Abstract
Background
Streptococcus mutans is studied for its acidogenic and aciduric characteristics, notably its biofilm formation in the presence of sucrose, toward its role in the caries process. Variations in both genotype and phenotype have been reported among clinical isolates of S. mutans. This study aimed to examine genotypic and phenotypic characteristics of S. mutans obtained from Thai children with varying caries statuses.
Methods
We determined the presence of S. mutans and caries status in 395 children aged 3–4 years. From 325 children carrying S. mutans, we selected 90 with different caries statuses—caries-free (CF; n = 30), low severity of caries (LC; n = 30), or high severity of caries (HC; n = 30). Three isolates of S. mutans were taken from each child, thus, a total of 270 isolates were obtained. Multilocus sequence typing (MLST) was used to genotype the isolates and assess their clonal relationships. The properties, including biofilm formation, collagen binding, and acid production and tolerance were also evaluated.
Results
Children with carious lesions showed a higher detection rate and number of S. mutans in saliva than those without caries. S. mutans from individuals with HC status showed the lowest biofilm formation ability, while this group had the highest detection rate of collagen-binding isolates. There was no difference in acid production or tolerance by caries status. Genotyping by MLST did not reveal any clone of S. mutans specific to CF status. This result remained even when we included MLST data from the open-access PubMLST database. MLST did identify clones containing only strains from caries-affected hosts, but tests of their phenotypic properties did not reveal any differences between S. mutans from these clones and clones that were from both caries-free and caries-affected children.
Conclusions
The clonal relationships of S. mutans indicated by MLST were not associated with the status of dental caries in the host.
Similar content being viewed by others
Introduction
Dental caries is a biofilm-mediated, diet-modulated, multifactorial disease where microbes also play a role in its occurrence [1]. Carious lesions arise from an imbalance in the oral microbiota, leading to dysbiosis and the formation of an acidic environment near tooth surfaces. Rather than being caused by a specific pathogen, the caries process involves the interplay of diverse species of microorganisms, including a well-known species, Streptococcus mutans, which was first isolated from a carious lesion [2,3,4]. The characteristics of S. mutans, studied for its role in the caries process, include the ability to efficiently colonize tooth surfaces (adhesion), along with acid production and tolerance [5]. These properties have also been investigated and demonstrated in other oral microbes associated with dental caries [6,7,8].
Although S. mutans has been found with a high prevalence in humans with carious lesions, this bacterium has also been reported in the oral cavities of caries-free individuals [4, 9, 10]. Various methodologies have confirmed variability in both genotypic and phenotypic characteristics of clinical isolates of S. mutans [11,12,13]. Consequently, some researchers have explored the differences between S. mutans strains from caries-free and caries-active hosts [11, 14,15,16,17]. Nevertheless, changes in microbial compositions and properties are also influenced by factors such as dietary habits, oral hygiene, and environmental changes during caries formation [7, 18, 19].
Multilocus sequence typing (MLST) has been proposed as a reproducible method to characterize and identify clonal relationships of bacteria [20]. Because it is a molecular typing method based on the nucleotide sequences of multiple housekeeping genes, MLST creates a standardized approach for gathering and comparing data via web-based databases. PubMLST (https://pubmlst.org/) is a website dedicated to the collection of open-access and curated MLST data [21]. PubMLST provides sequence data with provenance and phenotype information for > 130 microbial species and genera. MLST has been used widely to distinguish genotypes and study both prokaryotic and eukaryotic microorganisms in the oral cavity, including the population structure of microbes [22, 23] and to identify potential disease-related clones or clones associated with virulence factors and antimicrobial resistance traits [24,25,26]. Beginning in 2007, our research group has developed an MLST scheme for S. mutans [12]. This scheme was advantageous in specifying S. mutans clones with particular serotypes and virulence potential regarding bacteremia and infective endocarditis [27,28,29].
In the present study, we used this MLST scheme to genotype S. mutans sampled from the oral cavity of Thai children and to study their clonal relationships. We undertook genotypic study of these S. mutans isolates, and examined their phenotypes, i.e., biofilm formation in the presence of sucrose, ability to bind with collagen, acid production, and acid tolerance. Additionally, we supplemented our MLST results with those deposited in the PubMLST database regarding caries. We aimed to find whether there were associations of genotypic and phenotypic characteristics of S. mutans with the status of dental caries in the host.
Materials and Methods
Subjects, saliva collection, and dental examination
Three hundred and ninety-five subjects (216 boys and 179 girls), 3–4 years of age, with no prior history of antibiotic use, were recruited from a kindergarten in Bangkok, Thailand. Samples were collected between 8: 30 and 10: 30 a.m. Participants were instructed not to brush their teeth before collection. Saliva was obtained by asking participants to expectorate 2–3 ml of saliva into a sterile 50-ml tube. The saliva was kept on ice for ≤ 3 h before bacterial isolation.
Data on caries experience were recorded by the decayed, missing, filled teeth/surfaces (dmft/s) indices based on modified World Health Organization (WHO) criteria, which cavitated lesions within the enamel were also included as part of decayed teeth and decayed surfaces. In addition, the numbers of decayed teeth and decayed surfaces were reported as dt and ds, respectively.
Oral hygiene status was assessed by the Silness and Löe plaque index, which records the amount of plaque (score 0–3) on six teeth (maxillary right second molar, maxillary right lateral incisor, maxillary left first molar, mandibular right first molar, mandibular left lateral incisor, and mandibular left second molar) [30]. The average of all scores from these teeth was presented as the plaque index for each child.
Some subjects in the present study were also studied and presented in Lapirattanakul et al. [10]. The research protocol for the present study was approved by the Ethics Committee of Mahidol University (MU-DT/PY-IRB 2015/DT048; COA.No.MU-DT/PY-IRB 2015/040.0109), and prior written informed consent was obtained from the parents or legal guardians of all participants.
Determination of numbers of S. mutans
The collected saliva specimens were tenfold serially diluted in saline solution and cultured on Mitis Salivarius agar (Difco Laboratories, Detroit, MI, USA) containing bacitracin (0.2 U/ml; Sigma Chemical Co., St. Louis, MO, USA), 0.001% (v/v) tellurite solution (Becton, Dickinson and Co., Sparks, MD, USA) and 15% (w/v) sucrose (MSB agar plates). After a 2-day incubation at 37 °C in a humidified atmosphere containing 5% CO2, the number of S. mutans was detected from MSB agar plates based on colony morphology (raised, convex, undulate, opaque, pale-blue colonies that are granular or frosted glass in appearance. Colonies may exhibit a glistening bubble on the surface due to glucan synthesis). Then, the number of S. mutans in the saliva samples was enumerated in colony-forming units (CFU) per milliliter of saliva.
Isolation of S. mutans from Thai children with different caries statuses
Based on microbiological data, we identified children carrying S. mutans, and through dental examination, we classified them into three groups: (i) caries-free (CF), (ii) low severity of caries (LC), or (iii) high severity of caries (HC). We then randomly selected 30 children from each group. The 30 children in the CF group were caries-free and had no experience with dental caries. The LC group contained 30 children with fewer than five decayed teeth (dt < 5), and the HC group was composed of 30 children having at least 10 decayed teeth (dt ≥ 10). The carious lesions in the LC group were limited to enamel breakdown without visible dentin. In contrast, the decayed teeth in the HC group also contained dentin caries.
Three colonies of S. mutans were randomly picked from each subject’s MSB agar plate based on colony morphology. Thus, collectively, 270 isolates of S. mutans were obtained for further evaluation. All S. mutans isolates were kept as stocks in brain heart infusion (BHI) broth containing 50% glycerol at − 80 °C.
Biofilm formation assay
Biofilm formation in the presence of sucrose was evaluated in 96-well flat-bottom plates (Thermo Fisher Scientific, Jiangsu, China). The procedure followed that in our previous publication with some modifications [31]. One-half strength Todd-Hewitt (TH) medium (Becton, Dickinson and Co.) with 0.25% sucrose was used as the culture medium. Pre-grown S. mutans cultured at 37 °C for 16–18 h were diluted 1:100 in the medium, then distributed at 100 μl per well. The plates were then incubated at 37 °C under 5% CO2 for 24 h. After discarding the medium from the wells, the plates were washed three times with distilled water. Formed biofilms were fixed with 100 µl of 25% formaldehyde at room temperature for 10 min. Following washing with distilled water, biofilms were then stained with 100 µl of 0.05% (w/v) crystal violet in water for 1 min, and washed with water. The absorbance at 590 nm (A590 nm) of the dye dissolved in 100 µl of 7% acetic acid was measured using an ELx800TM Absorbance Reader (BioTek Instruments Inc., Winooski, VT, USA). Three independent experiments were performed in triplicate.
Collagen-binding assay
Collagen-binding properties of S. mutans isolates were determined by a previously described procedure with some modifications [28]. In brief, 10 µg of type I collagen in 0.1 M acetic acid (Sigma) was coated onto each well of a 96-well (round-bottom) cell culture plate (Corning Incorporated, Corning, NY, USA) and incubated at 4 °C for 24 h. Plates were washed with phosphate-buffered saline (PBS) and incubated with 5% bovine serum albumin (Sigma) in PBS at 37 °C for 1.5 h, washed with PBS containing 0.01% Tween 20, and then 1 × 109 S. mutans cells in PBS were added to each well. After 1 h of incubation at 37 °C, adherent cells were washed with PBS, and treated with 25% formaldehyde, 0.05% crystal violet, and 7% acetic acid as described above in the section “Biofilm formation assay”. The collagen-binding ability of the tested isolates is presented as A590 nm of the crystal violet dye in 7% acetic acid measured by using the ELx800TM Absorbance Reader. S. mutans strain TLJ11–2 possessing collagen-binding ability and S. mutans strain TLJ1-1 lacking this ability [27] were used as positive and negative controls, respectively. Three independent experiments were conducted in triplicate.
Acid production assay
To compare acid production among the S. mutans isolates, an acid production assay was performed using our previously reported method [31]. Two milliliters of a seed culture of S. mutans (109 CFU/ml) were inoculated into 200 ml of phenol red broth (Difco) containing 1% glucose. The cultures were incubated in closed containers at 37 °C for 24 h. During incubation, 2 ml of the culture mixture was collected at 0, 4, 8, 12, 18, and 24 h, and the pH was determined using a pH meter (Ohaus Corporation, NJ, USA). Three independent experiments were performed for all tested S. mutans.
Acid tolerance assay
An acid tolerance assay based on our previous report was used [31]. S. mutans were cultured overnight in TH broth containing 0.3% yeast extract (THYE), then diluted tenfold in fresh THYE and incubated at 37 °C until reaching the mid-logarithmic growth phase (OD600nm = 0.4–0.5). Bacterial cells were harvested and resuspended in THYE (pH 5.0), then incubated at 37 °C for 2 h. Acid tolerance was detected by incubating the cells at 37 °C at a potential killing pH of 3.5 for 2 h. Cells were counted in triplicate by plating on THYE plates before and after incubation at the killing pH. Results are shown as the percentage survival rate, which was calculated using the formula: 100 × (number of cells following incubation at pH 3.5)/(number of cells before incubation at pH 3.5).
MLST analysis of S. mutans
S. mutans isolates were genotyped by our previous MLST scheme with some modifications [12]. Firstly, genomic DNA was extracted by the protocol for Gram-positive bacteria [27]. The obtained DNA was confirmed to be S. mutans genome using PCR with species-specific primers [32]. Then, internal fragments of eight housekeeping genes, namely, transketolase (tkt), glutamine synthetase type I (glnA), glutamate synthetase (gltA), glucose kinase (glk), shikimate 5-dehydrogenase (aroE), glutamate racemase (murI), signal peptidase I (lepC), and DNA gyrase A subunit (gyrA), were PCR amplified using primers shown in Table S1. The reaction mixture (20 µl) contained 0.5 U Ex Taq™ DNA polymerase (Takara Bio Inc., Shiga, Japan), 2 µl of 10 × Ex Taq buffer (Takara Bio) containing 20 mM Mg2+, 1.6 µl dNTPs (2.5 mM), 0.5 µl each of primer (20 µM), 40 ng DNA, and 13.3 µl sterilized water. Thermocycling was carried out in a Bioer Life Express thermocycler (Bioer, Hangzhou, China) as follows: 94 °C for 5 min; followed by 25 cycles (30 cycles for tkt) at 94 °C for 30 s, 55 °C (50 °C for tkt) for 30 s, and 72 °C for 30 s; with a final extension at 72 °C for 7 min. After checking PCR products by agarose gel electrophoresis visualized by staining with ethidium bromide, the products were purified with illustra ExoProStar 1-Step (GE Healthcare, Little Chalfont, UK), and sequenced (Macrogen Inc., Seoul, South Korea) using their respective PCR primers. All nucleotide sequences from these isolates determined in the process of MLST were registered in the GenBank database (accession numbers OQ809072–OQ809341 for tkt; OQ829640–OQ829909 for glnA; OQ829910–OQ830179 for gltA; OQ830180–OQ830449 for glk; OQ866634–OQ866903 for aroE; OQ938982–OQ939251 for murI; OQ939252–OQ939521 for lepC; and OQ972993–OQ973262 for gyrA).
Distinct nucleotide sequences in each housekeeping locus were assigned different allele numbers. For an S. mutans isolate, the allele numbers for each of the eight loci define the allelic profile and consequently the sequence type (ST). There is a public database gathering information on S. mutans studied by MLST; hence the nucleotide sequences of housekeeping gene fragments obtained in this study were compared with those deposited in the Oral Streptococcus PubMLST database (http://pubmlst.org/oralstrep/) [21]. The same allele numbers and STs were assigned for matched results, while new ones were submitted to the database for designation.
Allelic profiles of the 270 S. mutans isolates were analyzed using START (sequence type analysis and recombinational tests) [33] to produce a matrix of pairwise differences in the allelic profiles, and a dendrogram was constructed from the matrix using the unweighted pair group method with arithmetic mean. Related STs were grouped by using the goeBURST (global optimal enhanced based upon related sequence types) algorithm implemented in the PHYLOViZ program [34]. Two or more independent isolates sharing identical alleles at ≥ 6 loci were defined as members of a ‘clonal complex’ (CC) or ‘group’ of STs that have diversified from a common ancestor.
Moreover, the allelic profiles of the 270 S. mutans in the present study were further analyzed for relatedness together with profiles of 115 strains selected from the Oral Streptococcus PubMLST database (Table S2). The selection criteria were that these 115 strains of S. mutans were genotyped by the same MLST scheme as used in the present study (the Nakano scheme) [12], and that the strains had precise information regarding the caries status of the subject from whom they were isolated [35,36,37,38,39,40].
Statistical analysis
SPSS software v.18.0 (SPSS Corp., Chicago, IL, USA) and GraphPad Prism software v.5.0 (GraphPad Software Inc., La Jolla, CA, USA) were used in statistical analyses. The χ2 test was used to test association for categorical variables. Student’s t-test, the Mann–Whitney U test (Mann-U), and the Kruskal–Wallis test were used to compare continuous variables. Correlation analysis was performed by using Spearman’s rank correlation. The level of significance was set at p-value < 0.05.
Results
Dental examination and microbiological data
Among 395 kindergarten children recruited in Bangkok, Thailand, 284 had decayed teeth (71.9%); 71.3% of boys and 72.6% of girls had carious lesions (no statistically significant difference by sex). The group with dental caries had a higher plaque index compared to the group without caries (p-value < 0.001, Mann-U test) (Table 1). S. mutans was detected in 325 children (82.3%). The detection rate of this bacterium was higher in caries-affected children than in those with no caries (p-value < 0.001, χ2 test). In the subjects carrying S. mutans, the value of log CFU/ml of S. mutans was higher in the saliva of caries-affected children than in children without caries (p-value < 0.001, t-test). In addition, a positive correlation was found between log CFU/ml of S. mutans and all caries indices (p-value < 0.001, Spearman’s rank correlation); the correlation coefficients (rs) were 0.475 (dmft), 0.478 (dmfs), 0.488 (dt), and 0.493 (ds).
Data regarding the plaque index, the caries indices, and the number of bacteria in the saliva of the 90 children who were the sources of the 270 S. mutans isolates analyzed further in this study are shown in Table 2; the children were divided into three caries statuses: caries-free (CF), low severity of caries (LC), and high severity of caries (HC).
Biofilm formation and collagen binding of S. mutans from children with the three caries statuses
All 270 isolates of S. mutans could form biofilm in a culture medium with sucrose. The biofilm formation, represented by the A590 nm value, ranged from 0.073 to 2.369. When the ability to form biofilm in the presence of sucrose was compared based on the caries status of the host child, S. mutans isolated from children in the HC group showed the lowest biofilm formation (p-value < 0.01, Kruskal–Wallis test). In addition, isolates with low biofilm formation properties, indicated by A590 nm < 1.500, were mostly present in the HC group (Fig. 1a).
a Biofilm formation and b collagen-binding properties of S. mutans from Thai children. The comparison was based on caries status of the donor: caries-free (CF; n = 90 S. mutans isolates), low severity of caries (LC; n = 90), and high severity of caries (HC; n = 90). S. mutans from children in the HC group showed the lowest biofilm formation ability and the highest collagen-binding ability. **p-value < 0.01, Kruskal–Wallis test. c In addition, S. mutans with collagen-binding ability (n = 23) showed lower biofilm formation ability than the non-collagen binding isolates (n = 247). ***p-value < 0.001, Mann–Whitney U test
The collagen binding assay showed that only 23 of the 270 S. mutans isolates could bind to collagen (for which A590 nm ranged from 0.810 to 1.733) (Fig. 1b). These 23 isolates belonged to subjects in the HC (21 isolates) and CF (2 isolates) groups, thus there was an association between having collagen-binding ability and the caries status of the host (p-value < 0.001, χ2 test). Interestingly, S. mutans with collagen-binding ability showed lower biofilm formation than those without collagen-binding ability (p-value < 0.001, Mann-U test) (Fig. 1c).
Acid production and acid tolerance of S. mutans from children with the three caries statuses
Acid production in the presence of 1% glucose showed no significant difference considering pH values at any time point (0 to 24 h) among S. mutans from subjects with different caries statuses (Fig. S1). Moreover, acid tolerance was similar among the three groups analyzed; there was no difference in the percentage of surviving cells after treatment in an acid environment (median survival: CF = 43.18%, LC = 43.09%, HC = 42.41%; p-value = 0.208, Kruskal–Wallis test).
Genotyping of S. mutans isolates by MLST
A total of 270 S. mutans isolates from 90 children were classified into 81 unique STs, of which ST2 was the most common (Table S3). In 62 children (68.9%), the same ST was detected for all three isolates from the child. For the other 28 children (31.1%), MLST indicated mixed STs, i.e., two STs were found in 27 children, and three STs in one child (the child in HC status). The right-hand column of Table 2 shows the number of children with mixed STs of S. mutans in the CF, LC, and HC groups. No significant difference in the percentage of subjects with mixed STs was found according to caries status (p-value = 0.147, χ2 test).
Clonal relationships of S. mutans analyzed by MLST
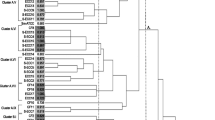
Determination of the clonal relationships of the 81 STs identified from the 270 S. mutans isolates revealed 32 STs as singletons unrelated to any other ST. The remaining 49 STs were grouped into 19 CCs (Fig. 2). No CC contained only strains from CF status individuals—the 12 CCs possessing S. mutans strains from CF subjects also contained strains from LC and HC status donors (Groups 1, 2, 4, 5, 6, 7, 8, 9, 12, 16, 17, and 18). The remaining seven CCs consisted of S. mutans from caries-affected subjects only (Groups 3, 10, 11, 13, 14, 15, and 19). Group 14 consisted of only S. mutans from children with HC status, while only strains from those with LC status were present in Group 19. The number of STs in most of these seven CCs was two, but Groups 10 and 13 contained three and five STs respectively. Considering collagen binding, Groups 2 and 13 contained 100% and 40% STs with collagen-binding ability (Fig. 2). No CC showed an association with ability to form biofilm or production or tolerance of acid. S. mutans in the seven CCs consisting only of S. mutans from caries-affected hosts showed no difference in phenotypic properties (biofilm formation, collagen binding, acidogenicity, and acid tolerance) from those in the 12 CCs that included strains from hosts with and without dental caries (Table 3).
Dendrogram of 270 S. mutans isolated from Thai children with different caries statuses, i.e., caries-free (CF; n = 90 S. mutans isolates), low severity of caries (LC; n = 90), or high severity of caries (HC; n = 90). The dendrogram was constructed from allelic profiles of 81 sequence types (STs) detected by multilocus sequence typing (MLST). MLST indicated 30, 31, and 39 STs from individuals with CF, LC, and HC status, respectively. Clonal complex groups and the number of strains in each ST are indicated based on caries status. Asterisks indicate S. mutans with collagen-binding ability. The gray diamond (◇) indicates a clonal complex group containing S. mutans from both caries-free and caries-affected hosts
When we included the data for 115 strains from the Oral Streptococcus PubMLST database in the evaluation of clonal relationships, the number of CCs increased to 36 (Table 4). The largest CC was Group 5, having 14 STs, with China, Sweden, Thailand, and the USA as the countries of origin. ST2, the most common ST among Thai children in the present study, was located in this CC (Table S3). From a total of 36 CCs, there were 20 CCs containing S. mutans from both caries-free and caries-affected hosts. We still did not find a CC related to S. mutans from caries-free individuals only.
Discussion
The caries prevalence of Thai children in the present study was 71.9%. The plaque index indicated that the oral hygiene status of children with caries was poorer than that of children without caries. Additionally, children with carious lesions showed a higher detection rate and number of S. mutans in their saliva than those without caries. This can be attributed to the fact that individuals with poorer oral hygiene and more plaque tend to develop higher levels of S. mutans in their saliva compared to children with less plaque. Our findings are consistent with previous reports showing the association of S. mutans with the cariogenic environment and poor oral hygiene [9, 19, 41]. In the children carrying S. mutans, we found a positive correlation between the number of salivary S. mutans and caries indices, though the correlation coefficients indicated only weak correlations (rs < 0.5). Notably, S. mutans was detected in more than 50% of children without caries. This is reasonable given the multifactorial etiology of dental caries as well as evidence suggesting the role of acidogenic and aciduric bacteria other than S. mutans in caries occurrence [2, 7]. Moreover, the presence of acidogenic and aciduric microorganisms alone cannot predict caries risk without the influence of a diet rich in sugars and frequent sugar consumption [18, 42].
Our study also evaluated the phenotypic traits of S. mutans from children with one of three caries statuses: CF, LC, or HC. Statistically significant differences were observed among the S. mutans sampled from these groups in terms of biofilm formation in the presence of sucrose and collagen binding. S. mutans from children having ≥ 10 decayed teeth, i.e., the HC group, showed the lowest biofilm formation ability, while the detection rate of S. mutans with collagen-binding ability was highest in this group. Either poor oral hygiene with a high amount of accumulated plaque or the high number of carious lesions in this HC group might help retain strains with different binding efficiencies and mechanisms. Therefore, S. mutans in the HC group exhibited a greater variety of biofilm formation in the presence of sucrose compared to both the CF and LC groups.
Both biofilm formation and collagen binding are related to the adhesion of, and colonization by, S. mutans [43]. All S. mutans in the present study could form biofilm in the medium containing sucrose, however, binding to collagen was not ubiquitous in our S. mutans isolates, approximately 10% of the evaluated S. mutans bound to type I collagen. This percentage was consistent with the previous findings [27]. Almost all S. mutans having collagen-binding ability were isolated from children with HC status, suggesting a potential link between collagen-binding ability, plaque accumulation, dentin access, and caries severity, though it did not establish causation. Some works have suggested an association between S. mutans strains with collagen-binding proteins and an increase in caries parameters and caries risk [44, 45]. In dentin caries, the ability to adhere to collagen may endow S. mutans with an alternative to the sucrose-dependent mechanism of colonization [43]. However, the risk of developing caries involves multiple factors beyond the severity and damage to the dentin. Fig. 1c showed that S. mutans with collagen-binding properties formed less monospecies biofilm in the presence of sucrose than the non-collagen binding isolates; thus, binding to collagen seemed to compensate for the low ability to form biofilm. In addition, the carious lesions exposing dentin collagen in the HC group might influence the high detection rate of collagen-binding S. mutans in this group.
In contrast to the biofilm formation and collagen-binding properties, the acidogenic and aciduric properties of S. mutans isolates were similar irrespective of the caries status of the host. S. mutans from donors with all three caries statuses (CF, LC, and HC) could decrease pH to < 5.5, the critical pH of enamel, within the experimental period, and nearly half of the vital cells of the isolates from all three statuses could survive at pH 3.5. This demonstrates the bacterium’s ability to thrive in acidic environments, highlighting its adaptability in conditions that contribute to the caries process. In multiple previous studies of acidogenic and aciduric properties of S. mutans, only some detected differences that were dependent on caries statuses [11, 14, 16, 17, 46,47,48]. Variations in both methodology and culture medium used can lead to such inconsistent findings. A study showed that the acid production in BHI broth was significantly greater for S. mutans from subjects with caries compared with those without caries, but this difference was not observed when a chemically defined medium was used [17].
For genotyping study of the S. mutans isolates, we used the allele-based MLST approach, which considers the allele as the unit of analysis [12]. Nucleotide sequence data were compared for each housekeeping locus. Distinct sequences were assigned different allele numbers. Because eight housekeeping genes are included in this MLST scheme, eight allele numbers define the allelic profile and the ST of the S. mutans isolate. The detection of the same ST among analyzed S. mutans isolates indicates that they are identical. In our present study, the genotyping of 270 S. mutans isolates by MLST found 81 STs. Because these S. mutans were from 90 children, we analyzed the number of STs in each child. In a majority of subjects (68.9%), the three isolates from the subject were classified in the same ST. These findings were consistent with results using the same MLST scheme to genotype S. mutans from Japanese and Chinese populations [39, 49]. Previous studies have evaluated whether there is an association between the number of S. mutans genotypes and caries activity. Most studies indicated more genotypes of S. mutans in children having severe caries compared with those who were caries-free [14, 37, 50, 51], but other authors found the opposite result [52], or no difference in genotypic diversity among children with different caries statuses [11, 16]. Certainly, the typing method used and the number of isolates analyzed could influence the results. In the present study, the children carrying multiple S. mutans STs were mostly found in the HC group, and one child in this group showed three STs. Thus, the genotypic diversity of S. mutans seemed higher in the HC group compared with the CF and LC groups. However, there was no statistically significant difference; thus, analyzing only three isolates of S. mutans per subject might limit statistical findings in this aspect.
The MLST scheme used for S. mutans in this study was based on eight housekeeping genes [12]. In analysis of relatedness among the analyzed strains, two or more isolates of S. mutans sharing identical alleles at least six housekeeping loci were grouped into a CC of STs that have diversified from a common ancestor. The clonal relationships of 270 S. mutans isolated from Thai children with three caries statuses (CF, LC, and HC) are shown in Fig. 2. We aimed to find specific clone(s) related to the dental caries status of the host. However, MLST indicated no specific CC related to caries-free status. Based on phenotypic properties examined, S. mutans from individuals in the CF group possessed biofilm formation, acidogenic, and aciduric properties not less than those from the LC and HC groups. Therefore, it is reasonable to suggest that the presence of S. mutans alone does not predict caries occurrence, as its impact is significantly influenced by host factors and dietary habits. According to the ecological plaque hypothesis, ecological stress, such as frequent exposure to a low pH from sugar fermentation, is essential for the shift from healthy to disease status [53]. Even when we added data on S. mutans from the Oral Streptococcus PubMLST database into the analysis, the findings confirmed no caries-free related clone in the S. mutans population.
Other genotyping methods performed to differentiate S. mutans derived from subjects with different caries statuses were gel-based genotyping techniques, such as arbitrary-primed PCR [16], chromosomal DNA fingerprinting with a restriction endonuclease [54], and pulsed-field gel electrophoresis [55]. In keeping with our results, no specific cluster of S. mutans associated with caries-free status was found in these studies. In addition, some studies aimed to identify parts of genomes specific to S. mutans strains from children with a high number of caries, compared with strains from those with a low number of caries or caries-free children. No differences in the distributions of putative virulence genes, genomic islands, or insertion sequences were found between S. mutans strains from children with severe early childhood caries and those isolated from caries-free children [54]. Similarly, comparative genome hybridization using microarray technology and whole genome comparison by in silico genome subtraction failed to indicate any genetic loci of S. mutans consistently associated with caries status [15, 56].
Esberg et al. [44] have linked the detection of some surface protein-encoding genes of S. mutans, i.e., adhesin SpaP variant B and collagen-binding protein (Cnm), with increased baseline caries and prospective caries development. Our study found two CCs of S. mutans with collagen-binding ability (Groups 2 and 13). However, not all S. mutans in these two CCs were from caries-affected hosts; consequently, there was no relatedness between clones of collagen-binding S. mutans and caries status. Although MLST indicated that seven CCs contained S. mutans only from children having dental caries (Groups 3, 10, 11, 13, 14, 15, and 19), S. mutans strains in these clones did not show higher abilities to form biofilm, bind to type I collagen, or produce/tolerate acid than those in the CCs from both caries-affected and caries-free children. On adding S. mutans from the Oral Streptococcus PubMLST database into the analysis, Table 4 shows that Group 10 changed to include S. mutans from both caries-free and caries-affected subjects. The other six CCs remained as having no S. mutans from caries-free individuals, while 10 more CCs of S. mutans from caries-affected hosts only were detected (Groups 20, 21, 24, 27, 28, 29, 31, 32, 33, and 34). However, the number of members in the CCs lacking strains from caries-free subjects was mostly two STs, which is insufficient to be regarded as potential disease-related clones. Because of the efficiency and relatively low cost of modern sequencing technology, standard MLST (7–8 loci) has been expanded to include analysis of more genes, for example, ribosomal MLST (rMLST, 53 loci), core-genome MLST (cgMLST, > 500 loci), and whole-genome MLST (wgMLST, all loci) [57]. Recently, a cgMLST method analyzing 594 core genes was developed for S. mutans and showed more discriminatory power than the traditional MLST method [39]. The data for S. mutans studied by this cgMLST were also deposited into the Oral Streptococcus PubMLST database. Further accumulation of such MLST data and strain analysis might improve understanding of the S. mutans population.
Conclusions
Overall, children with carious lesions showed a higher detection rate and a higher number of S. mutans in saliva than those without caries. We found no difference in acid production or acid tolerance in S. mutans isolates from Thai children with different caries statuses. Although S. mutans from children with high severity of dental caries showed the lowest biofilm formation ability, they had the highest detection rate of collagen-binding isolates. S. mutans isolates from caries-free hosts could adhere as well as produce and tolerate acid not less than that of isolates from caries-affected individuals. Genotyping by MLST did not reveal any S. mutans clones specific to caries-free individuals. It showed clones containing only S. mutans strains from individuals with caries, but the properties of these strains did not differ from those in clones from both caries-free and caries-affected children. Thus, the clonal relationships of S. mutans indicated by MLST were not associated with the status of dental caries in the host.
Availability of data and materials
All data generated or analyzed during this study are included in the article and its supplementary files. All nucleotide sequences determined in the process of MLST were registered in the GenBank database (accession numbers OQ809072–OQ809341 for tkt; OQ829640–OQ829909 for glnA; OQ829910–OQ830179 for gltA; OQ830180–OQ830449 for glk; OQ866634–OQ866903 for aroE; OQ938982–OQ939251 for murI; OQ939252–OQ939521 for lepC; and OQ972993–OQ973262 for gyrA). Further enquiries can be directed to the corresponding author.
Abbreviations
- CC:
-
Clonal complex
- CF:
-
Caries-free
- HC:
-
High severity of caries
- LC:
-
Low severity of caries
- MLST:
-
Multilocus sequence typing
- ST:
-
Sequence type
References
Machiulskiene V, Campus G, Carvalho JC, Dige I, Ekstrand KR, Jablonski-Momeni A, et al. Terminology of dental caries and dental caries management: Consensus report of a workshop organized by ORCA and Cariology Research Group of IADR. Caries Res. 2020;54(1):7–14.
Fakhruddin KS, Ngo HC, Samaranayake LP. Cariogenic microbiome and microbiota of the early primary dentition: A contemporary overview. Oral Dis. 2019;25(4):982–95.
Baker JL, Morton JT, Dinis M, Alvarez R, Tran NC, Knight R, Edlund A. Deep metagenomics examines the oral microbiome during dental caries, revealing novel taxa and co-occurrences with host molecules. Genome Res. 2021;31(1):64–74.
Bhaumik D, Manikandan D, Foxman B. Cariogenic and oral health taxa in the oral cavity among children and adults: a scoping review. Arch Oral Biol. 2021;129:105204. https://doi.org/10.1016/j.archoralbio.2021.105204.
Lemos J A, Palmer S R, Zeng L, Wen Z T, Kajfasz J K, Freires I A, et al. The biology of Streptococcus mutans. Microbiol Spectr. 2019;7(1):10–128.
Beighton D. The complex oral microflora of high-risk individuals and groups and its role in the caries process. Community Dent Oral Epidemiol. 2005;33(4):248–55.
Takahashi N, Nyvad B. Caries ecology revisited: microbial dynamics and the caries process. Caries Res. 2008;42(6):409–18.
Du Q, Ren B, Zhou X, Zhang L, Xu X. Cross-kingdom interaction between Candida albicans and oral bacteria. Front Microbiol. 2022;13.
Teanpaisan R, Thitasomakul S, Piwat S, Thearmontree A, Pithpornchaiyakul W, Chankanka O. Longitudinal study of the presence of mutans streptococci and lactobacilli in relation to dental caries development in 3–24 month old Thai children. Int Dent J. 2007;57(6):445–51.
Lapirattanakul J, Nomura R, Okawa R, Morimoto S, Tantivitayakul P, Maudcheingka T, et al. Oral lactobacilli related to caries status of children with primary dentition. Caries Res. 2020;54(2):194–204.
Lembo FL, Longo PL, Ota-Tsuzuki C, Rodrigues CR, Mayer MP. Genotypic and phenotypic analysis of Streptococcus mutans from different oral cavity sites of caries-free and caries-active children. Oral Microbiol Immunol. 2007;22(5):313–9.
Nakano K, Lapirattanakul J, Nomura R, Nemoto H, Alaluusua S, Gronroos L, et al. Streptococcus mutans clonal variation revealed by multilocus sequence typing. J Clin Microbiol. 2007;45(8):2616–25.
Bedoya-Correa CM, Rincon Rodriguez RJ, Parada-Sanchez MT. Genomic and phenotypic diversity of Streptococcus mutans. J Oral Biosci. 2019;61(1):22–31.
Napimoga MH, Kamiya RU, Rosa RT, Rosa EA, Hofling JF, de Oliveira M-G, et al. Genotypic diversity and virulence traits of Streptococcus mutans in caries-free and caries-active individuals. J Med Microbiol. 2004;53(Pt 7):697–703.
Argimón S, Konganti K, Chen H, Alekseyenko AV, Brown S, Caufield PW. Comparative genomics of oral isolates of Streptococcus mutans by in silico genome subtraction does not reveal accessory DNA associated with severe early childhood caries. Infect Genet Evol. 2014;21:269–78.
Valdez RMA, Duque C, Caiaffa KS, Dos Santos VR, Loesch MLA, Colombo NH, et al. Genotypic diversity and phenotypic traits of Streptococcus mutans isolates and their relation to severity of early childhood caries. BMC Oral Health. 2017;17(1):115.
Banas JA, Takanami E, Hemsley RM, Villhauer A, Zhu M, Qian F, et al. Evaluating the relationship between acidogenicity and acid tolerance for oral streptococci from children with or without a history of caries. J Oral Microbiol. 2020;12(1):1688449.
Bradshaw DJ, Lynch RJ. Diet and the microbial aetiology of dental caries: new paradigms. Int Dent J. 2013;63(Suppl 2):64–72.
Kaushal D, Kalra N, Khatri A, Tyagi R, Singh NP, Aggarwal A, Saha R. Oral health status and microbial load of Streptococcus mutans in children with Cerebral palsy in a tertiary care hospital in Delhi. J Indian Soc Pedod Prev Dent. 2021;39:214–20.
Maiden MC. Multilocus sequence typing of bacteria. Annu Rev Microbiol. 2006;60:561–88.
Jolley KA, Bray JE, Maiden MCJ. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018;3:124.
Moorhouse AJ, Moreno-Lopez R, Gow NAR, Hijazi K. Clonal evolution of Candida albicans, Candida glabrata and Candida dubliniensis at oral niche level in health and disease. J Oral Microbiol. 2021;13(1):1894047.
Kalizang’oma A, Kwambana-Adams B, Chan JM, Viswanath A, Gori A, Richard D, et al. Novel multilocus sequence typing and global sequence clustering schemes for characterizing the population diversity of Streptococcus mitis. J Clin Microbiol. 2023;61(1).
Enersen M, Olsen I, Kvalheim O, Caugant DA. fimA genotypes and multilocus sequence types of Porphyromonas gingivalis from patients with periodontitis. J Clin Microbiol. 2008;46(1):31–42.
McManus BA, Maguire R, Cashin PJ, Claffey N, Flint S, Abdulrahim MH, et al. Enrichment of multilocus sequence typing clade 1 with oral Candida albicans isolates in patients with untreated periodontitis. J Clin Microbiol. 2012;50(10):3335–44.
Penas PP, Mayer MP, Gomes BP, Endo M, Pignatari AC, Bauab KC, et al. Analysis of genetic lineages and their correlation with virulence genes in Enterococcus faecalis clinical isolates from root canal and systemic infections. J Endod. 2013;39(7):858–64.
Lapirattanakul J, Nakano K, Nomura R, Leelataweewud P, Chalermsarp N, Klaophimai A, et al. Multilocus sequence typing analysis of Streptococcus mutans strains with the cnm gene encoding collagen-binding adhesin. J Med Microbiol. 2011;60(Pt 11):1677–84.
Lapirattanakul J, Nomura R, Nemoto H, Naka S, Ooshima T, Nakano K. Multilocus sequence typing of Streptococcus mutans strains with the cbm gene encoding a novel collagen-binding protein. Arch Oral Biol. 2013;58(8):989–96.
Lapirattanakul J, Nomura R, Matsumoto-Nakano M, Srisatjaluk R, Ooshima T, Nakano K. Variation of expression defects in cell surface 190-kDa protein antigen of Streptococcus mutans. Int J Med Microbiol. 2015;305(3):383–91.
Silness J, Löe H. Periodontal disease in pregnancy. II. Correlation between oral hygiene and periodontal condtion. Acta Odontol Scand. 1964;22:131–5.
Lapirattanakul J, Takashima Y, Tantivitayakul P, Maudcheingka T, Leelataweewud P, Nakano K, et al. Cariogenic properties of Streptococcus mutans clinical isolates with sortase defects. Arch Oral Biol. 2017;81:7–14.
Chen Z, Saxena D, Caufield PW, Ge Y, Wang M, Li Y. Development of species-specific primers for detection of Streptococcus mutans in mixed bacterial samples. FEMS Microbiol Lett. 2007;272(2):154–62.
Jolley KA, Feil EJ, Chan MS, Maiden MC. Sequence type analysis and recombinational tests (START). Bioinformatics. 2001;17(12):1230–1.
Francisco AP, Vaz C, Monteiro PT, Melo-Cristino J, Ramirez M, Carrico JA. PHYLOViZ: phylogenetic inference and data visualization for sequence based typing methods. BMC Bioinformatics. 2012;13:87.
Momeni SS, Whiddon J, Moser SA, Cheon K, Ruby JD, Childers NK. Comparative genotyping of Streptococcus mutans by repetitive extragenic palindromic polymerase chain reaction and multilocus sequence typing. Mol Oral Microbiol. 2013;28(1):18–27.
Momeni SS, Whiddon J, Cheon K, Moser SA, Childers NK. Assessment of two multilocus sequence typing (MLST) schemes available for Streptococcus mutans. Arch Oral Biol. 2015;60(12):1769–76.
Momeni SS, Whiddon J, Cheon K, Ghazal T, Moser SA, Childers NK. Genetic diversity and evidence for transmission of Streptococcus mutans by DiversiLab rep-PCR. J Microbiol Methods. 2016;128:108–17.
Momeni SS, Whiddon J, Moser SA, Childers NK. Transmission patterns of Streptococcus mutans demonstrated by a combined rep-PCR and MLST approach. Clin Oral Investig. 2018;22(8):2847–58.
Liu S, Li X, Guo Z, Liu H, Sun Y, Liu Y, et al. A core genome multilocus sequence typing scheme for Streptococcus mutans. mSphere. 2020;5(4):e00348–20.
Elyassi M, Babaeekhou L, Ghane M. Streptococcus mutans and Streptococcus sobrinus contributions in dental caries in Iranian and Afghan children: A report from serotype distribution and novel STs. Arch Oral Biol. 2022;139.
Karayilmaz H, Yalcin-Erman H, Erken-Gungor O, Ozturk Z, Felek R, Kupesiz A. Evaluation the oral hygiene conditions, oral Candida colonization and salivary Streptococcus mutans and Lactobacilli density in a group of β-thalassemic children and adolescence. Med Oral Patol Oral Cir Bucal. 2019;24(6):e712–8.
Sandy LPA, Helmyati S, Amalia R. Nutritional factors associated with early childhood caries: A systematic review and meta-analysis. Saudi Dent J. 2024;36(3):413–9.
Aviles-Reyes A, Miller JH, Lemos JA, Abranches J. Collagen-binding proteins of Streptococcus mutans and related streptococci. Mol Oral Microbiol. 2017;32(2):89–106.
Esberg A, Sheng N, Marell L, Claesson R, Persson K, Boren T, et al. Streptococcus mutans adhesin biotypes that match and predict individual caries development. EBioMedicine. 2017;24:205–15.
Garcia BA, Acosta NC, Tomar SL, Roesch LFW, Lemos JA, Mugayar LRF, et al. Association of Candida albicans and Cbp(+) Streptococcus mutans with early childhood caries recurrence. Sci Rep. 2021;11(1):10802.
Moreira MJS, Klaus NM, Dall’Onder AP, Grando D, Parolo CCF, Faccini LS, et al. Genotypic diversity and acidogenicity of Streptococcus mutans in Down syndrome children. Spec Care Dentist. 2019;39(6):578–86.
Bedoya-Correa CM, Rincon-Rodriguez RJ, Parada-Sanchez MT. Acidogenic and aciduric properties of Streptococcus mutans serotype c according to its genomic variability. Eur J Oral Sci. 2021;129(6).
Liu S, Li H, Zhang K, Guo Z, Zheng Q, Hu F, et al. Phenotypic and genetic characteristics of Streptococcus mutans isolates from site-specific dental plaque in China. J Med Microbiol. 2021;70(3). https://doi.org/10.1099/jmm.0.001313. Epub 2021 Jan 18.
Lapirattanakul J, Nakano K, Nomura R, Hamada S, Nakagawa I, Ooshima T. Demonstration of mother-to-child transmission of Streptococcus mutans using multilocus sequence typing. Caries Res. 2008;42(6):466–74.
Alaluusua S, Matto J, Gronroos L, Innila S, Torkko H, Asikainen S, et al. Oral colonization by more than one clonal type of mutans streptococcus in children with nursing-bottle dental caries. Arch Oral Biol. 1996;41(2):167–73.
Zhou Q, Qin X, Qin M, Ge L. Genotypic diversity of Streptococcus mutans and Streptococcus sobrinus in 3-4-year-old children with severe caries or without caries. Int J Paediatr Dent. 2011;21(6):422–31.
Kreulen CM, de Soet HJ, Hogeveen R, Veerkamp JS. Streptococcus mutans in children using nursing bottles. ASDC J Dent Child. 1997;64(2):107–11.
Marsh PD. Are dental diseases examples of ecological catastrophes? Microbiology. 2003;149(Pt 2):279–94.
Argimón S, Caufield PW. Distribution of putative virulence genes in Streptococcus mutans strains does not correlate with caries experience. J Clin Microbiol. 2011;49(3):984–92.
Herczegh A, Ghidan A, Deseo K, Kamotsay K, Tarjan I. Comparison of Streptococcus mutans strains from children with caries-active, caries-free and gingivitis clinical diagnosis by pulsed-field gel electrophoresis. Acta Microbiol Immunol Hung. 2008;55(4):419–27.
Zhang L, Foxman B, Drake DR, Srinivasan U, Henderson J, Olson B, et al. Comparative whole-genome analysis of Streptococcus mutans isolates within and among individuals of different caries status. Oral Microbiol Immunol. 2009;24(3):197–203.
Tagini F, Greub G. Bacterial genome sequencing in clinical microbiology: a pathogen-oriented review. Eur J Clin Microbiol Infect Dis. 2017;36(11):2007–20.
Acknowledgements
The authors express their gratitude to all the child volunteers who enthusiastically participated in this research study.
Funding
Open access funding provided by Mahidol University The study was supported by the Faculty of Dentistry, Mahidol University (DTRS-EG-2021–07), Okayama University Graduate School of Medicine, Dentistry and Pharmaceutical Sciences, Osaka University Graduate School of Dentistry, and KAKENHI research project No. 21KK0160 from the Japan Society for Promotion of Science. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
J.L. conceived and designed the experiments, performed experiments, analyzed data, and wrote the paper. R.N., R.O., P.T., R.K., A.L., P.L., and D.B. performed experiments and contributed ideas. M.M. and K.N. contributed ideas and supervised the project. All authors have read, edited, and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was performed following the principles of the Declaration of Helsinki. The research protocol was approved by the Ethics Committee of Mahidol University (MU-DT/PY-IRB 2015/DT048; COA.No.MU-DT/PY-IRB 2015/040.0109), and prior written informed consent was obtained from the parents or legal guardians of all participants.
Consent for publication
Not applicable. This study did not contain any details related to individual persons that required agreement for publication.
Competing interests
Ryota Nomura is an Associate Editor on the Editorial Board of BMC Oral Health, while the other authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
12903_2024_4759_MOESM2_ESM.docx
Additional file 2: Table S2 Allelic profiles and STs of 115 S. mutans strains from the Oral Streptococcus PubMLST database.
12903_2024_4759_MOESM3_ESM.docx
Additional file 3: Table S3 Allelic profiles and STs of 270 S. mutans isolated from the kindergarten children in this study.
12903_2024_4759_MOESM4_ESM.docx
Additional file 4: Fig. S1 Acid production by S. mutans from Thai children. The comparison was based on caries status: caries-free (CF; n = 90 S. mutans isolates), low severity of caries (LC; n = 90), and high severity of caries (HC; n = 90). The data shown represent the median values of each group. No significant difference in pH values was observed at any time point.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Lapirattanakul, J., Nomura, R., Okawa, R. et al. Multilocus sequence typing and phenotypic properties of Streptococcus mutans from Thai children with different caries statuses. BMC Oral Health 24, 1063 (2024). https://doi.org/10.1186/s12903-024-04759-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12903-024-04759-9