Abstract
Background
Asthma is a common complex chronic, inflammatory polygenic disease with heterogeneous manifestations, affecting individuals of all age groups and posing an immense burden on healthcare resources. A number of studies have identified the association between a disintegrin and metalloprotease 33 (ADAM33) polymorphisms and asthma risk, however, the results still remain inconclusive. The objective of the present study was to identify the effect of ADAM33 variants in asthma susceptibility.
Methods
Eligible case-control studies published between January 2000 and June 2018 was searched and retrieved from online electronic databases. The odds ratio (OR) with its 95% confidence interval (CI) was employed to calculate the effect.
Results
A total of 63 case-control studies were finally screened out, including 13,280 asthma patients and 13,340 controls. Eleven SNPs of ADAM33 gene were identified. Our results detected a significant association between ADAM33 T2, Q1, F + 1 and AA genotype of T + 1 polymorphisms and asthma risk in total population. Subgroup analysis by ethnicities showed that the alleles and genotypes of T2, Q1 and F + 1 polymorphisms were associated with asthma susceptibility among Asian populations, while V4 polymorphism was associated with asthma among Caucasian populations. Subgroup analysis by ages showed that T2, F + 1 and ST + 4 polymorphisms were associated with childhood asthma, while Q1 and V4 polymorphisms were associated with asthma risk in adults. Subgroup analysis by asthma severity showed that only the G allele of ADAM33 T1 polymorphism was associated with the severity of asthma when compared with the controls. In addition, T2, Q1 and F + 1 polymorphisms of ADAM33 were significantly associated with increased the asthma risk in Chinese asthma patients.
Conclusions
Our results found that T2, Q1 and F + 1 polymorphisms of ADAM33 gene might contribute to asthma risk. Future well-designed case-control studies with large population and more ethnicities are still needed to estimate the association.
Similar content being viewed by others
Background
Asthma is a lifelong respiratory disease of the lung airways, characterized by variable and recurring symptoms, reversible airflow obstruction and bronchospasm [1]. It involves inflammation and swelling of the airways, resulting in permanent remodeling/scarring of the airways. The symptoms of asthma include episodes of wheezing, coughing, chest tightness, and shortness of breath, which may occur a few times a day or a few times per week [2]. Depending on the individuals, the symptoms may become worse at night or with exercise, posing significant burden on quality of life, work productivity, activity impairment, and healthcare resources use [3]. Both the genetic factors (genetic polymorphisms, family history) and environmental factors (tobacco smoke, air pollution, allergens) contribute to the development of asthma [4, 5]. Prevalence rates increase and mortality rates decline for asthma in many countries over the past few decades: in 2016, 360 million people globally had asthma, up from 183 million in 1990 [6]; an estimated 420,000 people died from asthma worldwide in 2016, compared to 474,000 in 1990. Although many medicines are now widely used due to their greater effectiveness and fewer side effects in treatment of asthma, unfortunately, none of these medications are available without a prescription [7, 8]. In addition, a remarkably high frequency of poorly controlled asthma has been identified, and for some 5–10% of asthma patients, the disease is refractory to corticosteroid treatment and often leads to hospital admissions caused by respiratory viral infection with rhinovirus. What’s more, asthma symptoms beginning in adults might have originated in childhood [9], and if lung function no longer returns normal when not having an attack, asthma moves into the category of disease known as chronic obstructive pulmonary disease (COPD). Therefore, there is an urgent need to identify some significant biomarkers which can predict this disease and guild the therapeutic strategies.
A large number of genetic studies have been conducted to identify genetic polymorphisms associated with asthma susceptibility. A disintegrin and metalloprotease 33 (ADAM33) gene, located on human chromosome 20p13, was one of the first asthma candidate genes identified by positional cloning. It is one member of the ADAM family of zinc-dependent metalloproteases, and plays an important biological role as an activator of growth factors and Th2 cytokines [10]. ADAM33 consists of 22 exons that encode a signal sequence, pre-domain, catalytic domain, disintegrin domain, cysteine-rich domain, EGF domain, transmembrane domain, and cytoplasmic domain with a long 3′-untranslated region (UTR) [11]. These different domains translate into different biological functions of ADAM33, involving cell activation, proteolysis, adhesion, fusion, and intracellular signaling [12]. Genetic studies have demonstrated that ADAM33 may be involved in determining lung function throughout life, associated with increased the risk of therapeutic intervention decline in asthma [13]. Soluble ADAM33 (sADAM33) is identified to promote angiogenesis, defining it as a tissue remodeling gene with potential to affect airflow obstruction and lung functions independently of inflammation [14]. Evidences have shown that ADAM33 functions as a susceptibility target gene for asthma [15], and has an important role in the natural history and possibly the origins of asthma [16]. Besides, the preferential expression of ADAM33 mRNA in smooth muscle, fibroblasts, and myofibroblasts suggest that the abnormalities of its function may link to bronchial hyperresponsiveness (BHR) and airway wall “remodelling” which contributes to the early life origins of asthma. Moreover, a higher expression of ADAM33 protein was detected in asthma patients compared to controls [17]. The ADAM33 expression was also increased as asthma severity increases, which may contribute to the remodeling process that occurs with asthma progression and may have implications for future treatment in severe disease [18]. Genetic polymorphisms might alter the gene function, thus playing a role in disease progression. At present, more than 100 single-nucleotide polymorphisms (SNPs) of the ADAM33 gene have been reported to be associated with asthma and related traits: V4 (rs2787094, 3’UTR, C/G), T + 1 (rs2280089, intron, G/A), T2 (rs2280090, cytoplasmatic domain, G/A), T1 (rs2280091, cytoplasmatic domain, A/G), S2 (rs528557, transmembrane domain, G/C), S1 (rs3918396, transmembrane domain, G/A), Q1 (rs612709, intron, G/A), F + 1 (rs511898, intron, C/T), S + 1 (rs2853209, intron, A/T) and so on. Several polymorphic sites were shown to be associated with asthma risk in different populations [19].
A serial of case-control studies have identified the role of ADAM33 polymorphisms in asthma risk, however, the results still remain inconclusive. For example, Awasthi et al. demonstrated that the V4 C/G of the ADAM33 gene was associated with asthma risk [20], while Anand et al. found that there was no association of ADAM33 V4 polymorphism with asthma risk [21]. Fedorova et al. detected a significant association between A allele of ST + 4 and asthma susceptibility in Russian patients [22], while Dmitrievazdorova et al. found that probably ADAM33 ST + 4 gene was not directly involved in predisposition to asthma in Russian patients [23]. In addition, asthma is a heterogeneous disorder with a complex etiology, and the frequency and severity of asthma differs between different racial and ethnic groups, ages, and economic conditions [24, 25]: the average annual asthma prevalence is higher in children (9.5%) than adults (7.7%); the prevalence of asthma is higher in black persons than white persons; asthma prevalence increases with each successive lower poverty level group [26]. Therefore, we conducted the present meta-analysis to evaluate the ADAM33 gene polymorphisms in asthma risk based on all the available published data to obtain a more reliable result.
Methods
Literature search and identification of eligible studies
A comprehensive literature search was conducted using the electronic databases of English (PubMed, Embase, LISTA, Web of science and Library of Congress) and Chinese (Wanfang and China National Knowledge Infrastructure) to search relevant articles published between January 2000 and June 2018. The Medical Subject Heading (MeSH) terms were: “asthma or asthmatic”, “a disintegrin and metalloprotease 33 or ADAM33”, and “polymorphism or SNP or variant”. The equivalent Chinese words were used in the Chinese databases. The references of retrieved articles were manually checked to obtain more data. When the same department published more than one article on the same subject among the same participants, only the most recent full-text article was included into the final analysis.
Inclusion and exclusion criteria for article screening
Eligible articles must meet the following inclusion criteria: 1) case-control study evaluating the association between ADAM33 polymorphisms and asthma susceptibility; 2) the patients were diagnosed with asthma by chest physicians according to Global Initiative for Asthma (GINA) guidelines [27]; 3) the controls were age-matched unrelated participants of the same ethnicity with no history of asthma or other chronic pulmonary diseases; 4) the alleles and genotypes for each SNP were available to extract, and the results were expressed as odds ratio (OR) with its corresponding 95% confidence interval (CI); and 5) the genotypes in controls must be consistence with the Hardy-Weinberg equilibrium (HWE, P > 0.05). The exclusion criteria were: 1) conference papers, reports, comments or review articles; 2) with duplicated data; 3) without the control group; and 4) the data couldn’t be extracted.
Quality assessment and data extraction
Two of our authors independently estimated the information extracted from case-control studies based on our inclusion and exclusion criteria. Any disagreement was resolved by discussing with the third author to reach a final consensus. The following information was retrieved: the name of first author, publication year, country, ethnicity, mean age, number of cases and controls, genotyping methods and SNPs.
Statistical analysis
All the statistical analyses were carried out using the RevMan software (version 5.3). The OR with 95% CI was employed to calculate the association between ADAM33 polymorphisms and asthma risk. The pooled OR was determined using the Z test with P-value less than 0.05 (considered as significant). For each SNP, the allelic model, homozygous model, heterozygous model, dominant model and recessive model were examined to evaluate its effect. For subgroup analysis, only the allele and genotype differences from control subjects were analyzed. The between-study heterogeneity was assessed by the Q test and the I2 test. The fixed-effect model was used when the P-value of the Q test was more than 0.10 and I2 of the I2 test was less than 50%; otherwise, the random-effect model was used. To estimate whether our results were substantially influenced by the presence of any individual study, a sensitivity analysis was performed by systematically omitting each included case-control study and recalculating the significance of the result. The funnel plot was used to examine the publication bias. In the absence of publication bias, it assumes that studies with high precision (large study effects) will be plotted near the average, and studies with low precision (small study effects) will be spread evenly on both sides of the average, creating a roughly funnel-shaped distribution. Deviation from this shape can indicate publication bias.
Results
Main characteristics of included studies
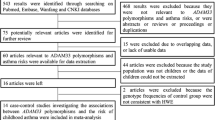
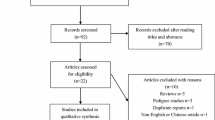
According to combination of the key searching MeSH terms and the online databases, we firstly obtained 375 available case-control studies. After applying the inclusive and exclusive criteria, a total of 63 articles were finally screened out, including 13,280 asthma patients and 13,340 controls. Figure 1 showed the flow chart of literature search and study selection. The 63 included articles were conducted in twenty countries: China [28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61], Korea [62], Japan [63, 64], Germany [65], Thailand [66], Brazil [67], India [68,69,70,71,72,73], Colombia [74], Saudi Arabia [75], Czech Republic [76], Turkey [77], Egypt [78, 79], Portugal [80], Jordan [81], Iran [82,83,84], Pakistan [85,86,87], Iraq [88], Venezuela [89], Mixed (Dutch and America) [90]. Of the retrieved articles, forty-one were written in English and twenty-two were in Chinese; fifty-three were performed in Asian populations, seven were in Caucasian populations and two were in African populations; twenty-one were conducted in children, fifteen were in adults, and others were in mixed populations or not mention the study populations. The sample size ranged from 89 to 1872. 11 SNPs of ADAM33 gene were finally identified: T + 1 (rs2280089, G/A), T1 (rs2280091, A/G), T2 (rs2280090, G/A), Q1 (rs612709, G/A), S1 (rs3918396, G/A), S2 (rs528557, G/C), S + 1 (r2853209, A/T), F + 1 (rs511898, C/T), ST + 4 (rs44707, C/A), ST + 5 (rs597980, C/T) and V4 (rs2787094, C/G). The main characteristics of included studies were presented in Table 1.
Meta-analysis of association between ADAM33 genetic polymorphisms and asthma risk
If the number of available case-control articles for each SNP or subgroup analysis was equal to or more than three, we will conduct the statistical analyses. Table 2 listed the meta-analysis results of association between ADAM33 genetic variants and asthma risk in total population and subgroup analysis by ethnicities and ages.
T + 1 (rs2280089, G/A)
Ten articles included 1943 asthma patients and 2940 controls. Eight articles were conducted in Asian population and two were in Caucasian populations. Our result found that only AA genotype was associated with asthma risk when compared with the GG genotype (AA vs. GG: OR = 1.55, 95% CI = 1.01–2.38, P = 0.04) in the fixed-effect model. Subgroup analysis by ethnicities showed that the A allele and AA+AG genotype of ADAM33 T + 1 polymorphism was not associated with asthma risk in Asians population (P > 0.05). This genetic polymorphism did not increase the risk of childhood asthma as well (A vs. G: OR = 1.16, 95% CI = 0.54–2.49, P = 0.71; AA+AG vs. GG: OR = 1.17, 95% CI = 0.51–2.69, P = 0.71) in the random-effect model.
T1 (rs2280091, A/G)
Thirty-four case-control studies included 7873 asthma patients and 8537 controls. A significant between-study heterogeneity was observed, and the random-effect model was used. Overall, we did not detect a significant association between T1 polymorphism and asthma risk in total population. Subgroup analysis by ethnicities and ages did not find a significant association between this genetic polymorphism and asthma susceptibility in any comparison groups.
T2 (rs2280090, G/A)
Twenty-nine case-control studies included 6799 cases and 7998 controls. Our result detected a significant association between this genetic polymorphism and asthma risk under all the genetic models (A vs. G: OR = 1.32, 95% CI = 1.08–1.60, P = 0.006; AA vs. GG: OR = 1.77, 95% CI = 1.09–2.87, P = 0.02; AG vs. GG: OR = 1.25, 95% CI = 1.03–1.52, P = 0.02; AA+AG vs. GG: OR = 1.30, 95% CI = 1.06–1.61, P = 0.01; AA vs. AG + GG: OR = 1.62, 95% CI = 1.07–2.46, P = 0.02). Figure 2 showed the T2 polymorphism in asthma risk under the allelic model. Subgroup analysis by ethnicities showed that both the A allele and AA+AG genotypes of ADAM33 T2 polymorphism was associated with increased the asthma risk in Asian populations (A vs. G: OR = 1.44, 95% CI = 1.13–1.85, P = 0.004; AA+AG vs. GG: OR = 1.45, 95% CI = 1.11–1.88, P = 0.006), not in Caucasian populations (A vs. G: OR = 0.93, 95% CI = 0.80–1.09, P = 0.38; AA+AG vs. GG: OR = 0.88, 95% CI = 0.67–1.15, P = 0.34). Subgroup analysis by ages demonstrated that A allele and AA+AG genotypes of T2 variant was associated with increased the childhood asthma risk (A vs. G: OR = 1.45, 95% CI = 1.11–1.89, P = 0.006; AA+AG vs. GG: OR = 1.43, 95% CI = 1.07–1.92, P = 0.02), not with adult patients (A vs. G: OR = 1.04, 95% CI = 0.83–1.29, P = 0.75; AA+AG vs. GG: OR = 1.03, 95% CI = 0.81–1.32, P = 0.79).
Q1 (rs612709, G/A)
Eight relevant articles contained 1892 patients and 2896 controls. All the 8 articles were conducted in Asian population. Our result found Q1 polymorphism was related with increased the risk of asthma under four genetic comparison models (A vs. G: OR = 1.26, 95% CI = 1.10–1.45, P = 0.0007; AA vs. GG: OR = 1.78, 95% CI = 1.11–2.84, P = 0.02; AA+AG vs. GG: OR = 1.27, 95% CI = 1.09–1.48, P = 0.002; AA vs. AG + GG: OR = 1.65, 95% CI = 1.04–2.61, P = 0.03) as shown in Fig. 3. Subgroup analysis by ages showed that Q1 variant was not associated with childhood asthma.
S1 (rs3918396, G/A)
Sixteen articles included 3756 asthma patients and 3630 controls. The statistical analysis showed that there was no significant association between S1 polymorphism and asthma risk under any genetic comparison models. This insignificance was also detected in Asians and Caucasians in subgroup analysis by ethnicities.
S2 (rs528557, G/C)
Twenty-nine articles included 6492 patients and 7512 controls. Our analysis did not found a significant association between S2 polymorphism and asthma risk under any comparison models. Subgroup analysis by ethnicities and ages also did not find a significant relationship.
S + 1 (r2853209, A/T)
Five articles included 1006 asthma patients and 2112 controls. No significant association was detected between S + 1 polymorphism and asthma risk.
F + 1 (rs511898, C/T)
Twenty-two articles contained 5149 patients and 4795 controls. A significant association between F + 1 polymorphism and asthma risk was found under each comparison models (T vs. C: OR = 1.17, 95% CI = 1.06–1.30, P = 0.002; TT vs. CC: OR = 1.43, 95% CI = 1.14–1.80, P = 0.002; TC vs. CC: OR = 1.11, 95% CI = 1.01–1.21, P = 0.02; TT + TC vs. CC: OR = 1.15, 95% CI = 1.06–1.26, P = 0.001; TT vs. TC + CC: OR = 1.29, 95% CI = 1.05–1.59, P = 0.02). Subgroup analysis by ethnicities showed this genetic variant was associated with asthma in Asians (T vs. C: OR = 1.15, 95% CI = 1.03–1.28, P = 0.009; TT + TC vs. CC: OR = 1.13, 95% CI = 1.03–1.24, P = 0.01). Subgroup analysis by ages showed it was related with childhood asthma (T vs. C: OR = 1.26, 95% CI = 1.05–1.52, P = 0.01; TT + TC vs. CC: OR = 1.22, 95% CI = 1.07–1.39, P = 0.003).
ST + 4 (rs44707, C/A)
Fifteen articles included 4096 patients and 3767 controls. Our result did not detect a significant association between ST + 4 variant and asthma risk in total population and subgroup analysis by ethnicities. However, subgroup analysis by ages showed that the A allele and AA+CA genotype was associated with childhood asthma (A vs. C: OR = 0.82, 95% CI = 0.69–0.97, P = 0.02; AA+CA vs. CC: OR = 0.81, 95% CI = 0.69–0.95, P = 0.010).
ST + 5 (rs597980, C/T)
Six articles included 1535 patients and 1683 controls. No significant association was found between this genetic polymorphism and asthma risk.
V4 (rs2787094, C/G)
Twenty-eight articles included 7108 patients and 7794 controls. Our result did not detect a significant association between V4 polymorphism and asthma risk. Subgroup analysis by ethnicities showed that it was associated with asthma risk in Caucasians (G vs. C: OR = 1.12, 95% CI = 1.00–1.26, P = 0.04; GG + CG vs. CC: OR = 1.17, 95% CI = 1.01–1.36, P = 0.04). Subgroup analysis by ages showed it was associated with asthma in adults (G vs. C: OR = 1.39, 95% CI = 1.04–1.84, P = 0.02; GG + CG vs. CC: OR = 1.52, 95% CI = 1.04–2.21, P = 0.03).
Association between ADAM33 genetic polymorphisms and asthma severity
According to the GINA guidelines, the confirmed asthma patients were divided into two group: the low-severity group included patients with intermittent or mild persistent asthma attacks; the high-severity group consisted of patients with moderate or severe persistent asthma attacks. Several included case-control studies considered the ADAM33 polymorphisms in asthma severity.
For T1 variant, 7 articles contained 1315 patients and 1223 control. Our result found that only the G allele of ADAM33 T1 polymorphism was associated with low-severity (G vs. A: OR = 1.52, 95% CI = 1.02–2.25, P = 0.04) and high-severity asthma (G vs. A: OR = 1.70, 95% CI = 1.05–2.77, P = 0.03) when compared with the controls as shown in Fig. 4. No significant difference was noted between the high- and low- severity groups in the allele frequencies in this genetic polymorphism. The GG + GA genotypes was not associated with asthma severity in low-severity group (GG + GA vs. AA: OR = 1.60, 95% CI = 0.87–2.92, P = 0.13) or high-severity group (GG + GA vs. AA: OR = 1.77, 95% CI = 0.94–3.35, P = 0.08).
For T2 variant, 8 articles contained 1539 patients and 1466 controls. No significant association was found between the allele and genotype of T2 variant and asthma severity (A vs. G:OR = 1.53, 95% CI = 0.86–2.70, P = 0.15; AA+AG vs. GG: OR = 1.42, 95% CI = 0.74–2.73, P = 0.29 for the low-severity group and A vs. G:OR = 2.05, 95% CI = 0.80–5.26, P = 0.14; AA+AG vs. GG: OR = 2.37, 95% CI = 0.80–6.99, P = 0.12 for the high-severity group).
For S2 variant, 8 articles contained 1398 patients and 1434 controls. Our result showed that this genetic variant was not associated with asthma severity (C vs. G: OR = 1.44, 95% CI = 0.79–2.62, P = 0.24; CC + GC vs. GG: OR = 1.51, 95% CI = 0.57–3.95, P = 0.41 for the low-severity group; C vs. G: OR = 1.47, 95% CI = 0.86–2.51, P = 0.16; CC + GC vs. GG: OR = 1.27, 95% CI = 0.60–2069, P = 0.53 for the high-severity group).
For V4 variant, 8 articles contained 1269 patients and 1347 controls. No significant association was detected (G vs. C: OR = 0.91, 95% CI = 0.60–1.37, P = 0.65; GG + CG vs. CC OR = 1.00, 95% CI = 0.66–1.51, P = 0.99 for the low-severity group; G vs. C: OR = 1.13, 95% CI = 0.60–2.15, P = 0.70; GG + CG vs. CC OR = 1.11, 95% CI = 0.54–2.27, P = 0.78 for the high-severity group).
Association between ADAM33 polymorphisms and asthma risk in Chinese population
Thirty-four case-control studies were conducted in Chinese population, including 6136 asthma patients and 5016 controls. Table 3 listed the summary analysis of ADAM33 genetic polymorphisms and asthma risk among Chinese population. Our result showed that the alleles and genotypes of ADAM33 T2 (A vs. G: OR = 1.70, 95% CI = 1.33–2.17, P < 0.0001; AA+AG vs. GG: OR = 1.71, 95% CI = 1.33–2.21, P < 0.0001), Q1 (A vs. G: OR = 1.45, 95% CI = 1.01–2.07, P = 0.04; AA+AG vs. GG: OR = 1.58, 95% CI = 1.02–2.43, P = 0.04), and F + 1 (T vs. C: OR = 1.13: OR = 1.03–1.24, P = 0.01; TT + TC vs. CC: OR = 1.15: OR = 1.02–1.30, P = 0.02) variants were significantly associated with increased the asthma risk in Chinese asthma patients. This statistically significance was not detected in other genetic polymorphisms.
Sensitivity analysis and publication bias
We conducted a sensitivity analysis to assess if our results were substantially affected by the presence of any individual study. Each included case-control study was deleted at each time to recalculate the significance of the results. Our result suggested that the pooled ORs were not significantly changed. The funnel plot was applied to evaluate the publication bias, and the shape of the funnel plot was symmetrical, further indicating that no publication bias existed in this meta-analysis as shown in Fig. 5.
Discussion
In this meta-analysis, we totally screened out 63 case-control articles. Our results found that T2, Q1 and F + 1 polymorphisms of ADAM33 gene were associated with asthma risk in total population. Subgroup analysis by ethnicities showed that the alleles and genotypes of T2, Q1 and F + 1 polymorphisms were associated with asthma susceptibility among Asian populations, while V4 polymorphisms was related with asthma risk among Caucasian populations. Subgroup analysis by ages showed that T2, F + 1 and ST + 4 polymorphisms were associated with childhood asthma, while Q1 and V4 were associated with asthma risk in adults. Subgroup analysis by asthma severity showed that the G allele of ADAM33 T1 polymorphism was associated with the severity of asthma when compared with the controls. In addition, T2, Q1 and F + 1 polymorphisms of ADAM33 were significantly associated with increased the asthma risk in Chinese asthma patients. Our results were inconsistence with previous meta-analyses conducted by Liu et al. which found that S2 polymorphism may contribute to asthma susceptibility [91]; Deng et al. which suggested that T1 polymorphism might be a potential susceptible predictor of asthma for Asian children [92]; Zheng et al. which showed that ADAM33 V4 polymorphism contributes to the risk of asthma and may be utilized as a biomarker for the early diagnosis of asthma [93]; Sun et al. which demonstrated that ADAM33 polymorphisms T1, T2, V4, S2 and ST + 4 were significantly associated with a high risk of childhood asthma [94].
Asthma, caused by obstruction of the conducting airways of the lung, is a chronic inflammatory disease in which cells of the innate and adaptive immune systems act together with epithelial cells, leading to mucus overproduction, bronchial hyperreactivity, airway wall remodeling and airway narrowing [95, 96]. It is a complex disease influenced by the interaction of gene-gene, gene-protein and protein-protein, and the environmental issue, which plays a significant role in causing asthma pathogenesis and makes it difficult to define precisely in population studies. Because of differences in the influence of genes and environment, there is a wide range of disease heterogeneity and severity in asthma [97]. Identification and measurement of significant biomarkers are particularly crucial in clinical research of asthma to characterize the population and to associate the disease with therapeutic effects [98].
ADAM33, a membrane-anchored protease expressed in multiple airway cell types, is known to be an important gene involved in asthma pathogenesis. It is upregulated during acute or chronic lung inflammation, and recent functional and genetic analyses have linked it to disease development [99]. Studies have found that ADAM33 can be detected as sADAM33 in asthmatic airways in which high levels of sADAM33 are correlated with reduced lung function [100]. The expression of serum ADAM33 was up-regulated in asthmatic patients along with the severity of asthma [101]. In addition, ADAM33 was positively correlated with serum IL-4 and IL-13, implying that the expression of ADAM33 may be regulated by Th2 type cytokines [101]. The identification of ADAM33 as a major risk factor involved in the pathogenesis of BHR and airway wall remodeling provides insight into the pathogenesis of asthma and represents a novel therapeutic target [102]. Taken together, it can be concluded that ADAM33 mediates the airway inflammatory and remodeling effects and is a potential therapeutic target for asthma.
ADAM33 is one of the positionally cloned asthma associated genes, which has been identified as susceptibility gene for asthma in different populations. Genetic polymorphism is the occurrence of two or more clearly different morphs or forms, also referred to as alternative phenotypes, in the population of a species, thus involving in disease progression. Van Eerdewegh et al. performed a genome-wide scan on 460 Caucasian families and firstly identified the ADAM33, a putative asthma susceptibility gene identified by positional cloning in an outbred population, provided insights into the pathogenesis and natural history of asthma. Subsequently, a large number of related studies have been carried out. However, the results were inconclusive in different populations, even in the same population. The study conducted by Werner et al. evaluated the association between ADAM33 polymorphisms and asthma and related traits in two German populations, and replicated the results of Van Eerdewegh et al., but most of the associated SNPs were at non-identical positions in the German, UK and US samples [103]. Jongepier et al. found that ADAM33 S2 variant was shown to be associated with the development of asthma and disease progression, possibly related to enhanced airway remodeling in Dutch Caucasian individuals [104]. Noguchi et al. surveyed 155 families and confirmed the involvement of ADAM33 in the development of childhood asthma among the Japanese [105]. Lind et al. studied 659 mild or moderate-severe subjects, and concluded that the ADAM33 gene was not an important risk factor for asthma or asthma-associated phenotypes in Mexicans or in Puerto Ricans [106]. Kedda et al. investigated the role of 10 SNPs of ADAM33 gene in asthma risk and asthma severity in Australian Caucasian population, and did not detect significant association between any one of the SNPs and asthma or asthma severity, but there was a significant global haplotype association with asthma and disease severity [107]. Murk et al. found that SNPs in frequently replicated asthma risk genes ADAM33 revealed no association [108]. Blakey et al. conducted the analysis in a large, nationally representative, white ethnicity population (n = 7703) and found that S2, not F + 1 polymorphism of ADAM33 influenced asthma risk in the UK population [109]. Although several meta-analyses had been conducted to identify the role of ADAM33 variants in asthma risk, the results still remain inconclusive [110]. Therefore, we conducted the present meta-analysis.
China is the world’s largest developing country and has the largest population. The prevalence of asthma significantly varies among different regions of China. Although the incidence rate of asthma is lower in Chinese children and adults than that in developed countries, the prevalence has been on the rise during the past three decades. Besides, 40% of asthma patients in China are uncontrolled, resulting in the increase of hospitalization, emergency room visit and absence of work or school [111]. Even though asthma control has been improved in some cities, the level of asthma control in China is still far from ideal at present [112]. Environmental exposure and genetic predisposition were identified as risk factors for asthma in China [113]. In our study, we found that T2, Q1 and F + 1 polymorphisms of ADAM33 were significantly associated with increased the asthma risk in Chinese asthma patients, indicating that ADAM33 might be a promising biomarker and therapeutic option for asthma.
In addition, several studies focused on ADAM33 as a major hub and identified some proteins whose interaction with ADAM33 had been associated with asthma. Kong et al. found that the combination of ADAM33, Smad7, and LIGHT would be a reliable and useful childhood asthma model for prediction and diagnosis [114]. Vishweswaraiah et al. recognized amyloid β (A4) precursor protein, ataxin-7, α4-integrin, and α5-integrin, which were involved in airway hyperresponsiveness, and through the interaction with ADAM33, they might have potential relevance in asthma [115]. Sangeetha et al. identified few proteins, such as APP, ATXN7, ITGA4, and ITGA5, whose interaction with the ADAM33 might have been associated with asthma [116]. Besides, ADAM33 might also interact with other environmental factors. Bukvic et al. demonstrated several novel significant interactions between polymorphisms in 20p13-p12 and early-life environmental tobacco smoke exposure with asthma presence and, amongst asthmatics, a significant association with the severity of their disease [117].
Several limitations were presented in our meta-analysis. Firstly, half of the included articles were conducted in Chinese population, while other populations should be discussed. Secondly, the between-study heterogeneity was detected in most genetic comparison models, which might influence our results. It probably because these included articles were conducted in different geographical regions, and asthma patients might receive different treatment, which might influences the asthma phenotype, leading to heterogeneity. Thirdly, the haplotypes among these SNPs should be considered because identifying these statistical associations and few alleles of a specific haplotype sequence can facilitate identifying all other such polymorphic sites that are nearby on the chromosome. Lastly, interaction of gene-gene and gene-environmental factors should be considered in future studies.
Conclusions
In conclusions, our results demonstrated that T2, Q1 and F + 1 polymorphisms of ADAM33 gene might be risk factors for asthma susceptibility. Moreover, T2, Q1 and F + 1 polymorphisms were associated with asthma susceptibility among Asian populations, whereas V4 polymorphism was associated with asthma among Caucasian populations; T2, F + 1 and ST + 4 polymorphisms were associated with childhood asthma, whereas Q1 and V4 were associated with asthma risk in adults; T1 G allele polymorphism was associated with low-severity and high-severity asthma risk. Besides, ADAM33 T2, Q1 and F + 1 polymorphisms were significantly associated with increased the asthma risk in Chinese asthma patients. Future case-control studies with large populations are still needed.
Abbreviations
- ADAM33 :
-
A disintegrin and metalloprotease 33
- BHR:
-
Bronchial hyper-responsiveness
- CI:
-
Confidence interval
- MeSH:
-
Medical Subject Heading
- OR:
-
Odds ratio
- SNP:
-
Single nucleotide polymorphism
- UTR:
-
Untranslated region
References
Beuther DA, Krishnan JA. Finding asthma: building a Foundation for Care and Discovery. Am J Respir Crit Care Med. 2017;196:401–2.
Patadia MO, Murrill LL, Corey J. Asthma: symptoms and presentation. Otolaryngol Clin N Am. 2014;47:23–32.
Nunes C, Pereira AM, Morais-Almeida M. Asthma costs and social impact. Asthma Res Pract. 2017;3:1.
Martinez FD. Genes, environments, development and asthma: a reappraisal. Eur Respir J. 2007;29:179–84.
Toskala E, Kennedy DW. Asthma risk factors. In International Forum of Allergy & Rhinology. 2015:S11–6.
Vos T, Abajobir AA, Abate KH. Global, regional, and national incidence, prevalence, and years lived with disability for 328 diseases and injuries for 195 countries, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet. 2017;390:1211–59.
Michel O. Asthma: diagnosis and treatment in general pratice. Rev Med Brux. 2017;38:295–9.
Choby GW, Lee S. Pharmacotherapy for the treatment of asthma: current treatment options and future directions. Int Forum Allergy Rhinol. 2015;5(Suppl 1):S35–40.
Fuchs O, Bahmer T, Rabe KF, Von ME. Asthma transition from childhood into adulthood. Lancet Respir Med. 2017;5:224–34.
Shapiro SD, Owen CA. ADAM-33 surfaces as an asthma gene. N Engl J Med. 2002;347:936.
Van Eerdewegh P, Little RD, Dupuis J, Del Mastro RG, Falls K, Simon J, Torrey D, Pandit S, McKenny J, Braunschweiger K, et al. Association of the ADAM33 gene with asthma and bronchial hyperresponsiveness. Nature. 2002;418:426–30.
Holgate ST, Davies DE, Powell RM, Holloway JW. ADAM33: a newly identified gene in the pathogenesis of asthma. Immunol Allergy Clin N Am. 2005;25:655–68.
Holgate ST, Davies DE, Powell RM, Holloway JW. ADAM33: a newly identified protease involved in airway remodelling. Pulm Pharmacol Ther. 2006;19:3–11.
Davies ER, Kelly JF, Howarth PH, Wilson DI, Holgate ST, Davies DE, Whitsett JA, Haitchi HM. Soluble ADAM33 initiates airway remodeling to promote susceptibility for allergic asthma in early life. JCI Insight. 2016;1(11):e87632.
Holgate ST, Yang Y, Haitchi HM, Powell RM, Holloway JW, Yoshisue H, Pang YY, Cakebread J, Davies DE. The genetics of asthma: ADAM33 as an example of a susceptibility gene. Proc Am Thorac Soc. 2006;3:440–3.
Cakebread JA, Haitchi HM, Holloway JW, Powell RM, Keith T, Davies DE, Holgate ST. The role of ADAM33 in the pathogenesis of asthma. Springer Semin Immunopathol. 2004;25:361–75.
Tripathi P, Awasthi S, Husain N, Prasad R, Mishra V. Increased expression of ADAM33 protein in asthmatic patients as compared to non-asthmatic controls. Indian J Med Res. 2013;137:507–14.
Foley SC, Mogas AK, Olivenstein R, Fiset PO, Chakir J, Bourbeau J, Ernst P, Lemiere C, Martin JG, Hamid Q. Increased expression of ADAM33 and ADAM8 with disease progression in asthma. J Allergy Clin Immunol. 2007;119:863–71.
Priya T, Shally A, Peisong G. ADAM metallopeptidase domain 33 (ADAM33): a promising target for asthma. Mediat Inflamm. 2014;2014:572025.
Awasthi S, Tripathi P, Husain N, Ganesh S. 291 association of Adam33 gene polymorphism with bronchial asthma. Pediatr Res. 2010;68:150.
Anand MP, Visweshwaraiah S, Nallur R, Biligere J, Sindaghatta C, Holla A. Association between genetic polymorphisms of ADAM33 with asthma and its phenotypes. Eur Respir J. 2015;46:PA1230.
Fedorova Y, Karunas A, Gimalova G, Guryeva L, Ramazanova N, Levashova S, Muhtarova L, Murzina R, Etkina E, Zagidullin S. Association of ADAM33 gene polymorphisms with asthma in Volga-Ural region of Russia. Eur Respir J. 2011;38(Suppl 55):P436.
Dmitrievazdorova E, Gabaeva M, Latysheva E, Voronko O. ADRB2, ADAM33 and AAT gene polymorphisms and IgE-mediated asthma in Russian patients. Eur Respir J. 2011.
Moraes TJ, Sears MR, Subbarao P. Epidemiology of asthma and influence of ethnicity. Semin Respir Crit Care Med. 2018;39:3–11.
Ortega VE, Meyers DA. Implications of population structure and ancestry on asthma genetic studies. Curr Opin Allergy Clin Immunol. 2014;14:381–9.
Loftus PA, Wise SK. Epidemiology of asthma. Curr Opin Otolaryngol Head Neck Surg. 2016;24:245–9.
Kroegel C. Global initiative for asthma (GINA) guidelines: 15 years of application. Expert Rev Clin Immunol. 2009;5:239–49.
Wang P, Liu QJ, Li JS, Li HC, Wei CH, Guo CH, Gong YQ. Lack of association between ADAM33 gene and asthma in a Chinese population. Int J Immunogenet. 2006;33:303–6.
Qiu YM, Luo YL, Lai WY, Qiu SJ. Association between ADAM33 gene polymorphism and bronchial asthma in South China Han population. J Southern Med Univ. 2007;27:485–7.
Hu Z. Lack of association between ADAM33 gene polymorphisms and asthma in a Huadong Chinese Han population [Master's degree]. Fudan university; 2008. Retrieved from China National Knowledge Infrastructure (2009019076).
Su D, Zhang X, Sui H, Lü F, Jin L, Zhang J. Association of ADAM33 gene polymorphisms with adult allergic asthma and rhinitis in a Chinese Han population. Bmc Med Genetics. 2008;9:82.
Hong-Chuan YU, Chen GQ, Yan-Qi LI, Wang LJ, Jiang CS, Meng LS. Association of T1 polymorphism in disintegrin and metalloprotease 33 gene with asthma. J Clin Pediatr. 2011;29:852–5.
Jie Z, Hu Z, Bai C, Jin M. ADAM33 gene polymorphisms associate with asthma susceptibility and severity in East China Han population. J Asthma Res. 2011;48:979–85.
Qu S, Sun D, Wang Y, Zhang C, Lv Y, Yao L. Association of ADAM33 polymorphisms with childhood asthma in a northern Chinese population. Exp Mol Pathol. 2011;91:775–9.
Chiang CH, Lin MW, Chung MY, Yang UC. The association between the IL-4, ADRβ2 and ADAM 33 gene polymorphisms and asthma in the Taiwanese population. J Chin Med Assoc. 2012;75:635–43.
Chi XY, Xiao W, Wang XQ, Qing LI, Wang J, Jiang HJ, Wang LC. Study on relationship between genetic polymorphisms of ADAM33 and susceptibility to bronchial asthma. Chin J Asthma. 2012;6:12–6.
Xie CL, Zhao YJ, Liao ZB, Yang L, Qiu FM, Chen HY, Wen YT, Yan JH, Cai J, Jia-Chun LV. The interaction between ADAM33 gene polymorphism and smoking in chronic obstructive pulmonary disease and asthma. Guangdong Med J. 2013;34:3402–6.
Xue W, Han W, Zhou ZS. ADAM33 polymorphisms are associated with asthma and a distinctive palm dermatoglyphic pattern. Mol Med Rep. 2013;8:1795–800.
Yilihamu N, Wushouer Q, Arkin K, Xin H, Yadav U. Association of a disintegrin and metalloprotease 33 gene polymorphisms with asthma. Mol Clin Oncol. 2014;2:1076–80.
Yao L, Cui S, Wushouer Q, Xia Y, Yan F, Simayi M, Yang Z, Wang J. Relationship between disintegrin and metalloproteinase gene polymorphism of bronchial asthma and its severity in Uygur population. Zhonghua Yi Xue Za Zhi. 2014;94:1866–9.
Wang J, Simayi M, Wushouer Q, Xia Y, He Y, Yan F, Zhang J, Cui S, Wen H. Association between polymorphisms in ADAM33, CD14, and TLR4 with asthma in the Uygur population in China. Genet Mol Res Gmr. 2014;13:4680–90.
Gong L, Xiang Hong LU, Bin YU. To investigate the relationship between ADAM33 gene polymorphism and dust mite allergic asthma susceptibility. Guide China Med. 2015;13:20–1.
Fan JG, Wang ZA, Zhao HX. The ADAM33 S2 polymorphism is associated with susceptibility to pediatric asthma in the Chinese Han population. Genet Testing Mol Biomarkers. 2015;19:573–8.
Xin HU, Simayi M, Cui S, Wang J, Yan F, Sun H, Huang Y, Wushouer Q. Association between polymorphism of S2,ST+4,ST+5 locus in ADAM33 gene and bronchial asthma in Xinjiang Uyghur population. J Xinjiang Med Univ. 2016;39:261–4.
Bo S, Rong L, Wang CC, Jing R, Yan S, Yang YL, Lin YY. ADAM33 gene polymorphisms identified to be associated with asthma in a Chinese Li population. Biomedical Rep. 2017;6:323–8.
Li H, Li Y, Zhang M, Xu G, Feng X, Xi J, Zhao B. Associations of genetic variants in ADAM33 and TGF-beta1 genes with childhood asthma risk. Biomed Rep. 2014;2:533–8.
Wang J, Wu Y, Duolikun M. Correlation of asthma with ADAM33 SNP in Uygur and Han children in Urumqi. J Clin Pediatr. 2016;34:926–9.
Zhu S, Naren G. Association between polymorphism of T1 locus allele in ADAM33 gene and bronchial asthma in Inner Mongolia Han and Mongolian population. Journal of Clinical Pulmonary Medicine. 2017;22:61–4.
Liang S, Deng J, Wei X, Chen Z, Gong C, Yang M, Wang Z. S2 and T2 ADAM33 polymorphisms and haplotype associated with asthma in the Zhuang population in Guangxi, China. Int J Clin Exp Med. 2016;9:23691–6.
Yu S, Qu S. Relationship between T2 locus polymorphism of ADAM33 gene and bronchial asthma. Chin J Biochem Pharm. 2017;37:412–4 416.
Ye H, Luo Y, Lai W. ADAM33 polymorphisms with asthma susceptibility and severity in adolescent asthma. J Clin Intern Med. 2009;26:383–5.
Jin J, Jiang Y, Liu W, Li C. Study on association between IL-4 R, IL-13 and ADAM33 gene mutation and asth matic patients of with bronchial asthma in Han nationality of Wannan area. Chin J Immunol. 2011;27:135–9.
Zhao B, Li H, Zhang H, Liu Q. Association of polymorphism of V4 and T2 locus alleles in ADAlM33 gene with asthma susceptibility. J Chin Physician. 2012;4:1596–9.
Ren L. Association between ADAM33, LTS pathway, ADRB2 genes polymorphisms and childhood asthma [Master's degree]. ChongQing Med Univ. 2011. Retrieved from China National Knowledge Infrastructure (1011174222).
Qiao L. Association of polymorphism of T2 locus allele in ADAM33 gene with bronchial asthma in Xinjiang Han population [Master's degree]. Xinjiang Med Univ. 2015. Retrieved from China National Knowledge Infrastructure (1015410579).
Zhang L. Association between ADAM33, IL-4R genes polymorphisms and 153 childhoods with asthma in Guiyang [Master's degree]. Guizhou Med Univ. 2016. Retrieved from China National Knowledge Infrastructure (1016242357).
Lv J. Correlation of ADAM33 V4 gene polymorphism、vitamin D levels with asthma children in Urumqi [Master's degree]. Xinjiang Med Univ. 2017. Retrieved from China National Knowledge Infrastructure (1017172454).
Huang Y. Association between polymorphism of F+1, S2 locus allele in ADAM33 gene and bronchial asthma in Xinjiang Kazak population [Master's degree]. Xinjiang Med Univ. 2017. Retrieved from China National Knowledge Infrastructure (1017173325).
Yang Z. Association study between the polymorphism of S2 and T2 loci of ADAM33 gene and Asthma in Henan Chinese Han population [Master's degree]. Xi'an Jiaotong Univ. 2011. Retrieved from China WanFang database (XWXJTU201306170000046194).
Wei X. The polymorphisms of T1 and V4 locuses of ADAM33 gene in asthma paients of Guangxi population [Master's degree]. Guangxi Medical Univ. 2012. Retrieved from China National Knowledge Infrastructure (1012349438).
Xiong JY, He QQ, Jiang ZQ, Li JF. Association of polymorphism of TI locus a allele in ADAM33 gene with bronchial asthma. J Appl Clin Pediatr. 2009;24:1241–3.
Lee JH, Park HS, Park SW, Jang AS, Uh ST, Rhim T, Park CS, Hong SJ, Holgate ST, Holloway JW. ADAM33 polymorphism: association with bronchial hyper-responsiveness in Korean asthmatics. Clin Exp Allergy. 2004;34:860–5.
Hirota T, Hasegawa K, Obara K, Matsuda A, Akahoshi M, Nakashima K, Shirakawa T, Doi S, Fujita K, Suzuki Y. Association between ADAM33 polymorphisms and adult asthma in the Japanese population. Clin Exp Allergy. 2006;36:884–91.
Miyake Y, Tanaka K, Arakawa M. ADAM33 polymorphisms, smoking and asthma in Japanese women: the Kyushu Okinawa maternal and child health study. Int J Tuberc Lung Dis. 2012;16:974–9.
Schedel M, Depner M, Schoen C, Weiland SK, Vogelberg C, Niggemann B, Lau S, Illig T, Klopp N, Wahn U. The role of polymorphisms in ADAM33, a disintegrin and metalloprotease 33, in childhood asthma and lung function in two German populations. Respir Res. 2006;7:91.
Thongngarm T, Jameekornrak A, Limwongse C, Sangasapaviliya A, Jirapongsananuruk O, Assawamakin A, Chaiyaratana N, Luangwedchakarn V, Thongnoppakhun W. Association between ADAM33 polymorphisms and asthma in a Thai population. Asian Pacific J Allergy Immunol. 2008;26:205–11.
de Faria IC, de Faria EJ, Toro AA, Ribeiro JD, Bertuzzo CS. Association of TGF-beta1, CD14, IL-4, IL-4R and ADAM33 gene polymorphisms with asthma severity in children and adolescents. J Pediatr. 2008;84:203–10.
Bijanzadeh M, Ramachandra NB, Mahesh PA, Mysore RS, Kumar P, Manjunath BS, Jayaraj BS. Association of IL - 4 and ADAM33 gene polymorphisms with asthma in an Indian population. Lung. 2010;188:415–22.
Awasthi S, Tripathi P, Ganesh S, Husain N. Association of ADAM33 gene polymorphisms with asthma in Indian children. J Hum Genet. 2011;56:188–95.
Tripathi P, Awasthi S, Prasad R, Husain N, Ganesh S. Association of ADAM33 gene polymorphisms with adult-onset asthma and its severity in an Indian adult population. J Genet. 2011;90:265.
Tripathi P, Awasthi S, Prasad R, Ganesh S. Haplotypic association of ADAM33 (T+1, S+1 and V - 3) gene variants in genetic susceptibility to asthma in Indian population. Ann Hum Biol. 2012;39:479–83.
Awasthi S, Tripathi P, Prasad R, Ganesh S. Association of a disintegrin and metalloprotease 33 gene polymorphisms and their haplotypes with asthma in the north indian population. Indian J Med Sci. 2016;68:54–62.
Sinha S, Singh J, Jindal SK. Significant association of ADAM33 V4C>G polymorphisms with asthma in a north Indian population. Bioresearch Commun. 2015;1:62–8.
Vergara CI, Acevedo N, Jiménez S, Martínez B, Mercado D, Gusmão L, Barnes KC, Caraballo L. A six-SNP haplotype of ADAM33 is associated with asthma in a population of Cartagena, Colombia. Int Arch Allergy Immunol. 2010;152:32–40.
Al-Khayyat AI, Al-Anazi M, Warsy A, Vazquez-Tello A, Alamri AM, Halwani R, Alangari A, Al-Frayh A, Hamid Q, Al-Muhsen S. T1 and T2 ADAM33 single nucleotide polymorphisms and the risk of childhood asthma in a Saudi Arabian population: a pilot study. Annals Saudi Med. 2012;32:479–86.
Godava M, Kopriva F, Bohmova J, Vodicka R, Dusek L, Cvanova M, Muzik J, Markova M, Schneiderova E, Vrtel R. Association of STAT6 and ADAM33 single nucleotide polymorphisms with asthma bronchiale and IgE level and its possible epigenetic background. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2012;156:236–47.
Bora E, Arikan-Ayyildiz Z, Firinci F, Çankaya T, Giray-Bozkaya Ö, Uzuner N, Ülgenalp A. ADAM33 gene polymorphisms are not associated with asthma in Turkish children. Pediatr Allergy Immunol Pulmonol. 2012;25:97–100.
El-Falaki MM, Wilson MM, Ezzat GM, Mokhtar DA, Baz MSE, Hamed DH. A disintegrin and metalloproteinase 33 (ADAM33) gene polymorphism association with asthma in Egyptian children. Egypt J Med Human Genet. 2013;14:55–62.
Shalaby SM, Abdul-Maksoud RS, Abdelsalam SM, Abdelrahman HM, Almalky MAA. ADAM33 and ADAM12 genetic polymorphisms and their expression inEgyptian children with asthma. Ann Allergy Asthma Immunol. 2016;116:31–6.
Berenguer AG, Fernandes AT, Oliveira S, Rodrigues M, Ornelas P, Romeira D, Serrão T, Rosa A, Câmara R. Genetic polymorphisms and asthma: findings from a case–control study in the Madeira island population. Biol Res. 2014;47:40–8.
Zihlif M, Zihlif N, Obeidat NM, Mahafza T, Froukh T, Ghanim MT, Al-Akhras FM, Naffa R. Association between ADAM33 polymorphisms and susceptibility with adult and childhood asthma among Jordanians. Genet Testing Mol Biomarkers. 2014;18:767–74.
Karimi MRZ, Faridhosseini R, Abbaszadegan MR, Azad FJ, Shirkani A, Riyahi A, Montazar M, Gholamin M. Association of ADAM33 gene polymorphisms with allergic asthma. Iranian J Basic Med Sci. 2014;17:716–21.
Zeinaly I, Sadeghi-Shabestrai M, Babaloo Z, Razavi A, Sajay-Asbaghi M, Sadigh-Eteghad S, Kazemi T. Investigating the association of ADAM33 single nucleotide polymorphisms (SNPs) with susceptibility to allergic asthma in Azerbaijan population of Iran: a case-control study. Iran J Allergy Asthma Immunol. 2017;16:378–85.
Farjadian S, Moghtaderi M, Hoseini-Pouya B-A, Ebrahimpour A, Nasiri M. ADAM33 gene polymorphisms in southwestern Iranian patients with asthma. Iranian J Basic Med Sci. 2018;21:813–7.
Raja GK, Khan M, Saba N, Saqlain RM, Ahmed N, Mansoor A. Association of Adam33 Gene SNPS with Asthma in a Local Pakistani Population. 2014;2:97–103.
Sabar MF, Ghani MU, Shahid M, Sumrin A, Ali A, Akram M, Tariq MA, Bano I. Genetic variants of ADAM33 are associated with asthma susceptibility in the Punjabi population of Pakistan. J Asthma Res. 2016;53:341–8.
Saba N, Yusuf O, Rehman S, Munir S, Noor A, Saqlain M, Mansoor A, Raja GK. Single nucleotide polymorphisms in asthma candidate genes TBXA2R , ADAM33 FCER1B and ORMDL3 in Pakistani asthmatics a case control study. Asthma Res Pract. 2018;4:4–9.
Khadhim MM, AL-Damerchi AT. 3 MAK: Evaluation of A Disintegrin and Metalloprotein33 Gene Polymorphism in Bronchial Asthma. Al-Qadisiah Med J. 2015;11:1–9.
Martínez D, Lema D, Del CMD, Hipólito GA, Valentina GJ, De Sanctis JB. Single nucleotide polymorphisms V4 and T1 of the ADAM33 gene in Venezuelan patients with asthma or chronic obstructive pulmonary disease. Investigacion Clinica. 2016;57:176–86.
Howard TD, Postma DS, Jongepier H, Moore WC, Koppelman GH, Zheng SL, Xu J, Bleecker ER, Meyers DA. Association of a disintegrin and metalloprotease 33 (ADAM33) gene with asthma in ethnically diverse populations ☆. J Allergy Clin Immunol. 2003;112:717–22.
Liu Y, Wang ZH, Zhen W, Lu SJ, Liu Z, Zou LY, Xu JJ. Association between genetic polymorphisms in the ADAM33 gene and Asthma risk: a meta-analysis. Dna Cell Biol. 2014;33:793–801.
Deng R, Zhao F, Zhong X. T1 polymorphism in a disintegrin and metalloproteinase 33 (ADAM33) gene may contribute to the risk of childhood asthma in Asians. Inflamm Res. 2017;66:1–12.
Zheng W, Wang L, Su X, Hu XF. Association between V4 polymorphism in the ADAM33 gene and asthma risk: a meta-analysis. Genet Mol Res Gmr. 2015;14:989–99.
Sun FJ, Zou LY, Tong DM, Lu XY, Li J, Deng CB. Association between ADAM metallopeptidase domain 33 gene polymorphism and risk of childhood asthma: a meta-analysis. Brazilian J Med Biol Res. 2017;50:1–7.
Lambrecht BN, Hammad H. The immunology of asthma. Nat Immunol. 2015;16:45–56.
Erle DJ, Sheppard D. The cell biology of asthma. J Cell Biol. 2014;205:621–31.
Carr TF, Bleecker E. Asthma heterogeneity and severity. World Allergy Organ J. 2016;9:41.
Agache I, Rogozea L. Asthma biomarkers: do they bring precision medicine closer to the clinic? Allergy, Asthma Immunol Res. 2017;9:466–76.
Dreymueller D, Uhlig S, Ludwig A. ADAM-family metalloproteinases in lung inflammation: potential therapeutic targets. Am J Physiol Lung Cell Mol Physiol. 2015;308:L325–43.
Lee JY, Park SW, Chang HK, Kim HY, Rhim T, Lee JH, Jang AS, Koh ES, Park CS. A disintegrin and metalloproteinase 33 protein in patients with asthma: relevance to airflow limitation. Am J Respir Crit Care Med. 2006;173:729–35.
FANG L, CHEN M, ZHANG F, Xin SU, Zhang P, Xin X, Shi Y. Expression of serum ADAM33 in patients with different severity of asthma and the relation to airway chronic inflammation. J Med Postgraduates. 2017;30:521–4.
Powell RM, Hamilton LM, Holgate ST, Davies DE, Holloway JW. ADAM33: a novel therapeutic target for asthma. Expert Opin Ther Targets. 2003;7:485–94.
Werner M, Herbon N, Gohlke H, Altmüller J, Knapp M, Heinrich J, Wjst M. Asthma is associated with single-nucleotide polymorphisms in ADAM33. Clin Exp Allergy. 2004;34:26–31.
Jongepier H, Boezen HM, Dijkstra A, Howard TD, Vonk JM, Koppelman GH, Zheng SL, Meyers DA, Bleecker ER, Postma DS. Polymorphisms of the ADAM33 gene are associated with accelerated lung function decline in asthma. Clin Exp Allergy. 2004;34:757–60.
Noguchi E, Ohtsuki Y, Tokunaga K, Yamaoka-Sageshima M, Ichikawa K, Aoki T, Shibasaki M, Arinami T. ADAM33 polymorphisms are associated with asthma susceptibility in a Japanese population. Clin Exp Allergy. 2010;36:602–8.
Lind DL, Choudhry S, Ung N, Ziv E, Avila PC, Salari K, Ha C, Lovins EG, Coyle NE, Nazario S. ADAM33 is not associated with asthma in Puerto Rican or Mexican populations. Am J Respir Crit Care Med. 2003;168:1312–6.
Kedda MA, Duffy DB, O'Hehir RE, Thompson PJ. ADAM33 haplotypes are associated with asthma in a large Australian population. Eur J Hum Genet. 2006;14:1027–36.
Murk W, Walsh K, Hsu LI, Zhao L, Bracken MB, Dewan AT. Attempted replication of 50 reported asthma risk genes identifies a SNP in RAD50 as associated with childhood atopic asthma. Hum Hered. 2011;71:97–105.
Blakey JD, Sayers I, Ring SM, Strachan DP, Hall IP. Positionally cloned asthma susceptibility gene polymorphisms and disease risk in the British 1958 Birth cohort. Thorax. 2009;64:381–7.
Tizaoui K, Hamzaoui K, Hamzaoui A. Update on asthma genetics: results from meta-analyses of candidate gene association studies. Curr Mol Med. 2017;17:647–67.
Wang G, Wang F, Gibson PG, Guo M, Zhang WJ, Gao P, Zhang HP, Harvey ES, Li H, Zhang J. Severe and uncontrolled asthma in China: a cross-sectional survey from the Australasian severe asthma network. J Thorac Dis. 2017;9:1333–44.
Lin JT, Wang W, Zhou X, Wang C, Huang M, Cai S, Chen P, Lin Q, Zhou J, Gu Y. Evaluation of asthma control from China national asthma control surveys. In: Ers International Congress; 2017. p. PA1125.
Yan C, Wong GW, Jing L. Environmental exposure and genetic predisposition as risk factors for asthma in China. Allergy, Asthma Immunol Res. 2016;8:92–100.
Kong Q, Li WJ, Huang HR, Zhong YQ, Fang JP. Differential gene expression profiles of peripheral blood mononuclear cells in childhood asthma. J Asthma. 2015;52:343–52.
Vishweswaraiah S, Veerappa AM, Mahesh PA, Jayaraju BS, Krishnarao CS, Ramachandra NB. Molecular interaction network and pathway studies of ADAM33 potentially relevant to asthma. Ann Allergy Asthma Immunol. 2014;113:418–24 e411.
V S. Molecular analysis of ADAM33 polymorphisms in relation to airway obstruction and asthma in South Indian population [Doctor's degree]: University of Mysore; 2015.
Bukvic BK, Blekic M, Simpson A, Marinho S, Curtin JA, Hankinson J, Aberle N, Custovic A. Asthma severity, polymorphisms in 20p13 and their interaction with tobacco smoke exposure. Pediatr Allergy Immunol. 2013;24:10–8.
Acknowledgements
This work was supported by Weifang Medical University and Weifang People’s Hospital, Weifang, China.
Availability of data and materials
The databases which we collected Literature include PubMed, Embase, LISTA, Web of science and Library of Congress, and Chinese datasets (WanFang, China National Knowledge Infrastructure) until June 30, 2018.
Author information
Authors and Affiliations
Contributions
HFL and WT conceived of the project and designed the study. HFL, LPY and WT wrote the manuscript. KW and XTL participated in selecting study and extracting data. LPY and HXL take part in Manuscript Revise. WT was in charge of quality control. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Li, Hf., Yan, Lp., Wang, K. et al. Association between ADAM33 polymorphisms and asthma risk: a systematic review and meta-analysis. Respir Res 20, 38 (2019). https://doi.org/10.1186/s12931-019-1006-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12931-019-1006-1