonas, and Stenotrophomonas, are commonly found in indoor environments [39, 40]. As an early colonizer and species bridge, Veillonella played an important role in organizing the creation of complex multi-species communities in the human oral environment. Its ability to form strong bonds with other components of the oral microbiome is a key component in maintaining a balanced and healthy oral environment. Leptococcus may function as an “auxiliary pathogen” that promotes the development of other pathogenic species in oral biofilms and highlights the complex link between these bacteria and biofilm-induced periodontitis [41]. Neisseria and Leptococcus are well-known indicators of dental health. Specifically, Leptococcus can absorb lactic acid produced by Streptococcus mutans, helping to prevent tooth decay [42]. Neisseria is a biomarker of a good periodontal environment because its number in the periodontal pocket is reduced from hc and the surface layer to the deep layer [43]. In clinical studies of periodontal therapy, it has been found that the relationship between leptococcus and treatment results is stronger [44].
Remarkably, we also discovered that toothbrushes had less Prevotella and Streptococcus than the oral cavity did. According to a recent study, Prevotella and Streptococcus are the main microorganisms that cause oral illnesses [45], Because of the oral cavity’s high fluidity, toothbrushes have more antimicrobial chemicals than the oral cavity, which makes the remaining components in toothbrushes more effective at preventing bacteria development. Significant improvements were also found in pathways like the Two-component system, Bacterial secretion system, Glyoxylate and dicarboxylate metabolism, Microbial metabolism in diverse environments, Glycine, serine, and threonine metabolism, and Oxidative phosphorylation, according to our functional prediction analysis. However, the PD group showed evidence of various metabolic pathways, including pyruvate metabolism, ribosome, purine, and cysteine metabolism, pyrimidine metabolism, amino acid biosynthesis, antibiotic biosynthesis, biosynthesis of secondary metabolites, glycolysis/gluconeogenesis, carbon metabolism, purine and cysteine metabolism, quorum sensing, ABC transporters, and purine metabolism (Fig. 4B). However, the shotgun metagenomic approach must be used to corroborate these findings further because high throughput sequencing only yields a limited amount of data. To summarize, the microbial communities found in toothbrushes display a range of characteristics, from strong subsets of the oral microbiota to mixed assemblages that contain more common strains that are particular to a given habitat or situation.
To characterize the saliva and toothbrush microbiome in the healthy and periodontal groups, our pilot analysis was limited by a small sample size (n = 35) and a lack of paired donor samples. Weak but potentially important trends identified in this pilot study, such as the relationship between oral hygiene habits and the microbiome, tooth loss, adenoids and tonsillectomy, the presence of bathroom Windows, and the location of toothbrush storage, may benefit from a larger cohort. Ultimately, although environmental factors have a significant influence on indoor microbial communities, correlations between these characteristics and community measures are often weak or nonexistent [46]. In addition, the comparison between the toothbrush microbiome and its human-related and built environment counterparts was “high” due to site-specific microbial strains and potentially confusable variables across the dataset (e.g., differences in treatment of actual washbasin water versus tap water in the reference dataset for subjects’ households, batch effects in sequencing preparation). For example, some so-called oral microbiota may be exaggerated and come from other parts of the human body. Source Tracker’s human score predictions are based on the most prevalent members of the toothbrush microbiome in the oral metagenome and are thought to be generated by the human microbiome [47]. The gut, vaginal, and skin microbiota all include many of the same microbial species (e.g., Leptococcus and Streptococcus). In the future, longitudinal studies of the microbiome of toothbrushes and donors may be needed to draw more definitive conclusions about community clustering of oral and toothbrush microbiome characteristics. This tracking may involve a comparison of sequence variations at the genome level. However, given that differences between the toothbrush microbiota and the human microbiota may indicate local selection, these microbiotas appear to be important reservoirs of periodontal disease. Whether this tap water in turn leads to the spread of harmful bacteria in the human oral niche is unknown. It is recommended to brush your teeth at least three to four times a day, for three to five minutes after each meal [48]. Frequent brushing is beneficial to oral health, and oral microorganisms and toothbrush microorganisms can better maintain ecological balance, which may have an impact on oral diseases and other systemic diseases [49]. Therefore, brushing your teeth at night is very important to reduce the risk of cardiovascular disease and related diseases. There is a need to raise public awareness of proper brushing times [50]. It is vital to encourage individuals to consistently change their toothbrushes regularly at intervals of no more than three to four months.
Limitations
This study was limited by sample size and may not adequately represent the overall microbial characteristics of populations at high altitudes. In addition, in addition to the single factor of altitude, other environmental, dietary, genetic, or behavioral variables should be considered that may contribute to the observed microbial diversity. Follow-up studies will expand the sample size and fully consider the influencing factors.
Conclusions
In conclusion, this study compared the microbial diversity found in toothbrush and oral samples from those in good health and those who have been diagnosed with periodontal disease (PD). The study found that toothbrushes included many bacteria, including several diseases. The results point to the possibility that these bacteria enter the oral cavity through toothbrushes and spread, raising the risk of cardiovascular disease, cancer, and periodontal illnesses. The study reveals unique microbial compositions in toothbrush and oral samples from communities living at high altitudes, suggesting possible adaptations to difficulties encountered in these settings. As such, the study highlights how crucial it is to customize dental care plans according to ethnicity and altitude to effectively maintain periodontal health in these communities. It also emphasizes the necessity of greater investigation into certain microbial pathways and the creation of focused therapies to maximize oral health outcomes for populations living in various altitudes.
Data availability
Availability of data and materialsThe datasets utilized in this study are available online at http://www.ncbi.nlm.nih.gov/bioproject/1134913, under the accession number PRJNA1134913.
References
Verma D, Garg PK, Dubey AK. Insights into the human oral microbiome. Arch Microbiol. 2018;200:525–40. https://doi.org/10.1007/s00203-018-1505-3.
Curtis MA, Diaz PI, Van Dyke TE. (2020) The role of the microbiota in PD. Periodontol 2000 83: 14–25. https://doi.org/10.1111/prd.12296.
Lamont RJ, Koo H, Hajishengallis G. The oral microbiota: dynamic communities and host interactions. Nat Rev Microbiol. 2018;16:745–59. https://doi.org/10.1038/s41579-018-0089-x.
Shetty P, Shetty S, Rai P, Kumar BK, Bhat R. Role of oral microbiota in irreversible pulpitis - current strategies and future perspectives. Acta Microbiol Immunol Hung. 2023;70:177–86. https://doi.org/10.1556/030.2023.02082.
Buonavoglia A, Zamparini F, Lanave G, Pellegrini F, Diakoudi G, Spinelli A, Lucente MS, Camero M, Vasinioti VI, Gandolfi MG, Martella V, Prati C. Endodontic Microbial communities in Apical Periodontitis. J Endod. 2023;49:178–89. https://doi.org/10.1016/j.joen.2022.11.015.
Lin D, Yang L, Wen L, Lu H, Chen Q, Wang Z. Crosstalk between the oral microbiota, mucosal immunity, and the epithelial barrier regulates oral mucosal disease pathogenesis. Mucosal Immunol. 2021;14:1247–58. https://doi.org/10.1038/s41385-021-00413-7.
Tuominen H, Rautava J. Oral microbiota and Cancer Development. Pathobiology. 2021;88:116–26. https://doi.org/10.1159/000510979.
Hou K, Wu ZX, Chen XY, Wang JQ, Zhang D, Xiao C, Zhu D, Koya JB, Wei L, Li J, Chen ZS. Microbiota in health and diseases. Signal Transduct Target Ther. 2022;7:135. https://doi.org/10.1038/s41392-022-00974-4.
Zhou J, Li M, Chen Q, Li X, Chen L, Dong Z, Zhu W, Yang Y, Liu Z, Chen Q. Programmable probiotics modulate inflammation and gut microbiota for inflammatory bowel disease treatment after effective oral delivery. Nat Commun. 2022;13:3432. https://doi.org/10.1038/s41467-022-31171-0.
Vogtmann E, Hua X, Yu G, Purandare V, Hullings AG, Shao D, Wan Y, Li S, Dagnall CL, Jones K, Hicks BD, Hutchinson A, Caporaso JG, Wheeler W, Sandler DP, Beane Freeman LE, Liao LM, Huang WY, Freedman ND, Caporaso NE, Sinha R, Gail MH, Shi J, Abnet CC. The oral Microbiome and Lung Cancer Risk: an analysis of 3 prospective cohort studies. J Natl Cancer Inst. 2022;114:1501–10. https://doi.org/10.1093/ji/djac149.
Olsen I, Hicks SD. Oral microbiota and autism spectrum disorder (ASD). J Oral Microbiol. 2020;12:1702806. https://doi.org/10.1080/20002297.2019.1702806.
Wang W, Yan Y, Yu F, Zhang W, Su S. Role of oral and gut microbiota in childhood obesity. Folia Microbiol (Praha). 2023;68:197–206. https://doi.org/10.1007/s12223-023-01033-3.
Giannella L, Grelloni C, Quintili D, Fiorelli A, Montironi R, Alia S, Delli Carpini G, Di Giuseppe J, Vignini A, Ciavattini A. Microbiome changes in pregnancy disorders. Antioxid (Basel). 2023;12. https://doi.org/10.3390/antiox12020463.
Jungbauer G, Stähli A, Zhu X, Auber Alberi L, Sculean A, Eick S. Periodontal microorganisms and Alzheimer disease - A causative relationship? Periodontol 2000. 2022;89:59–82. https://doi.org/10.1111/prd.12429.
Chu XJ, Cao NW, Zhou HY, Meng X, Guo B, Zhang HY, Li BZ. The oral and gut microbiome in rheumatoid arthritis patients: a systematic review. Rheumatology (Oxford). 2021;60:1054–66. https://doi.org/10.1093/rheumatology/keaa835.
Petersen PE, Ogawa H. The global burden of PD: towards integration with chronic disease prevention and control. Periodontol 2000. 2012;60:15–39. https://doi.org/10.1111/j.1600-0757.2011.00425.x.
Könönen E, Gursoy M, Gursoy UK. Periodontitis: a multifaceted disease of tooth-supporting tissues. J Clin Med. 2019;8. https://doi.org/10.3390/jcm8081135.
Tonetti MS, Chapple IL, Jepsen S, Sanz M. Primary and secondary prevention of periodontal and peri-implant diseases: introduction to, and objectives of the 11th European workshop on periodontology consensus conference. J Clin Periodontol. 2015;42(Suppl 16):S1–4. https://doi.org/10.1111/jcpe.12382.
Al-Harthi LS, Cullinan MP, Leichter JW, Thomson WM. The impact of periodontitis on oral health-related quality of life: a review of the evidence from observational studies. Aust Dent J. 2013;58:274–7. https://doi.org/10.1111/adj.12076. quiz 384.
Peres MA, Macpherson LMD, Weyant RJ, Daly B, Venturelli R, Mathur MR, Listl S, Celeste RK, Guarnizo-Herreño CC, Kearns C, Benzian H, Allison P, Watt RG. Oral diseases: a global public health challenge. Lancet. 2019;394:249–60. https://doi.org/10.1016/s0140-6736(19)31146-8.
Gil-Montoya JA, de Mello AL, Barrios R, Gonzalez-Moles MA, Bravo M. Oral health in the elderly patient and its impact on general well-being: a nonsystematic review. Clin Interv Aging. 2015;10:461–7. https://doi.org/10.2147/cia.S54630.
Isola G, Santonocito S, Lupi SM, Polizzi A, Sclafani R, Patini R, Marchetti E. (2023) Periodontal Health and Disease in the Context of Systemic Diseases. Mediators Inflamm 2023: 9720947. https://doi.org/10.1155/2023/9720947.
Mohd-Dom T, Ayob R, Mohd-Nur A, Abdul-Manaf MR, Ishak N, Abdul-Muttalib K, Aljunid SM, Ahmad-Yaziz Y, Abdul-Aziz H, Kasan N, Mohd-Asari AS. Cost analysis of periodontitis management in public sector specialist dental clinics. BMC Oral Health. 2014;14:56. https://doi.org/10.1186/1472-6831-14-56.
Leite FRM, Nascimento GG, Scheutz F, López R. Effect of smoking on Periodontitis: a systematic review and Meta-regression. Am J Prev Med. 2018;54:831–41. https://doi.org/10.1016/j.amepre.2018.02.014.
Pulikkotil SJ, Nath S, Dharamarajan L, Jing KT, Vaithilingam RD. Alcohol consumption is associated with periodontitis. A systematic review and meta-analysis of observational studies. Community Dent Health. 2020;37:12–21. https://doi.org/10.1922/CDH_4569Pulikkotil10.
O’Connor JP, Milledge KL, O’Leary F, Cumming R, Eberhard J, Hirani V. Poor dietary intake of nutrients and food groups are associated with increased risk of PD among community-dwelling older adults: a systematic literature review. Nutr Rev. 2020;78:175–88. https://doi.org/10.1093/nutrit/nuz035.
Ferreira RO, Corrêa MG, Magno MB, Almeida A, Fagundes NCF, Rosing CK, Maia LC, Lima RR. Physical activity reduces the prevalence of PD: systematic review and Meta-analysis. Front Physiol. 2019;10:234. https://doi.org/10.3389/fphys.2019.00234.
Zhang J, Lin S, Luo L, Zhang Q, Jiao Y, Liu W. Psychological stress: neuroimmune roles in PD. Odontology. 2023;111:554–64. https://doi.org/10.1007/s10266-022-00768-8.
Genco RJ, Ho AW, Grossi SG, Dunford RG, Tedesco LA. Relationship of stress, distress and inadequate coping behaviors to PD. J Periodontol. 1999;70:711–23. https://doi.org/10.1902/jop.1999.70.7.711.
Blaustein RA, Michelitsch LM, Glawe AJ, Lee H, Huttelmaier S, Hellgeth N, Ben Maamar S, Hartmann EM. Toothbrush microbiomes feature a meeting ground for human oral and environmental microbiota. Microbiome. 2021;9:32. https://doi.org/10.1186/s40168-020-00983-x.
Jiang H, Zhang Y, Xiong X, Harville EW, Qian OK. Salivary and serum inflammatory mediators among pre-conception women with periodontal disease. BMC Oral Health. 2016;16:1–7.
Lourenço TGB, de Oliveira AM, Tsute Chen G, Colombo APV. Oral-gut bacterial profiles discriminate between periodontal health and diseases. J Periodontal Res. 2022;57:1227–37. https://doi.org/10.1111/jre.13059.
Kumari M, Bhushan B, Eslavath MR, Srivastava AK, Meena RC, Varshney R, Ganju L. Impact of high altitude on composition and functional profiling of oral microbiome in Indian male population. Sci Rep. 2023;13:4038. https://doi.org/10.1038/s41598-023-30963-8.
Rizzatti G, Lopetuso LR, Gibiino G, Binda C, Gasbarrini A. Proteobacteria: a common factor in Human diseases. Biomed Res Int. 2017;2017:9351507. https://doi.org/10.1155/2017/9351507.
Lloyd-Price J, Mahurkar A, Rahnavard G, Crabtree J, Orvis J, Hall AB, Brady A, Creasy HH, McCracken C, Giglio MG, McDonald D, Franzosa EA, Knight R, White O, Huttenhower C. Strains, functions and dynamics in the expanded human Microbiome Project. Nature. 2017;550:61–6. https://doi.org/10.1038/nature23889.
Mark Welch JL, Rossetti BJ, Rieken CW, Dewhirst FE, Borisy GG. Biogeography of a human oral microbiome at the micron scale. Proc Natl Acad Sci U S A. 2016;113:E791–800. https://doi.org/10.1073/pnas.1522149113.
Lang JM, Coil DA, Neches RY, Brown WE, Cavalier D, Severance M, Hampton-Marcell JT, Gilbert JA, Eisen JA. A microbial survey of the International Space Station (ISS). PeerJ. 2017;5:e4029. https://doi.org/10.7717/peerj.4029.
Hartmann EM, Hickey R, Hsu T, Betancourt Román CM, Chen J, Schwager R, Kline J, Brown GZ, Halden RU, Huttenhower C, Green JL. Antimicrobial Chemicals Are Associated with elevated antibiotic resistance genes in the indoor dust Microbiome. Environ Sci Technol. 2016;50:9807–15. https://doi.org/10.1021/acs.est.6b00262.
Lax S, Sangwan N, Smith D, Larsen P, Handley KM, Richardson M, Guyton K, Krezalek M, Shogan BD, Defazio J, Flemming I, Shakhsheer B, Weber S, Landon E, Garcia-Houchins S, Siegel J, Alverdy J, Knight R, Stephens B, Gilbert JA. Bacterial colonization and succession in a newly opened hospital. Sci Transl Med. 2017;9. https://doi.org/10.1126/scitranslmed.aah6500.
Coil DA, Neches RY, Lang JM, Brown WE, Severance M, Cavalier D, Eisen JA. Growth of 48 built environment bacterial isolates on board the International Space Station (ISS). PeerJ. 2016;4:e1842. https://doi.org/10.7717/peerj.1842.
Zhou P, Manoil D, Belibasakis GN, Kotsakis GA. Veillonellae: beyond bridging species in oral Biofilm Ecology. Front Oral Health. 2021;2:774115. https://doi.org/10.3389/froh.2021.774115.
Sanz M, Beighton D, Curtis MA, Cury JA, Dige I, Dommisch H, Ellwood R, Giacaman RA, Herrera D, Herzberg MC, Könönen E, Marsh PD, Meyle J, Mira A, Molina A, Mombelli A, Quirynen M, Reynolds EC, Shapira L, Zaura E. Role of microbial biofilms in the maintenance of oral health and the development of dental caries and PDs. Consensus report of group 1 of the Joint EFP/ORCA workshop on the boundaries between caries and PD. J Clin Periodontol. 2017;44(Suppl 18):S5–11. https://doi.org/10.1111/jcpe.12682.
Cai Z, Lin S, Hu S, Zhao L. Structure and function of oral Microbial Community in Periodontitis based on Integrated Data. Front Cell Infect Microbiol. 2021;11:663756. https://doi.org/10.3389/fcimb.2021.663756.
Colombo AP, Bennet S, Cotton SL, Goodson JM, Kent R, Haffajee AD, Socransky SS, Hasturk H, Van Dyke TE, Dewhirst FE, Paster BJ. Impact of periodontal therapy on the subgingival microbiota of severe periodontitis: comparison between good responders and individuals with refractory periodontitis using the human oral microbe identification microarray. J Periodontol. 2012;83:1279–87. https://doi.org/10.1902/jop.2012.110566.
Shang Q, Gao Y, Qin T, Wang S, Shi Y, Chen T. Interaction of oral and Toothbrush Microbiota affects oral Cavity Health. Front Cell Infect Microbiol. 2020;10:17. https://doi.org/10.3389/fcimb.2020.00017.
Stephens B. What have we learned about the microbiomes of indoor environments? mSystems. 2016;1. https://doi.org/10.1128/mSystems.00083-16.
Knights D, Kuczynski J, Charlson ES, Zaneveld J, Mozer MC, Collman RG, Bushman FD, Knight R, Kelley ST. Bayesian community-wide culture-independent microbial source tracking. Nat Methods. 2011;8:761–3. https://doi.org/10.1038/nmeth.1650.
Akhter Y, Rastogi S, Kaithwas G. Frequent brushing of teeth inhibits the dissemination of the SARS-CoV-2: the biochemical mechanism. Environ Sustain (Singap. 2023;1–4. https://doi.org/10.1007/s42398-023-00279-4.
Hirano K, Shimbo T, Komatsu Y, Kobayashi D. Frequency of tooth brushing as a predictive factor for future kidney function decline. J Nephrol. 2022;35:191–9. https://doi.org/10.1007/s40620-021-00987-2.
Isomura ET, Suna S, Kurakami H, Hikoso S, Uchihashi T, Yokota Y, Sakata Y, Tanaka S. Not brushing teeth at night may increase the risk of cardiovascular disease. Sci Rep. 2023;13:10467. https://doi.org/10.1038/s41598-023-37738-1.
Acknowledgements
We are thankful to all participants for their cooperation.
Funding
The study was funded by the Natural Science Foundation of Gansu Province (Grant No. 22JR5RA190), the Fundamental Research Funds for the Central Universities (Grant No.31920240113), Gansu Provincial Key Laboratory of Oral Diseases (Grant No.SZD202101), Key R&D Project of Gansu Province (Grant No.23YFFA0072). There is no commercial relationship between the foundations and each author. The funding bodies did not participate in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Author information
Authors and Affiliations
Contributions
L.Z., S.L., and J.Z. participated in the design of the study. I.K., Z.X., and T.C. designed the data collection instruments. S.L., I.K., and X.X. performed the statistical analysis and drafted the manuscript. I.K. and J.Z. helped to draft the manuscript and provided critical comments. L.Z. provided key comments on intellectual content, and reviewed and revised the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and informed consent
The Medical Ethics Committee of the Northwest Minzu University, Lanzhou, Gansu, China, had approved the study. This study followed the ethical principles of the Declaration of Helsinki for Medical Research involving persons. All participants gave their written informed consent to use the data and biological material for research work.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Lei, S., Khan, I., Zhang, X. et al. Assessing oral and toothbrush microbial profiles among high-altitude individuals with and without periodontal disease: a case-control study. BMC Oral Health 24, 993 (2024). https://doi.org/10.1186/s12903-024-04603-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12903-024-04603-0