Abstract
Background
The kidney is exceptionally vulnerable during sepsis, often resulting in sepsis-associated acute kidney injury (SA-AKI), a condition that not only escalates morbidity but also significantly raises sepsis-related mortality rates. Circular RNA circ_001653 has been previously reported to be upregulated in the serum of SA-AKI patients, while the role and underlying mechanism of circ_001653 in SA-AKI remains unknown. In this study, we aimed to explore the functional role and the molecular mechanism of circ_001653 in the pathogenesis of SA-AKI.
Methods
LPS-stimulated HK-2 cells and ligation and perforation of cecum (CLP)-induced rats were used as in vitro and in vivo models of SA-AKI. The target gene expression levels were measured using qRT-PCR and western blot. Renal function (BUN, sCr, uNGAL, and uKIM-1), and renal pathological changes were detected in septic mice. TUNEL and EdU assays were conducted to measure apoptosis and proliferation rates in vitro. DCFH-DA staining was used to detect ROS levels in vitro and in vivo. Oxidative stress markers (SOD, GSH-Px, MDA, and SOD), and inflammation markers (IL-1β, IL-6, and TNF-α) were determined using commercial kits both in vitro and in vivo. Additionally, gain-and-loss-of-function assays and mechanistic experiments were conducted to explore the regulatory role of circ_001653 in SA-AKI pathogenesis.
Results
Data showed that circ_001653 expression was high in LPS-stimulated HK-2 cells and CLP-induced rat renal tissue and was mainly localized in the cytoplasm. Notably, circ_001653 silencing alleviated SA-AKI by reducing apoptosis and alleviating oxidative stress and inflammation in HK-2 cells and renal tissue of rats. Mechanistically, it was found that circ_001653 alleviated SA-AKI by recruiting BUD13 to activate the KEAP1/Nrf2/HO-1 signaling pathway.
Conclusions
To summarize, our study is the first to reveal elevated expression of circ_001653 in sepsis-associated AKI, and its downregulation effectively attenuates AKI by reducing apoptosis, inflammation, and oxidative stress. Mechanistically, circ_001653 exerts its effects by recruiting BUD13 to activate the KEAP1/Nrf2/HO-1 signaling pathway. These findings suggest circ_001653 as a potential therapeutic target for the drug development of sepsis-associated AKI.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sepsis is a condition of dysregulated inflammatory response to infection, causing organ damage with life-threatening risks [1]. Acute kidney injury (AKI) is usually diagnosed as a complication of sepsis. It is reported that 50% of AKI patients are also diagnosed with sepsis, while up to 60% of septic patients have AKI [1]. Sepsis-associated acute kidney injury (SA-AKI) is correlated with extremely high mortality and morbidity [2]. Currently, the underlying pathophysiological mechanism of SA-AKI remains unclear, and therapies for SA-AKI are often ineffective, which calls for a deepening understanding of the pathogenesis for developing new therapeutic strategies for SA-AKI.
CircRNA is a class of non-coding RNA generated by reverse splicing, and several studies have shown that circRNAs are involved in AKI pathogenesis [3, 4]. Meng et al. revealed that Circ-Snrk promoted AKI development and is associated with the activation of the MAPK signaling pathway [5]. Wang et al. reported that circ_0074371 boosted LPS-induced HK-2 cells apoptosis, inflammation, and oxidative stress via regulating miR-330-5p/ELK1 axis [6]. Circ_001653 was found in 2020 with the function of upregulating NR6A1 and promoting gastric cancer via binding to microRNA-377 [7]. More recently, Chang et al. [8] conducted circRNA microarray analysis using SA-AKI patient serum samples and reported 132 circRNAs that were differentially expressed among which 80 were upregulated and 52 were downregulated, respectively. Among these, circ_001653 was found to be highly expressed in the SA-AKI patient serum samples (p < 0.05, FC > 2). However, the functional role and the molecular mechanisms by which circ_001653 participates in SA-AKI progression have not been reported yet.
Several studies have identified various mechanisms implicated in AKI’s pathological process, including an uncontrolled inflammatory response, severe oxidative stress, maladaptive apoptosis, and aberrant endoplasmic reticulum stress, yet controversy surrounds their precise roles [9,10,11]. Excessive reactive oxygen species (ROS) reduce the bioavailability of nitric oxide, causing oxidative stress (OS), local tissue hypoxia, and mitochondrial dysfunction, thus leading to AKI [11,12,13]. Under different pathological conditions, there is substantial evidence demonstrating a significant correlation between oxidative stress injury, inflammation, and the nuclear factor E2-related factor 2 (Nrf2) [14, 15]. During normal conditions, the cap ‘n’ collar subfamily of basic region-leucine zipper transcription factor Nrf2 remains sequestered in the cytoplasm through its interaction with the ligand Kelch-like ECH associating protein 1 (KEAP1) [16]. Upon exposure to oxidative stress, the KEAP1/Nrf2 complexes undergo dissociation, allowing Nrf2 to translocate into the nucleus. Once in the nucleus, Nrf2 facilitates the expression of heme-oxygenase 1 (HO-1), which plays a crucial role in modulating molecules associated with oxidative stress, such as superoxide dismutase (SOD), malonaldehyde (MDA), reactive oxygen species (ROS), and glutathione peroxidase (GSH-Px) [17]. In prior studies, the regulation of KEAP1/Nrf2 and its downstream genes has proven effective in reducing the release of inflammatory factors and mitigating oxidative stress in kidney injury [18, 19]. Hence, activating the KEAP1/Nrf2 pathway to curb oxidative stress and inflammation could present a potential therapeutic approach for addressing SA-AKI. Nevertheless, whether circ_001653 could regulate this pathway remained unknown.
In this study, we examined the expression of circ_001653 in LPS-stimulated HK-2 cells and kidney tissues of sepsis-induced AKI rat models and explored the functional role and the molecular mechanism of circ_001653 in the development of SA-AKI, which could lighten new direction for treatment and reduce the mortality and infection rate of SA-AKI.
Materials and methods
Animals and establishment of CLP model
All animal experiments in this study were conducted in accordance with the guidelines for the Care and Use of Laboratory Animals and approved by the Animal Ethics Committee of Yancheng No.1 People’s Hospital. Wistar rats (160–200 g; 8–10 weeks old) were purchased from the SLAC Laboratory Animal Co., Ltd, China, and were used to establish an in vivo model through ceacal ligation and puncture (CLP). The CLP model was constructed according to a previous report [20]. Briefly, all rats were anesthetized with isoflurane (2.5%, Sigma-Aldrich, USA) inhalation, and a 2-cm midline laparotomy was made to expose the caecum under aseptic conditions. The caecum was then ligated with a 4 − 0 silk suture and pierced twice with a 20-gauge needle to mimic the sepsis-associated AKI clinical symptoms. The rats in the Sham group had their caecum exposed without perforation. After surgery, rats were placed in individual cages.
To explore the effects of circ_001653 knockdown on SA-AKI progression in vivo, adenovirus carrying shRNA against circ_001653 or comparable control (sh-NC) was constructed and obtained from Ribobio (Guangzhou, China). The rats were injected with adenovirus carrying sh-circ_001653 (1 × 109 TU) or sh-NC via the tail vein [20]. Two days after the surgery, rats were euthanized and the serum, urine, and kidney tissue samples were collected for subsequent experiments.
Hematoxylin and eosin (H&E) staining
Kidney tissues were fixed in 4% paraformaldehyde, embedded with paraffin, and then cut into 5 μm thick sections. The sections were then stained with Hematoxylin and Eosin Staining Kit (Beyotime, Shanghai, China) following the manufacturer’s protocol, respectively. Briefly, sections were dewaxed with xylene for 10 min, and immersed in 100%, 90%, 80%, and 70% gradient alcohol for 5 min each. Then the sections were stained with hematoxylin for 10 min and eosin for 2 min at room temperature. Next, sections were dehydrated with gradient alcohol, permeabilized with xylene, and mounted using neutral balsam. The degree of kidney injury was then evaluated by morphological changes detected under a light microscope. The scoring criteria from 0 to 5 were as follows [21]: 0 = normal morphology; 1 = Only degeneration without necrosis; 2 (25%) = necrosis; 3 (50%) = vacuolar degeneration; 4 (75%) = tubular dilatation; 5 (75%) = hemorrhage.
Cell culture and treatment
Human kidney-2 (HK-2) cells were purchased from ATCC (CRL-2190, USA) and were cultured in DMEM/F12 (Dulbecco’s modified Eagle’s medium; Gibco, USA) supplemented with 10% fetal bovine serum (FBS, DMEM, Gibco, Grand Island, USA) and 1% penicillin-streptomycin and incubated at 37 °C with 5% CO2 atmosphere.
To construct an in vitro cell model of SA-AKI, HK-2 cells (5 × 105 cells/well) were seeded into a 96-well plate and cultured for 24 h until 80% confluence. Subsequently, the cells were stimulated with lipopolysaccharide (LPS, SMB00610, Sigma-Aldrich) for 24 h at different concentrations (0, 2.5, 5, and 10 µg/mL) [22,23,24]. HK-2 cells without LPS treatment were used as the control group.
Cell viability
Cell viability of HK-2 cells was assessed using a CCK-8 assay kit (Beyotime, Shanghai, China). Briefly, cells were grown in a 96-well plate and incubated for 24 h. Then each well was added with 10 µL of CCK8 reagent and cultured for another 2 h at 37 °C. The absorbance was determined at 450 nm using a microplate reader (Promega, USA). Our initial result showed that 10 µg/mL of LPS had more prominent suppression on HK-2 cell viability, thus we used this dose in subsequent experiments.
Cell transfection
To evaluate the role of circ_001653, BUD13, and KEAP, sh-circ_001653 (-1, -2 and − 3) (OBiO Technology, China), sh-BUD13 (OBiO Technology, China), or oe-KEAP1 (GenePharma, China) vectors or the corresponding negative control vectors (sh-NC or oe-NC) were designed and synthesized by GenePharma (Shanghai, China), respectively. HK-2 cells were seeded into a 6-well plate at a density of 1 × 105 cells/well and then transfected with these vectors using Lipofectamine 2000 (11668027, ThermoFisher Scientific (CA, USA) following the manufacturer’s protocol. The cells were then cultured for 48-hours in the incubator at 37 °C with 5% CO2. After 48 h of transfection, cells were then left untreated or treated with 10 µg/mL of LPS for another 24 h. Finally, the cells were harvested for subsequent assays.
Quantitative real-time polymerase chain reaction (qRT-PCR)
The total RNA was extracted from HK-2 cells or kidney tissues using TRIzol® Reagent (15596026, ThermoFisher, CA, USA). iScript™ cDNA Synthesis Kit (1708890, Bio-Rad, USA) was used for reverse transcription into cDNA. The PCR reaction was performed in triplicate using a 7500 Real-Time PCR System with SYBR™ Green PCR Master Mix (4309155, ThermoFisher, USA). The target gene expression level was normalized to β-actin or U6 and calculated using the 2−ΔΔCT method. Primer sequences used in this study are shown in Table 1.
Ribonuclease (RNase) R treatment
RNase R assay was used to assess the stability of circ_001653 and DUSP22 mRNA in HK-2 cells. Briefly, 4 µg of RNAs extracted from HK-2 cells were incubated with RNase R (3 U/µg, RNR07250, Biosearch) for 1 h at 37 °C. After treatment, RNA was extracted with TRIzol, and qRT-PCR was conducted to determine the expression of circ_001653 and circ_001653 linear mRNA (DUSP22) levels in HK-2 cells.
Actinomycin D treatment
To block transcription, 2 mg/ml actinomycin D (SBR00013, Sigma-Aldrich) or negative control DMSO was added into the cell culture medium and cultured for 0, 6, 12, 18, and 24 h. After that, qRT-PCR was carried out to determine the expression level of circ_001653 and KEAP1.
Subcellular localization
Cytoplasmic and nuclear extracts of HK-2 cells were separated using a Cytoplasmic and Nuclear RNA Purification Kit (Norgen Biotek, USA) according to the manufacturer’s protocol. U6 was utilized as the nuclear endogenous control and β-actin as the cytoplasmic endogenous control. The distribution of RNA (U6, β-actin, and circSEMA5A) in the nucleus and cytoplasm of HK-2 cells was measured using qRT-PCR.
Fluorescence in situ hybridization (FISH) assay
FAM-labeled circ_001653 probes were designed and synthesized by Foco (Guangzhou, China). The subcellular localization of circ_001653 in HK-2 cells was determined using a FISH Tag™ RNA Red Kit (F32954, ThermoFisher, USA) following the manufacturer’s protocols. Briefly, cells were seeded on slides at the bottom of a 24-well plate and cultured to reach 70% confluence. Then cells were fixed with 4% paraformaldehyde at room temperature for 10 min, and incubated with pre-hybridization buffer at 37 °C for 30 min, followed by incubation with probes in hybridization buffer at 37 °C overnight in the dark. After that, cells were washed, and dehydrated, and DAPI was used to stain the nuclei. Images were taken with a laser scanning confocal microscope (Eclipse E6000; Nikon, Corporation, Tokyo, Japan).
TUNEL assay
TUNEL assay was used to determine the cell apoptotic rate. In brief, HK-2 cells (2 × 105 per group) were seeded on coverslips in a 24-well plate. The cells were then fixed with 4% paraformaldehyde before being treated with methanol and 30% H2O2 at a 50:1 ratio at room temperature for 30 min. After that, a One-step TUNEL In Situ Apoptosis Kit (E-CK-A320, Elabscience, USA) was used to detect TUNEL-positive cells in each group, according to the manufacturer’s procedure. Slides were incubated with 100 µL TdT Equilibration Buffer at 37 °C for 20 min. After removing the TdT Equilibration Buffer, cells were then stained with 50 µL working solution containing 35 µL TdT Equilibration Buffer, 10 µL Labeling Solution, and 5 µL TdT Enzyme at 37 °C for 60 min in the dark. Thereafter, the cell nuclei were stained with DAPI at room temperature in the dark and rinsed with PBS thrice. Coverslips were mounted with mounting medium and images were taken using a Zeiss Axioplan 2 microscope.
EdU assay
The proliferation rate of HK-2 cells was determined using an EdU Staining Proliferation Kit (iFluor 647) (ab222421, Abcam, USA). Briefly, HK-2 cells were grown in 96-well plates and cultured for one day. Then cells were stained with 100 µL of EdU solution (50 µM) for 2 h under optimal growth conditions. Following a washing step, cells were fixed with 4% paraformaldehyde (Sigma-Aldrich) for 15 min and then mixed with 0.5% Triton-X-100 (Sigma-Aldrich) for 20 min. After that, the nucleus was stained with DAPI (Sigma-Aldrich). Finally, the EdU-positive cells were observed under a fluorescence-inverted microscope (Olympus, Tokyo, Japan) and the percentage of EdU-positive cells was calculated.
Immunofluorescence measurement of ROS levels
A DCFDA/H2DCFDA-Cellular ROS Assay Kit (ab113851, Abcam) was used to determine the oxidative stress level in HK-2 cells. Briefly, HK-2 cells were seeded at a density of 3 × 105 cells/well into a 6-well plate. Following the indicated treatment, HK-2 cells were washed with PBS before being stained for 30 min at 37 °C with a 5 µmol/L solution of 2′,7′-dichlorofluorescin diacetate (DCFDA) in the dark. Finally, an Array Scan VII microscope (BX53, Japan) was used to scan the signal with the excitation wavelength at 488 nm and the emission wavelength at 525 nm. Ten randomly selected fields were chosen, and the target area of each section was quantified independently using the Indica Labs - Area Quantification FL v2.1.2 module in Halo v3.0.311.314 analysis software, and the mean intensity was calculated.
Determination of oxidative stress parameters
Lithium-heparin tubes were used to collect blood samples, which were then centrifuged for 10 min at 1,500 × g to separate the serum. The supernatant of cells after indicated transfection or treatment was collected from the medium. The contents of malondialdehyde (MDA, #A003-1-2), glutathione (GSH, #A006-2-1), superoxide dismutase (SOD, #A001-3-2), and catalase (CAT, #A007-1-1) in rat serum or cell supernatants were determined with relevant commercial kits according to the manufacturer’s protocol (Nanjing Jiancheng Bioengineering Institute). The optical density (OD) was measured using a spectrophotometer (SpectraMax® M5).
Evaluation of renal functional markers
Rat blood samples were obtained and subjected to centrifugation at 5000 g to separate the serum. To assess kidney function, levels of blood urea nitrogen (BUN) and serum creatinine (sCr) were measured using a colorimetric technique (#C013-1-1, Nanjing Jiancheng) and a creatinine test kit (#C011-2-1, Nanjing Jiancheng, China), following established protocols.
The urine samples were obtained and subjected to centrifugation at 600 g for 5 min. The levels of urinary neutrophil gelatinase-associated lipocalin (uNGAL, #E-EL-R3055, Elabscience) and urinary kidney injury molecule‐1 (uKIM‐1, #CSB-E08808r, Cusabio Biotech, Zhengzhou, China) were then measured using the commercial ELISA kits according to the provided instructions.
Enzyme-linked immunosorbent assay (ELISA) to measure inflammation indexes
The levels of IL-1β, IL-6, and TNF-α proinflammatory cytokines in the serum and supernatant of cultured HK-2 cells were measured using IL-1β ELISA kit (#CSB-E08055r, #CSB-E08053h, Cusabio Biotech, Wuhan, China), IL-6 ELISA kit (#CSB-E04640r, #CSB-E04638h, Cusabio Biotech), and TNF-α ELISA kit (#CSB-E11987r, CSB-E04740h, Cusabio Biotech), following the manufacturer’s instruction, respectively.
RNA immunoprecipitation (RIP) assay
Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (17–701, Sigma-Aldrich, USA) was used for RIP assay as the manual instructed. In brief, following HK-2 cell lysis with lysis buffer (Thermo Fisher) for 5 min, and centrifuged for 10 min at 14,000 rpm at 4 °C to remove the supernatant. Then the lysate was cocultured with magnetic beads coupled to anti-BUD13 (A303-320 A, ThermoFisher, USA), anti-EIF4A3 (PA5-117930, ThermoFisher, USA), anti-RANGAP1 (MA5-34762, ThermoFisher, USA), anti-SLTM (A302-834 A, ThermoFisher, USA), and negative control anti-IgG (Millipore, Massachusetts, USA) overnight at 4 °C. After digestion using Proteinase K buffer for 30 min at 55 °C, immunoprecipitated RNA was extracted and subject to qRT-PCR analysis.
Western blot (WB)
Protein in HK-2 cells or kidney tissue samples was extracted with RIPA lysis buffer (Sigma-Aldrich). The concentration of protein was measured using a Pierce™ BCA Protein Assay Kit (23225, Thermo Fisher, USA). A total of 20 µg of protein was loaded and separated on sodium dodecyl sulfate-polyacrylamide gel and then wet transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, USA). The membranes were incubated with primary antibodies at 4 °C overnight. One day later after washing, the membranes were incubated with goat anti-rabbit IgG secondary antibody (ab7090, Abcam, Cambridge, UK). Finally, the proteins were visualized using a BM Chemiluminescence Western Blotting Kit (11520709001, Sigma-Aldrich, USA). The primary antibodies were all purchased from Abcam (Cambridge, UK), including KEAP1 (ab227828, 1/2000), Nrf2 (ab62352, 1/1000), HO-1 (ab52947, 1/2000) and β-actin (ab6276, 1/5000).
Statistical analysis
SPSS 22.0 software (SPSS Inc., Chicago, US) was used for statistical analysis. The data were presented as the mean ± SD and all assays were carried out in triplicate. For the comparison of two groups, a student’s two-tailed t-test was used while one-way or two-way ANOVA followed by a post-hoc test was used for the comparison of more than two groups. P value < 0.05 was regarded as statistically significant.
Results
Characteristics of circ_001653 expression in in-vitro HK-2 cell model
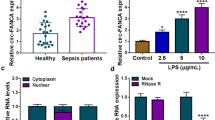
We first constructed an in vitro cell model of SA-AKI by treating HK-2 cells with different concentrations of LPS. CCK-8 assay showed that the viability of HK-2 cells was decreased by LPS exposure in a concentration-dependent manner compared with the HK-2 cells without LPS treatment (0 µg/mL). Notably, the concentration at 10 µg/mL most significantly reduced the viability of HK-2 cells (Fig. 1A; P < 0.05). Next, we evaluated the expression level of circ_001653 under the treatment of increasing concentration of LPS. qRT-PCR results showed that circ_001653 expression increased in LPS-treated HK-2 cells in a dose-dependent manner compared with the non-treated cells (Fig. 1B; P < 0.05). Particularly, it was observed that the expression of circ_001653 in HK-2 cells was the highest at the concentration of 10 µg/mL, therefore; we used this dose of LPS for subsequent experiments. We then evaluated circ_001653 expression after treatment with LPS at 10 µg/mL at different time points and found that circ_001653 expression was the highest at 24 h of LPS stimulation (Fig. 1C; P < 0.001). Therefore, LPS treatment at 10 µg/mL for 24 h was used for subsequent experiments. Since circRNAs are known to be resistant to RNase R stimulation, we validated the expression of circ_001653 in HK-2 cells by performing an RNase R digestion experiment. The findings further supported the circular character of the circ_001653 transcript by showing that the linear RNA transcript (DUSP22) was significantly degraded in comparison to circ_001653 (Fig. 1D; P < 0.001). Additionally, we assessed the stability of these two transcripts by treating HK-2 cells with actinomycin D, a transcription inhibitor. qRT-PCR revealed that the half-life of DUSP22 mRNA was 6–12 h, whereas that of circ_001653 exceeded 24 h (Fig. 1E; P < 0.001), indicating that the circ_001653 transcript had greater stability than the linear transcript. Subsequently, we evaluated circ_001653 cellular distribution in HK-2 cells. We discovered that circ_001653 was primarily localized in the cytoplasm of HK-2 cells (Fig. 1F-G). These results suggest that circ_001653 is abundantly and stably present in the cytoplasm of HK-2 cells.
Circ_001653 expression is high in the LPS-induced SA-AKI HK-2 cell model. (A) CCK-8 assay was used to detect the viability of HK-2 cells exposed to different concentrations (0, 2.5, 5, 10 µg/mL) of LPS for 24 h. (B) Relative expression level of circ_001653 in HK-2 cells stimulated with or without LPS at different concentrations (0, 2.5, 5, 10 µg/mL) for 24 h detected by qRT-PCR. (C) Relative expression level of circ_001653 detected at 0, 6, 12, and 24 h in HK-2 cells stimulated with 10 µg/mL LPS using qRT-PCR. (D) For RNase R digestion assay, HK-2 cells were treated with or without 3 U/µg RNase R for 1 h at 37 °C, and qRT-PCR was conducted to detect circ_001653 and its linear mRNA (DUSP22) expression in HK-2 cells. (E) Circ_001653 and linear mRNA (DUSP22) expression levels in HK-2 cells at different time points (0, 6, 12, 18, 24 h) after treatment with 2 mg/ml actinomycin D. (F-G) The localization of circ_001653 in HK-2 cells was detected by subcellular fractionation and FISH assays, respectively. **P < 0.01, ***P < 0.001. LPS: lipopolysaccharide; SA-AKI: sepsis-associated acute kidney injury; DUSP22: dual specificity phosphatase 22; DAPI: 4’,6-diamidino-2-phenylindole
Knock-down of circ_001653 alleviates LPS-induced injury in HK-2 cell
To determine the regulatory function of circ_001653 in LPS-induced injury in HK-2 cells, sh-circ_001653, sh-NC (negative control) was transfected into HK-2 cells, with its interference efficiency shown in Fig. 2A. TUNEL assay showed that LPS stimulation resulted in increased percentage of apoptotic cells compared to the unstimulated control group (P < 0.01). Interestingly, this increase in the number of TUNEL-positive cells by LPS stimulation was significantly decreased by the knock-down of circ_001653 relative to the LPS + sh-NC group, indicating that inhibiting circ_001653 can significantly reduce apoptosis in LPS-treated HK-2 cells (Fig. 2B; P < 0.01). Furthermore, the EdU assay demonstrated that the reduction in proliferation capacity of HK-2 cells induced by LPS was significantly increased after circ_001653 knock-down (Fig. 2C; P < 0.01).
Functional role of circ_001653 in LPS-induced HK-2 cells. (A) Expression of circ_001653 in HK-2 cells after transfection with sh-circ_001653-1/2/3 and negative control sh-NC for 48 h as measured by qRT-PCR. To explore the effects of circ_001653 knockdown on LPS-induced HK-2 cells, HK-2 cells were treated with LPS (10 µg/mL) for 24 h alone or after transfection with sh-NC or sh-circ_001653 for 48 h. HK-2 cells without LPS treatment were used as the control (Ctrl) group. (B) TUNEL assay was performed to measure apoptosis of HK-2 cells in each group. (C) EdU assay was used to detect the proliferation of HK-2 cells after the indicated treatment. (D) Relative fluorescence level of ROS in HK-2 cells from corresponding groups detected by DCFH-DA staining. (E) MDA, (F) SOD, (G) GSH activities, and (H) CAT levels in the supernatant of HK-2 cells in indicated groups were detected using the corresponding kits. (I) The contents of IL-1β, IL-6, and TNF-α in the supernatant of HK-2 cells from corresponding groups were detected by ELISA. **P < 0.01, ***P < 0.001. Ctrl: Control; LPS: lipopolysaccharide; DAPI: 4’,6-diamidino-2-phenylindole; TUNEL, terminal deoxynucleotidyl transferase dUTP nick-end labeling; EdU, Ethynyl-2ʹ-Deoxyuridine; ROS: reactive oxygen species; MDA: malondialdehyde; SOD: superoxide dismutase; GSH: glutathione; CAT: catalase; IL-1β: Interleukin (IL)-1beta; IL-6: Interleukin (IL)-6; TNF-α: tumor necrosis factor (TNF)-alpha; ELISA: Enzyme-Linked Immunosorbent Assay
Next, we evaluated ROS levels using molecular probe DCFH-DA staining. The images showed that LPS stimulation significantly promoted ROS levels in HK-2 cells compared to the control (HK-2 cells without LPS treatment), which were prominently reduced after circ_001653 knock-down (Fig. 2D; P < 0.01). We then evaluated oxidative stress-related parameters by measuring the levels of MDA, and determined the activities of SOD, GSH, and CAT, respectively. Results showed that LPS stimulation markedly increased the level of MDA while reducing the activities of SOD, GSH, and CAT in K-2 cells compared with cells without LPS treatment (Fig. 2E-H; P < 0.001). Interestingly, when we inhibited circ_001653 expression, it notably reduced the oxidative damage induced by LPS (Fig. 2E-H; P < 0.01). We also found that LPS stimulation induced inflammation in the HK-2 cells as evident from the increased production of inflammatory cytokines such as IL-1β, IL-6, and TNF-α, which were reduced by inhibiting circ_001653 expression (Fig. 2I; P < 0.001). Overall, these results demonstrate that inhibiting circ_001653 attenuates LPS-induced injury in HK-2 cells.
Circ_001653 inhibition alleviates renal dysfunction, oxidative stress, and inflammation in CLP-induced SA-AKI rat model
In the next step, we evaluated the expression and functional role of circ_001653 using a CLP-induced SA-AKI rat model. Rats in the Sham group received caecum exposure without ligation. qRT-PCR results revealed a remarkably upregulated expression of circ_001653 in kidney tissues from the CLP + sh-NC group compared to the control sham group, which was significantly reduced after silencing circ_001653 expression (Fig. 3A; P < 0.001). As evident from H&E staining, SA-AKI produced significant pathological changes, including proximal tubule dilation, brush border damage, proteinaceous casts, interstitial widening, and necrosis. However, circ_001653 silencing alleviated sepsis-induced pathological changes (Fig. 3B; P < 0.001). We then evaluated the renal functional markers such as BUN, sCr, uNGAL, and uKIM-1 in rats subjected to CLP, a model for sepsis-associated AKI. The CLP + sh-NC group demonstrated significantly higher levels of BUN, sCr, uNGAL, and uKIM-1 compared to the sham group. Interestingly, silencing circ_001653 expression in CLP rats significantly reduced SA-AKI relative to the CLP + sh-NC group (Fig. 3C-F; P < 0.001). These results reveal that circ_001653 expression is high in SA-AKI and inhibiting circ_001653 alleviates renal dysfunction induced by CLP.
Circ_001653 inhibition alleviates renal dysfunction, oxidative stress, and inflammatory response in the CLP-induced SA-AKI rat model. Wistar rats were divided into the sham, CLP + sh-NC, and CLP + sh-circ_001653 groups (n = 6 per group). Rats in the CLP + sh-NC group or CLP + sh-circ_001653 group received ceacal ligation and puncture. The rats received injections of adenovirus carrying sh-NC or sh-circ_001653 via the tail vein. Rats in the sham group received caecum exposure without ligation. (A) qRT-PCR was performed to measure the relative expression level of circ_001653 in kidney tissues in indicated groups. (B) H&E staining was performed to evaluate the pathological morphology of renal tissues in indicated groups (left panel). A semi-quantitative histopathological score of renal tissue on a scale of 0-5 based on corresponding images (Right panel). Levels of (C) BUN and (D) sCr in serum from corresponding groups were detected using corresponding kits. Levels of urinary (E) NGAL and (F) KIM-1 were detected by ELISA. (G) Relative fluorescence levels of ROS in kidney tissues were detected by DCFH-DA staining. (H) Serum MDA levels in indicated groups were measured using the corresponding kit. Serum (I) SOD, (J) GSH activities, and (K) CAT levels in indicated groups were measured using the corresponding kits. Relative mRNA expression levels of (L) IL‐1β, (M) IL‐6, and (N) TNF‐α in kidney tissues were measured with quantitative PCR. (O) The serum levels of IL-1β, IL-6, and TNF-α in indicated groups were detected by ELISA. ***P < 0.001. CLP: cecal ligation and puncture; H&E: hematoxylin and eosin; BUN: blood urea nitrogen; sCr: serum creatinine; NGAL: Neutrophil gelatinase-associated lipocalin; KIM-1: Kidney Injury Molecule-1; MDA: malondialdehyde; SOD: superoxide dismutase; GSH: glutathione; CAT: catalase; IL-1β: Interleukin (IL)-1beta; IL-6: Interleukin (IL)-6; TNF-α: tumor necrosis factor (TNF)-alpha; ELISA: Enzyme-Linked Immunosorbent Assay
Next, we explored the effects of circ_001653 silencing on CLP-induced oxidative stress and inflammation in renal tissues. First, we measured the changes in the oxidative stress status of renal tissues since oxidative stress is one of the key factors mediating AKI. As shown in Fig. 3G-K, ROS and MDA levels were markedly up-regulated while the levels of GSH, SOD, and CAT activities were significantly down-regulated in CLP + sh-NC group compared to the sham group (P < 0.001). Notably, circ_001653 silencing significantly inhibited the oxidative damage caused by sepsis (P < 0.001). To confirm the injury indeed happened in the kidney while sepsis occurred, we detected IL-1β, IL-6, and TNF-α in the kidney tissues. qRT-PCR data showed that the gene expression level of these inflammatory markers was significantly increased in the renal tissues of CLP + sh-NC group rats when compared to the sham group (P < 0.001), which were significantly reduced after inhibiting circ_001653 expression (Fig. 3L-N; P < 0.001). ELISA results further confirm this data showing that the increased serum levels of TNF-α, IL-6, and IL-1β in the CLP + sh-NC group were markedly reduced upon circ_001653 silencing (Fig. 3O; P < 0.001).
Circ_001653 regulates KEAP1 to activate the KEAP1/Nrf2 pathway
Previous studies have shown that KEAP1 regulates the transcription factor Nrf2 that in turn controls HO-1 and that KEAP1/Nrf2/HO-I signaling axis plays an important role in the pathogenesis of AKI [16, 25]. Therefore, we hypothesized that circ_001653 might regulate KEAP1 to mediate its function by activating the Nrf2/HO-1 pathway. qRT-PCR results demonstrated that LPS decreased KEAP1 and increased Nrf2 and HO-1 levels (P < 0.01), whereas circ_001653 silencing led to a further decrease in KEAP1 and an increase in Nrf2 and HO-1 levels in LPS-induced HK-2 cells, suggesting that circ_001653 may modulate the KEAP1/Nrf2/HO-1 signaling pathway by controlling KEAP1 (Fig. 4A; P < 0.01). To test this assumption, we overexpressed KEAP1 (oe-KEAP1) and transfected the overexpression vector into LPS treated HK-2 cells. The transfection efficiency is shown in Fig. 4B. Data showed that KEAP1 overexpression significantly reduced the elevated protein and gene expression levels of Nrf2 and HO-1 induced by circ_001653 silencing, confirming that the KEAP1/Nrf2 signaling pathway was modulated by circ_001653 via KEAP1 (Fig. 4C-D; P < 0.05). Collectively, these results suggest that circ_001653 could modulate the Nrf2/HO-1 pathway via regulating KEAP1.
Circ_001653 regulates KEAP1 to activate the KEAP1/Nrf2 pathway. (A) qRT-PCR was used to detect the relative mRNA expression levels of KEAP1, Nrf2, and HO-1 in HK-2 cells after transfection with sh-NC or sh-circ_001653 for 48 h and treated with LPS (10 µg/mL) for 24 h. HK-2 cells without LPS treatment were used as the control (Ctrl) group. (B) Expression of KEAP1 in HK-2 cells after transfection with KEAP1 overexpressing plasmid and negative control oe-NC for 48 h as measured by qRT-PCR. (C) Protein bands of KEAP1, Nrf2, HO-1 in HK-2 cells after indicated transfection were detected by Western blot. β-actin served as a loading control. (D) Relative expression of KEAP1, Nrf2, and HO-1 in HK-2 cells was detected by qRT-PCR. *P < 0.05, **P < 0.01, ***P < 0.001. Ctrl: Control; LPS: lipopolysaccharide; KEAP1, kelch like erythroid cell-derived protein with CNC homology [ECH] associated protein 1; Nrf2, nuclear factor erythroid 2 [NF-E2] related factor 2; HO-1: heme oxygenase-1
Circ_001653 silencing attenuates oxidative damage and inflammation in LPS-induced HK-2 cells by regulating KEAP1
To determine the regulatory effect of KEAP1 on LPS-stimulated injury, oe-KEAP1 was transfected together with sh-circ_001653 into HK-2 cells treated with LPS. TUNEL assay showed that the number of TUNEL positive cells decreased by sh-circ_001653 was significantly increased by oe-KEAP1, indicating that the apoptosis inhibited by sh-circ_001653 was reversed by oe-KEAP1 (Fig. 5A; P < 0.01). Similarly, EdU assay revealed that the number of EdU positive cells increased by sh-circ_001653 were decreased back by oe-KEAP1 (Fig. 5B; P < 0.01). Furthermore, the molecular probe DCFH-DA was used to detect ROS production. Results showed that the ROS suppressed by sh-circ_001653 was noticeably restored by oe-KEAP1 (Fig. 5C; P < 0.01), causing oxidative stress. We also noticed that the reduction in MDA levels induced by silencing circ_001653 was reversed when KEAP1 was overexpressed (Fig. 5D; P < 0.01). Subsequently, the elevated level of GSH and increased activities of SOD and CAT induced by circ_001653 knock-down were reversed back by overexpressing KEAP1 (Fig. 5E-G; P < 0.01). Additionally, ELISA results suggested that the levels of inflammatory factors (IL-1β, IL-6, and TNF-α) remarkably decreased by sh-circ_001653 were increased back by oe-KEAP1 (Fig. 5H; P < 0.001). These outcomes indicate that circ_001653 exacerbates oxidative damage and inflammation in HK-2 cells by regulating KEAP1.
Circ_001653 silencing attenuates oxidative damage and inflammation in LPS-induced HK-2 cells by regulating KEAP1. Rescue assays were performed in HK-2 cells after transfection with sh-NC, sh-circ_001653, or sh-circ_001653 + oe-KEAP1 for 48 h and then treated with LPS (10 µg/mL) for 24 h. sh-NC was used as the negative control. (A) TUNEL assay was performed to measure apoptosis of HK-2 cells in corresponding groups. (B) The proliferation of HK-2 cells in the corresponding groups was detected by EdU assay. (C) Relative fluorescence levels of ROS in HK-2 cells from corresponding groups were detected by the DCFH-DA probe. (D) MDA, (E) SOD, (F) GSH activities and (G) CAT levels in the supernatant of HK-2 cells in indicated groups were detected using the corresponding kits. (H) The contents of IL-1β, IL-6 and TNF-α in the supernatant of HK-2 cells from corresponding groups were detected by ELISA. **P < 0.01, ***P < 0.001. Ctrl: Control; LPS: lipopolysaccharide; DAPI: 4’,6-diamidino-2-phenylindole; TUNEL, terminal deoxynucleotidyl transferase dUTP nick-end labeling; EdU, Ethynyl-2ʹ-Deoxyuridine; ROS: reactive oxygen species; MDA: malondialdehyde; SOD: superoxide dismutase; GSH: glutathione; CAT: catalase; IL-1β: Interleukin (IL)-1beta; IL-6: Interleukin (IL)-6; TNF-α: tumor necrosis factor (TNF)-alpha; ELISA: Enzyme-Linked Immunosorbent Assay
Circ_001653 recruits BUD13 to stabilize KEAP1 expression
Finally, to explore the mechanism of circ_001653 regulating KEAP1, KEAP1-bound RNA-binding proteins (RBPs) were analyzed. Through overlapping results from circBank (http://www.circbank.cn/), and starBase (http://starbase.sysu.edu.cn/index.php), we identified RBPs that could bind to circ_001653 including BUD13, EIF4A3, RANGAP1 and SLTM (Fig. 6A). RIP results showed that among the four RBPs, BUD13 bound to circ_001653 with the highest enrichment, therefore; BUD13 was selected for subsequent experiments (Fig. 6B; P < 0.01). Since BUD13 was found to bound to circ_001653 and circ_001653 regulated KEAP1, we speculated that BUD13 might bind to KEAP1 to regulate its function. RIP results showed that in HK-2 cells, the anti-BUD13 group showed significantly higher enrichment than the control group, indicating that BUD13 could indeed bind to KEAP1 (Fig. 6C; P < 0.01). To further investigate the regulatory role of BUD13 between circ_001653 and KEAP1, sh-BUD13 was transfected into HK-2 cells with interference efficiency shown in Fig. 6D. qRT-PCR results showed that in HK-2 cells, KEAP1 expression was significantly decreased by sh-BUD13 (Fig. 6E; P < 0.01). Furthermore, relative expression of KEAP1 was detected at 0, 6, 12, 18, and 24 h after treatment with actinomycin-D to explore whether BUD13 could stabilize KEAP1 mRNA. KEAP1 showed relatively lower expression in the sh-BUD13 group than in the sh-NC group, indicating that BUD13 promotes the stability of KEAP1 mRNA (Fig. 6F; P < 0.01). Additionally, we evaluated the binding of BUD13 to KEAP1 after silencing circ_001653 in HK-2 cells. As detected by RIP assay, the binding of BUD13 to KEAP1 was significantly suppressed in HK-2 cells transfected with sh-circ_001653 (Fig. 6G). These results suggest that circ_001653 recruits BUD13 to stabilize KEAP1 mRNA.
Circ_001653 stabilizes KEAP1 expression by recruiting BUD13. (A) Bioinformatic prediction of potential RNA-binding proteins (RBPs) that could bind to circ_001653. (B) Relative enrichment of circ_001653 in HK-2 cells reacted with different antibodies, detected by RIP assay. Anti-IgG served as the negative control. (C) Relative enrichment of KEAP1 in HK-2 cells treated with anti-BUD13, detected by RIP assay. Anti-IgG served as the negative control. (D) Interference efficiency of sh-BUD13-1/2/3 in HK-2 cells was determined using qRT-PCR. (E) Relative expression of KEAP1 in HK-2 cells transfected with sh-BUD13/NC was detected using qRT-PCR. sh-NC served as the negative control. (F) Relative expression of KEAP1 at 0, 6, 12, 18 and 24 h in HK-2 cells transfected with sh-BUD13/NC. sh-NC served as the negative control. (G) RIP assay detected the relative enrichment of KEAP1 in HK-2 cells treated with anti-BUD13/IgG and transfected with sh-circ_001653/NC. **P < 0.01, ***P < 0.001
Discussion
Acute kidney injury (AKI) induced by sepsis is a prevalent complication in critically ill patients. Prevention and treatment of AKI remain a challenge [26]. The study of the roles of circular RNAs dysregulated in the injured kidney may provide clues for identifying therapeutic targets. In this study, we found that circ_001653 was highly expressed in LPS-stimulated HK-2 cells and CLP-induced rat renal tissue, and circ_001653 knockdown mitigated SA-AKI by decreasing apoptosis, oxidative stress, and inflammation both in vitro and in vivo. Mechanistically, it was discovered that circ_001653 attenuated SA-AKI by recruiting BUD13 to activate the KEAP1/Nrf2/HO-1 signaling pathway, which might provide a novel target for AKI therapy.
In our study, LPS was used to mimic the sepsis-associated AKI symptoms in HK-2 cells as an in vitro model as reported previously [27]. Also, we used the CLP method to construct an in vivo animal model to mimic SA-AKI, which is a gold standard method [21, 25]. SA-AKI is characterized by increased apoptosis, excessive oxidative stress, and inflammation [21, 28]. In our study, we found that there was increased apoptosis, less proliferation, and elevated levels of ROS in LPS-stimulated HK-2 cells and CLP-induced rat kidney tissues. Moreover, oxidative stress marker such as MDA was increased while SOD, GSH, and CAT activities were decreased and the concentration of IL-1β, IL-6, and TNF-α was increased in LPS-incubated HK-2 cells and CLP-induced rat kidney tissues. These results indicate that the septic AKI models were successfully replicated and are consistent with the above-mentioned reports.
CircRNAs belong to non-coding RNAs, which are closed-loop structures formed by back-splicing and are known to play an important role as potential diagnostic, and prognostic biomarkers in various diseases [29,30,31]. For example, circTLK1 is highly expressed in the CLP-induced rat model and promotes SA-AKI by modulating inflammation and oxidative stress via the miR-106a-5p/HMGB1 axis [20]. Wei et al. [32] demonstrated that hsa_circ_0068888 has a protective effect on AKI by sponging miR-21-5p. Similarly, silencing circ-Gatad1 mitigates LPS-induced injury in HK-2 cells by targeting the miR-22-3p/TRPM7 axis in septic acute kidney injury [33]. Accordingly, circ_001653 upregulates NR6A1 expression and promotes gastric cancer via binding to microRNA-377 [7]. Nevertheless, there is no report analyzing its function in SA-AKI. In our work, we found that circ_001653 was upregulated in response to LPS stimulation in HK-2 cells and CLP-induced rat renal tissue. Moreover, silencing circ_001653 was demonstrated to reverse the LPS-induced elevated apoptosis, oxidative stress, inflammatory response, and decreased proliferation of HK-2 cells in vitro. Furthermore, circ_001653 knock-down also alleviated renal dysfunction, oxidative stress, and inflammation in the CLP-induced SA-AKI rat model. These findings are consistent with the results of previous studies [20, 27, 33].
The KEAP1/Nrf2/HO-1 signaling pathway is reported to play a key role in the defense against oxidative damage and inflammation [14, 34, 35]. KEAP1/Nrf2 signaling pathway was proved to be associated with AKI via regulating autophagy [36]. Hu et al. demonstrated that the KEAP1/Nrf2 signaling pathway could eliminate oxidative stress and abnormal inflammatory responses to alleviate traumatic lung injury [37]. It was also confirmed that in ischemia AKI, urolithin A could meliorate oxidative stress and promote autophagy via KEAP1/Nrf2 signaling pathway [38]. Furthermore, Sun et al. revealed that the activation of the Keap1-Nrf2-ARE signaling pathway exerted an anti-apoptotic effect in renal tubular cells [39]. In agreement with these reports, we also found that LPS resulted in decreased KEAP1 and increased Nrf2 and HO-1 levels, whereas circ_001653 silencing leads to a further decrease in KEAP1 and an increase in Nrf2 and HO-1 levels in HK-2 cells induced by LPS, suggesting that circ_001653 activates KEAP1/Nrf2/HO-1 signaling pathway.
In 2015, Laston et al. discovered that BUD13 was one of the genetic factors related to chronic kidney disease [40]. Later, it was shown that BUD13 stabilizes FN1 mRNA to promote FN1 expression in diffuse large B-cell lymphoma [41]. In 2021, Xing et al. proved that in prostate cancer cells, the downregulated BUD13 could shorten the half-life of SERPINA3, indicating that BUD13 could stabilize SERPINA3 mRNA [42]. However, the regulatory role of BUD13 in SA-AKI and whether it has the same effect on mRNA stability remained unknown. We found that BUD13 was bound to KEAP1 and enhanced the stability of KEAP1 mRNA. Furthermore, circ_001653 was revealed to recruit BUD13 to stabilize KEAP1 mRNA.
This study also possesses some limitations. First, the sample size of animal experiments is relatively small, and only HK-2 cells were used for in vitro analysis, which might influence the generalization of the data. Second, the KEAP1/Nrf2/HO-1 signaling was demonstrated to be regulated by circ_001653 in LPS-treated HK-2 cells, while its implication in the CLP rat models with circ_001653 knockdown was not elucidated. Moreover, future studies are suggested to investigate the expression of circ_001653 in AKI patients with large sample sizes. Additionally, whether the KEAP1/Nrf2/HO-1 signaling is essential for circ_001653-mediated harmful effects on AKI still requires further exploration.
Conclusions
In summary, our study is the first to demonstrate that circ_001653 expression is high in sepsis-associated AKI and its silencing could alleviate AKI by reducing the levels of apoptosis, inflammation, and oxidative stress. Mechanistically, circ_001653 mediates its function by recruiting BUD13 to activate KEAP1/Nrf2/HO-1 signaling pathway. Our findings imply that circ_001653 could be a promising therapeutic target for the treatment of sepsis-associated AKI.
Data availability
No datasets were generated or analysed during the current study.
References
Poston JT, Koyner JL. Sepsis associated acute kidney injury. BMJ. 2019;364:k4891.
Qiao J, Cui L. Multi-omics techniques make it possible to Analyze Sepsis-Associated Acute kidney Injury comprehensively. Front Immunol. 2022;13:905601.
Li Z, Xing J. Potential therapeutic applications of circular RNA in acute kidney injury. Biomed Pharmacother. 2024;174:116502.
So BYF, Yap DYH, Chan TM. Circular RNAs in Acute kidney Injury: roles in pathophysiology and implications for Clinical Management. Int J Mol Sci, 2022. 23(15).
Meng F, et al. Inhibition of circ-snrk ameliorates apoptosis and inflammation in acute kidney injury by regulating the MAPK pathway. Ren Fail. 2022;44(1):672–81.
Wang QY et al. Silencing circ_0074371 inhibits the progression of sepsis-induced acute kidney injury by regulating miR-330-5p/ELK1 axis. Mamm Genome, 2022.
Zhou W, et al. hsa_circ_001653 up-regulates NR6A1 expression and elicits gastric cancer progression by binding to microRNA-377. Exp Physiol. 2020;105(12):2141–53.
Tian C, et al. Exosomal hsa_circRNA_104484 and hsa_circRNA_104670 may serve as potential novel biomarkers and therapeutic targets for sepsis. Sci Rep. 2021;11(1):14141.
Cheng C, et al. Acute kidney injury: exploring endoplasmic reticulum stress-mediated cell death. Front Pharmacol. 2024;15:1308733.
Manrique-Caballero CL, Rio-Pertuz GD, Gomez H. Sepsis-Associated Acute kidney Injury. Crit Care Clin. 2021;37(2):279–301.
Ow CPC et al. Targeting oxidative stress in septic acute kidney Injury: from theory to practice. J Clin Med, 2021. 10(17).
Zhao M, et al. Mitochondrial ROS promote mitochondrial dysfunction and inflammation in ischemic acute kidney injury by disrupting TFAM-mediated mtDNA maintenance. Theranostics. 2021;11(4):1845–63.
Qiongyue Z, et al. Post-treatment with Irisin attenuates acute kidney Injury in Sepsis mice through anti-ferroptosis via the SIRT1/Nrf2 pathway. Front Pharmacol. 2022;13:857067.
Saha S et al. An overview of Nrf2 Signaling Pathway and its role in inflammation. Molecules, 2020. 25(22).
Dinkova-Kostova AT, Copple IM. Advances and challenges in therapeutic targeting of NRF2. Trends Pharmacol Sci. 2023;44(3):137–49.
Wang Y, et al. Resveratrol ameliorates sepsis-induced acute kidney injury in a pediatric rat model via Nrf2 signaling pathway. Exp Ther Med. 2018;16(4):3233–40.
Lee S, Hu L. Nrf2 activation through the inhibition of Keap1-Nrf2 protein-protein interaction. Med Chem Res. 2020;29(5):846–67.
Lin DW et al. Insights into the Molecular mechanisms of NRF2 in kidney Injury and diseases. Int J Mol Sci, 2023. 24(7).
Wang T, et al. Hyperoside inhibits EHV-8 infection via alleviating oxidative stress and IFN production through activating JNK/Keap1/Nrf2/HO-1 signaling pathways. J Virol. 2024;98(4):e0015924.
Xu HP, Ma XY, Yang C. Circular RNA TLK1 promotes Sepsis-Associated Acute kidney Injury by regulating inflammation and oxidative stress through miR-106a-5p/HMGB1 Axis. Front Mol Biosci. 2021;8:660269.
Shi Y, et al. Circular RNA VMA21 ameliorates sepsis-associated acute kidney injury by regulating miR-9-3p/SMG1/inflammation axis and oxidative stress. J Cell Mol Med. 2020;24(19):11397–408.
Zhang Y, et al. Sirtuin 6 overexpression relieves sepsis-induced acute kidney injury by promoting autophagy. Cell Cycle. 2019;18(4):425–36.
Li H, et al. Knockdown of circ-FANCA alleviates LPS-induced HK2 cell injury via targeting miR-93-5p/OXSR1 axis in septic acute kidney injury. Diabetol Metab Syndr. 2021;13(1):7.
Zhang Y, et al. HSP70 ameliorates septic acute kidney Injury via binding with TRAF6 to inhibit of inflammation-mediated apoptosis. J Inflamm Res. 2022;15:2213–28.
Gao F, et al. USP10 alleviates sepsis-induced acute kidney injury by regulating Sirt6-mediated Nrf2/ARE signaling pathway. J Inflamm. 2021;18(1):25.
Peerapornratana S, et al. Acute kidney injury from sepsis: current concepts, epidemiology, pathophysiology, prevention and treatment. Kidney Int. 2019;96(5):1083–99.
Zhang J, et al. Loganin attenuates septic Acute Renal Injury with the participation of AKT and Nrf2/HO-1 signaling pathways. Drug Des Devel Ther. 2021;15:501–13.
Tomsa AM, et al. Oxidative stress as a potential target in acute kidney injury. PeerJ. 2019;7:e8046.
Liu X, et al. Circular RNA: an emerging frontier in RNA therapeutic targets, RNA therapeutics, and mRNA vaccines. J Control Release. 2022;348:84–94.
Kristensen LS, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–91.
Beltrán-García J et al. Circular RNAs in Sepsis: Biogenesis, function, and clinical significance. Cells, 2020. 9(6).
Wei W, et al. Circular RNA circ_0068,888 protects against lipopolysaccharide-induced HK-2 cell injury via sponging microRNA-21–5p. Biochem Biophys Res Commun. 2021;540:1–7.
Zhang P, et al. Knockdown of circ-Gatad1 alleviates LPS induced HK2 cell injury via targeting miR-22-3p/TRPM7 axis in septic acute kidney. BMC Nephrol. 2024;25(1):79.
Zheng F, et al. Sevoflurane reduces lipopolysaccharide-induced apoptosis and pulmonary fibrosis in the RAW264.7 cells and mice models to ameliorate acute lung injury by eliminating oxidative damages. Redox Rep. 2022;27(1):139–49.
Huang CY et al. Attenuation of Lipopolysaccharide-Induced Acute Lung Injury by Hispolon in mice, through regulating the TLR4/PI3K/Akt/mTOR and Keap1/Nrf2/HO-1 pathways, and suppressing oxidative stress-mediated ER stress-Induced apoptosis and autophagy. Nutrients, 2020. 12(6).
Liao W, et al. p62/SQSTM1 protects against cisplatin-induced oxidative stress in kidneys by mediating the cross talk between autophagy and the Keap1-Nrf2 signalling pathway. Free Radic Res. 2019;53(7):800–14.
Hu LY, et al. Expression of Nrf2-Keap1-ARE signal pathway in traumatic lung injury and functional study. Eur Rev Med Pharmacol Sci. 2018;22(5):1402–8.
Zhang Y, et al. Urolithin A alleviates acute kidney injury induced by renal ischemia reperfusion through the p62-Keap1-Nrf2 signaling pathway. Phytother Res. 2022;36(2):984–95.
Sun D, et al. Salvianolate ameliorates renal tubular injury through the Keap1/Nrf2/ARE pathway in mouse kidney ischemia-reperfusion injury. J Ethnopharmacol. 2022;293:115331.
Laston SL, et al. Genetics of kidney disease and related cardiometabolic phenotypes in Zuni indians: the Zuni kidney project. Front Genet. 2015;6:6.
Song Y, et al. Long non-coding RNA DBH-AS1 promotes cancer progression in diffuse large B-cell lymphoma by targeting FN1 via RNA-binding protein BUD13. Cell Biol Int. 2020;44(6):1331–40.
Xing Z, et al. CircSERPINA3 regulates SERPINA3-mediated apoptosis, autophagy and aerobic glycolysis of prostate cancer cells by competitively binding to MiR-653-5p and recruiting BUD13. J Transl Med. 2021;19(1):492.
Acknowledgements
None.
Funding
This work was not supported by any financial funding.
Author information
Authors and Affiliations
Contributions
HZ, and CH conceived and designed the experiments. CH, and LC contributed significantly to the experiments and arranging data. CH, and ZL performed data analyses. CH wrote the draft manuscript. HZ revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
All animal-related procedures were carried out in accordance with the ARRIVE guidelines and this study was approved by the Ethics Committee of the Chenzhou First People’s Hospital.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, X., Zhou, W., Chen, J. et al. Circ_001653 alleviates sepsis associated-acute kidney injury by recruiting BUD13 to regulate KEAP1/NRF2/HO-1 signaling pathway. J Inflamm 21, 37 (2024). https://doi.org/10.1186/s12950-024-00409-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12950-024-00409-7