Abstract
Background
Extracellular vesicles (EVs) are membrane-enclosed structures containing lipids, proteins, and RNAs that play a crucial role in cell-to-cell communication. However, the precise mechanism through which circulating EVs disrupt hepatic glucose homeostasis in gestational diabetes mellitus (GDM) remains unclear.
Results
Circulating EVs isolated from human plasma were co-cultured with mammalian liver cells to investigate the potential induction of hepatic insulin resistance by GDM-EVs using glucose output assays, Seahorse assays, metabolomics, fluxomics, qRT-PCR, bioinformatics analyses, and luciferase assays. Our findings demonstrated that hepatocytes exposed to GDM-EVs exhibited increased gluconeogenesis, attenuated energy metabolism, and upregulated oxidative stress. Particularly noteworthy was the discovery of miR-1299 as the predominant miRNA in GDM-EVs, which directly targeting the 3′-untranslated regions (UTR) of STAT3. Our experiments involving loss- and gain‐of‐function revealed that miR-1299 inhibits the insulin signaling pathway by regulating the STAT3/FAM3A axis, resulting in increased insulin resistance through the modulation of mitochondrial function and oxidative stress in hepatocytes. Moreover, experiments conducted in vivo on mice inoculated with GDM-EVs confirmed the development of glucose intolerance, insulin resistance, and downregulation of STAT3 and FAM3A.
Conclusions
These results provide insights into the role of miR-1299 derived from circulating GDM-EVs in the progression of insulin resistance in hepatic cells via the STAT3/FAM3A axis and downstream metabolic reprogramming.
Similar content being viewed by others
Introduction
Gestational diabetes mellitus (GDM) is a prevalent complication during pregnancy, characterized by impaired glucose intolerance typically identified in the second or third trimester of pregnancy, without overt diabetes manifestations [1]. The prevalence of GDM has risen over the last two decades, ranging from 1% to over 30%, posing significant risks for both maternal and fetal health [2]. Although the precise pathophysiology of GDM remains poorly understood, it shares similarities with type 2 diabetes, particularly in terms of systemic insulin resistance. Insulin resistance is characterized by diminished sensitivity to insulin in peripheral organs including the liver, skeletal muscle, and adipose tissue, leading to dysregulation of glucose and energy homeostasis within the body [3]. Thus, the pronounced peripheral insulin resistance, brought about through endocrine effects, may play a crucial role in the development of GDM.
Our previous studies have shown that mice fed on a high-fat diet during pregnancy exhibit impairment of glucose metabolism, peripheral insulin resistance, and suppressed insulin signaling, mimicking the characteristics of GDM [4, 5]. In addition, cell-to-cell communication potentially contributes to the pathophysiology of insulin resistance [6]. During pregnancy, diabetogenic autacoids, such as placental hormones, TNF-α and leptin, are predominantly released by the placenta and are thought to play a role in GDM pathogenesis [7]. Nonetheless, the alterations in placental hormones do not establish a direct correlation with maternal insulin resistance. Therefore, alternative types of cell-to-cell communication or inter-organ crosstalk may be key to understanding how insulin resistance develops during GDM pregnancy. Recent studies have highlighted the role of extracellular vesicles (EVs) as a novel pathway for intercellular communication and as significant modulators of physiological and pathological processes [8, 9]. EVs are nanoscale membrane vesicles enclosed by a lipid bilayer that are secreted by almost all cell types and released into the extracellular fluid compartment [10]. These nano-vesicles, which contain proteins, lipids, mRNA, and miRNA, can be selectively targeted to specific cells, transported to recipient cells via the bloodstream, and effect changes in recipient cell functions [11]. There is evidence that the concentration of circulating EVs was more than 50-fold greater in pregnant women than in non- pregnant women and increased gradually throughout pregnancy [12]. Remarkably, patients with diabetes or GDM have higher concentrations of circulating EVs than their normoglycemic counterparts [13, 14]. Hyperglycemia and insulin resistance have also been reported to stimulate the release of EVs and alter miRNA content which affects insulin signaling pathways during the development of diabetes [15, 16]. Consequently, miRNA carried by EVs may serve as promising metabolic regulators in the context of GDM-induced insulin resistance.
miRNAs are small non-coding RNA molecules approximately 21 nucleotides in length, which play a crucial role in the regulation of mRNA translation in cells [17]. An miRNA seed sequence consists of six to eight nucleotides, corresponding to complementary sequences in the 3ʹ-untranslated region (3ʹUTR) of its target mRNA [18]. Upon miRNA binding to the mRNA, RNA-induced silencing complexes (RISC) are recruited, leading to target mRNA translational arrest or mRNA degradation [19]. As a key component of EV content, miRNAs are effectively protected during transport to the specific target cells [20]. Several studies have identified maternal circulating EVs miRNA signatures in GDM, suggesting their potential for mediating insulin resistance [21,22,23]. However, there is still a great deal of uncertainty as to how EVs and their miRNAs regulate peripheral metabolic signaling in GDM. Due to the liver’s significant metabolic role, the knockout of insulin receptors in the liver results in extremely high insulin resistance and severe glucose intolerance [24]. Thomou et al. have shown that the majority of circulating EVs-associated miRNAs in mice have the ability to alter gene expression in the liver [25]. These findings led us to postulate that hepatic insulin resistance, modulated by circulating EVs, may play an important role in mediating the physiological disturbances associated with GDM.
The present study demonstrates that circulating EVs derived from GDM patients can be internalized by liver cells, both in vitro and in vivo, leading to peripheral insulin resistance and glucose intolerance. This study emphasizes the role of circulating EVs as a bloodborne regulator of hepatic glucose homeostasis and insulin sensitivity in GDM, primarily through their miRNA components. Specifically, we report that miR-1299 is one of the differentially expressed miRNAs in GDM-EVs, and its overexpression results in insulin resistance and glucose intolerance, likely mediated by the miR-1299/STAT3/FAM3A axis.
Materials and methods
Subjects and samples
Pregnant women between 24 and 28 weeks’ gestation were recruited from The First Affiliated Hospital of Chongqing Medical University, China. Women were invited to participate in the study if they were between 20 and 40 years of age and had a singleton pregnancy. The exclusion criteria for the study were individuals with a history of diabetes or GDM. A 75 g oral glucose tolerance test (OGTT) was conducted during recruitment to determine case or control status. The diagnosis of GDM was according to the International Association of Diabetes and Pregnancy Study Groups (IADPSG) guidelines [26]. A total of 35 women participated in the study as cases diagnosed with GDM, and 35 women participated as healthy controls. Both groups were matched for age and body mass index (BMI). Fasting blood samples were collected into EDTA blood collection tubes, and plasma was isolated. Ethical approval was granted by the Ethics committee of Chongqing Medical University (No. 2020 − 567), and written informed consent was obtained from all participants.
Isolation and characterization of EVs
EVs were isolated from plasma (1 ml) using differential ultracentrifugation as previously described [27] with modifications. In brief, plasma was diluted with an equal volume of Phosphate Buffered Saline (PBS) and centrifuged at 2,000×g for 30 min, and 12,000 ×g for 45 min. The supernatant fluid was transferred to an ultracentrifuge tube and centrifuged at 110,000×g for 120 min (CP70ME, Hitachi, Japan). The pellet was resuspended in PBS and passed through a 0.22 μm sterile filter (Millipore, USA) and then centrifuged at 110,000×g for 70 min. The resultant pellet was resuspended in 100 μl PBS and stored − 80 °C. To monitor EVs trafficking, EVs were labeled with PKH67 dye (PKH67 Green Fluorescent Cell Linker Mini Kit, Sigma) according to the manufacturer’s instructions. After PKH67 staining, the EVs were washed in PBS and collected by ultracentrifugation (110,000×g for 70 min). Finally, PKH67-labeled EVs were resuspended in PBS. The characterization of EVs was confirmed by measuring the expression of EV-associated protein markers, CD63, CD81, and TSG101 by western blot analysis. Nanoparticle Tracking Analysis (NTA) was applied to analyze the size distribution and particle concentration of EVs using a Zeta View (Particle Metrix, Germany). For transmission electron microscopy (TEM) analysis, a 10 μl aliquot of EVs was added to a formvar/carbon-coated copper grid for 5 min. The sample was negatively stained with 2% uranyl acetate for 1 min and placed at 37 °C for 20 min. Grids were imaged using Tecnai G2 Spirit BioTwin (FEI, USA) transmission electron microscope.
Cell culture and treatment
The normal human hepatic cell line (THLE-2, CL-0833) was kindly provided by Wuhan Pricella Biotechnology Co.,Ltd. THLE-2 cells were cultured in THLE-2 Specific Culture Medium (CM-0833) at 37 °C with 5% CO2. Primary mouse hepatocytes were isolated and cultured following perfusion and collagenase digestion of the liver, as described previously [28]. Two micrograms of EVs based on protein measurement were added to 1 × 105 recipient cells (GDM-EVs group and Normal-EVs group) for 24 h. The equivalent amount of PBS was utilized as a control vehicle (Vehicle group). To validate that GDM-EVs induced insulin resistance by inhibiting STAT3, we co-incubated EVs-treated THLE-2 cells with 1 μM STAT3 activator Colivelin (MCE, USA). To study the regulation of insulin in vitro, the cells were subjected to serum starvation and subsequently treated with insulin (100 nM).
Internalization of EVs by THLE-2 cells
THLE-2 cells were seeded into 12-well microtiter plates with cell climbing slices at 1 × 105 cells/well and incubated with PKH67-stained EVs for 24 h at 37 °C. The cell climbing slices were washed three times with PBS, and then fixed with 4% formaldehyde for 10 min. After washing with PBS three times, the cells were stained with DAPI (Beyotime, China). The samples were analyzed using a Leica fluorescence microscope (Leica, Germany).
Glucose output assay
Following the intervention of EVs for the indicated times, glucose production in hepatocytes was quantified according to the established methods [29, 30]. THLE-2 cells were incubated in a glucose production buffer (Krebs–Henseleit–HEPES buffer supplemented with sodium lactate 20 mM and sodium pyruvate 2 mM). Primary hepatocytes were incubated in a glucose production buffer comprising glucose-free DMEM, devoid of phenol red, and supplemented with 20 mM lactate and 2 mM pyruvate. Subsequent to a 6 h incubation under both insulin-stimulated (100 nM) and insulin-free conditions, the glucose released to the media was measured using a glucose detection assay kit (E1010, Applygen Company).
Western blot analysis
Cells or tissues were homogenized in RIPA buffer (Beyotime, China) supplemented with protease and phosphatase inhibitors. The concentration of the extracted protein was determined using a BCA assay kit (Beyotime, China). Equal amounts of cell lysate proteins from each biological replicate were transferred to a PVDF membrane (Millipore, USA) after SDS-PAGE. The membranes were blocked with TBST containing 5% skimmed milk powder for 1 h and then incubated with primary antibodies (described in Supplementary Table S1) at 4 °C overnight. After incubating with appropriate horseradish peroxidase-conjugated secondary antibodies for 1 h, band density was detected using a Fusion FX7 image analyzer (Vilber, France).
Energy metabolism assay
The energy metabolism of cells was measured using the Seahorse XFp Extracellular Flux Analyzer (Agilent, USA) according to the manufacturer’s user guide. The Seahorse XFp Cell Mito Stress Test Kit (Agilent, USA) was used to evaluate the oxygen consumption rate (OCR), which reflected the mitochondrial function. Mitochondrial parameters (basal respiration, maximal respiration, ATP production, and spare respiratory capacity) were evaluated using OCR indexes.
Metabolomics analysis
Intracellular metabolite extraction, chemical derivatization, isotope tracer analysis, GC-MS analysis, metabolite identification, metabolite quantification, and data normalization were performed as previously described [31]. For the isotope tracer experiment, the effect of 13C-labeled tracer (U-13C6 glucose) on flux estimation precision was undertaken [32]. In brief, 5 ml of liquid nitrogen was added to each plate of THLE-2 cells before intracellular metabolite extraction. Then 1.5 ml cold methanol/chloroform (9:1), containing the standard internal 2,3,3,3-d4-alanine, was used to extract metabolites from THLE-2 cells. The supernatant was obtained by centrifuged at 15,000 g for 15 min at 4 °C and dried in a SpeedVac (Labconco Corp., Missouri, USA) for 5 h at room temperature. The samples were derivatized using the methyl chloroformate (MCF) method and analyzed with an Agilent GC7890B coupled to an MSD5977A mass selective detector (EI) set at 70 eV. A ZB-1701 GC capillary column (30 m × 250 μm id × 0.15 μm with 5 m guard column, Phenomenex) was used for metabolite analysis. GC-MS data were analyzed by MassOmics XCMS R to extrapolate the relative abundance of the metabolites through the peak height of the most enriched ion mass (https://zenodo.org/record/4961895). The corresponding concentration of the identified metabolites were normalized to an internal standard (D4-alanine), total ion concentration of the cellular metabolome.
Cellular reactive oxygen species (ROS) detection
ROS in the THLE-2 cells was measured using a Reactive Oxygen Species Assay Kit (Beyotime, China). After EVs intervention for 24 h at 37 °C, the cell medium was removed, and the cells were incubated with DCFH-DA (10 μM) for 20 min. Subsequently, fluorescence was measured using a microplate reader at 485 nm (excitation) and 528 nm (emission).
miRNA-seq dataset of EVs and bioinformatics analysis
The miRNA expression profiles for circulating GDM EVs were obtained from the Gene Expression Omnibus database (GEO; http://www.ncbi.nlm.nih.gov/geo). We chose miRNA-seq dataset of the circulating GDM EVs in the second trimester from the GSE114860 dataset obtained by high throughput sequencing. The differential expression of miRNAs was analyzed using the DESeq2 package (version 1.38.3) with 1.5-fold changes and p-values under 0.05 as the threshold. To visualize the systems-level interpretation of miRNA functions and gene regulation, an miRNA-centric network visual analytics platform (miRNet 2.0; https://www.mirnet.ca) was used. TargeScan (https://www.targetscan.org), miRDB (http://mirdb.org), RNA22 (https://cm.jefferson.edu/data-t), and TarBase (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=tarbasev8/index) were used to predict potential targets of miRNAs. Only target genes predicted by all the programs were accepted and visualized with a Venn diagram.
RNA extraction and quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
Based on previous findings, RNaseA treatment (37 °C for 10 min) was used to select only RNA encapsulated within EVs [20]. RNA was extracted using the TRIzol LS Reagent (Invitrogen, USA). Reverse transcription was performed on 300 ng of total RNA using the miRNA first strand cDNA synthesis Kit (Accurate Biology, China). U6 was used as the internal reference gene. Total RNA was extracted from cells or tissues using AG RNAex Pro Reagent (Accurate Biology, China) following the manufacturer’s instructions. Reverse transcription for mRNA detection was performed with the Evo M-MLV RT Kit with gDNA Clean for qPCR kit (Accurate Biology, China), using β-actin as an internal reference control. qRT-PCR was conducted using the SYBR Green Premix Pro Taq HS qPCR Kit (Accurate Biology, China) on a CFX96 Real-Time System (Bio-Rad, USA). The primer sequences employed are detailed in Supplementary Table S2. The relative RNA expression levels were calculated using the 2−ΔΔCt method.
miRNA transfection
miRNA mimics (for gain-of-function experiments), inhibitors, and negative controls (Tsingke Biotechnology, China) were transfected into THLE-2 cells using a Lipofectamine 3000 transfection reagent (Thermo Fisher, USA). The miR-1299 mimic sequences were: 5’UUCUGGAAUUCUGUGUGAGGGA 3’ (forward) and 5’ CCUCACACAGAAUUCCAGAAUU 3’ (reverse). The miR-1299 inhibitor sequences were: 5’ CCUCACACAGAAUUCCAGAAUU 3’. Transfection used miRNA mimics at a concentration of 100 nM, and miRNA inhibitors at a concentration of 150 nM. After 48 h of transfection, RNA and protein was extracted from cells using the AG RNAex Pro Reagent (Accurate Biology, China) and RIPA buffer (Beyotime, China), respectively.
Luciferase reporter assay
The binding site of miR-1299 on STAT3 was predicted by TargetScan. The wild type 3’UTR of STAT3 (STAT3 WT) and the corresponding mutant 3’UTR (STAT3 MUT) were amplified by PCR. Then, the STAT3 WT or STAT3 MUT was cloned into the pmirGLO Dual-Luciferase miRNA Target Expression Vector using Nhel and Xbal restriction endonuclease sites. THLE-2 cells were grown in a 96-well microtiter plate until they reached 60% confluence. The luciferase reporter plasmids were co-transfected with miR-1299 mimic or negative controls into cells using Lipofectamine 3000 transfection reagent (Thermo Fisher, USA). The relative luciferase activity was measured after 48 h transfection by using the Dual Luciferase Reporter Gene Assay Kit (Yeasen, China).
Mouse experiments
C57BL/6J (strain C57BL/6J Gpt, Strain NO. N000013) mice were purchased from Gempharmatech (Nanjing, China). The generation of GDM mice has been previously described [4]. The GDM mice were fed a high-fat diet (HFD; Research Diets D12451, consisting of 20% protein, 35% carbohydrate, and 45% fat), while control mice were fed a low-fat diet (LFD; Research Diets AIN-93G, consisting of 20.3% protein, 63.9% carbohydrate, and 15.8% fat). At day 0.5 of gestation (GD0.5), GD11.5, and GD16.5, the mice underwent an oral glucose tolerance test (OGTT) and insulin tolerance test (ITT). At GD18.5, the whole blood of mice was obtained by extracting the eyeball blood under anaesthesia. The circulating EVs of the mice were isolated as described above and stored in a -80 °C until use. For the EVs experiment, mice were randomly divided into three groups: vehicle control (PBS), GDM-EVs treatment (circulating EVs derived from HFD mice) and Normal-EVs treatment (circulating EVs derived from LFD mice, ). All mice were fed a normal chow diet (SPF Rat & Mouse Breeder Diet, BEIJING KEAO XIELI FEED CO., LTD., consisting of 24% protein, 63% carbohydrate, and 13% fat) and were pregnant as recipients. At GD6.5, circulating EVs (30 μg every 2 days for a total of six applications) or vehicles were adoptively transferred into recipient mice via tail vein injection. OGTT or ITT were conducted at GD0.5, GD11.5, and GD16.5. On GD18.5, the mice underwent a six-hour fasting period before being euthanized for blood and liver sample collection for subsequent analysis. Hepatic insulin sensitivity was assessed by quantifying insulin-induced AKT phosphorylation. After the fasting period, mice received an intraperitoneal injection of insulin (0.5 units/kg body weight), and liver samples were harvested 30 min post-injection for Western blot analysis to detect phosphorylated AKT. All animal procedures were approved by the Ethical Committee of Chongqing Medical University (No.2020 − 568).
OGTT and ITT
For oral glucose tolerance tests, mice received glucose (2 g/kg body weight) via gavage after six hours of fasting. Tail vein blood glucose concentrations were determined at t = 0, 30, 60, 90, and 120 min using a glucometer (ACCU-CHEK Performa, Roche, Switzerland). For insulin tolerance tests, mice were fasted for six hours and then insulin (0.5 units/kg body weight) was administered via an intraperitoneal injection. Blood glucose concentration was then measured at t = 0, 15, 30, 60, and 120 min.
Measurement of serum insulin
Insulin serum levels were measured using a Mouse Insulin (INS) ELISA Kit (JL 11459, Shanghai Jianglai Biotechnology, China) according to the manufacturer’s protocols. The insulin resistance score was determined using Homeostatic Model Assessment for Insulin Resistance (HOMA-IR), which was calculated as follows:
Immunofluorescence
Immunofluorescence staining was performed using 15 μm frozen sections of mice liver. Frozen sections were fixed with 4% paraformaldehyde for 15 min at room temperature. Samples were permeabilized with 0.3% Triton-X100 (Sigma) for 4 min, 0.5% Triton-X100 (Sigma) for 10 min, and blocked for 1 h with 5% BSA. Incubation of primary antibodies was performed overnight at 4 °C followed by incubation with fluorochrome-conjugated secondary antibody (Supplementary Table S1). Nuclei were stained with DAPI (Beyotime, China) and imaged with a fluorescence microscope (Leica, Germany).
Statistical analysis
The maternal clinical characteristics were analyzed using R (version 4.2.2). Student’s t-tests were conducted for normally distributed data, while Mann-Whitney tests were used for non-normally distributed data. The results obtained from experiments were presented as mean ± standard error of mean (SEM). Statistical analysis was performed using GraphPad Prism 8 (GraphPad Software, USA) and student’s t-tests were used for comparisons between two groups. The metabolome data analysis was undertaken with MetaboAnalyst 5.0 (https://www.metaboanalyst.ca). To calculate the significance of the differences in metabolites between the two groups, the student’s t-test and the false discovery rate were applied. The Partial least square-discriminant analysis (PLS-DA) was performed to compare cell metabolome profiles between GDM-EVs and Normal-EVs groups. Hierarchical clustering was performed with the hclust function in package stat. Based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database and the Small Molecule Pathway Database (SMPD), the pathway enrichment analysis method and Pathway Activity Profiling (PAPi) analysis were used to identify the most relevant pathways. Differences were regarded as statistically significant if *p < 0.05; **p < 0.01; ***p < 0.001.
Results
EVs characterization and internalization of EVs into hepatic cells
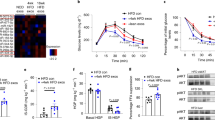
The demographic data of all the participants are summarized in Supplementary Table S3. No statistically significant differences could be found in maternal age and BMI between the GDM and normal groups. However, the GDM group tended to deliver earlier than women in the normal group. Additionally, the fasting, one-hour, and two-hour blood glucose levels following an OGTT were statistically significantly higher in women with GDM than in normal pregnant women. To elucidate the physiological significance of circulating EVs in GDM, we first purified EVs from the plasma of pregnant women by ultracentrifugation and visualized them by transmission electron microscopy after negative staining. Most of the particles appeared intact, approximately 100 nm in diameter, and had a characteristic morphology with a central depression (Fig. 1A). To further characterize the properties of EVs, particles with a diameter of 101.8 ± 28.2 nm (mean ± SD) were identified using Nanoparticle Tracking Analysis (Fig. 1B). Furthermore, the identification of EVs markers, including CD63, CD81, and TSG101, coupled with the absence of cellular marker Calnexin, substantiated that the isolated particles were predominantly EVs (Fig. 1C). To investigate whether the EVs could be internalized by liver cells, we incubated THLE-2 cells with PKH67-stained EVs isolated from the plasma of pregnant women. Within 24 h of co-incubation, immunofluorescence indicated the internalization of PKH67-stained EVs into the THLE-2 cells (Fig. 1D). In a subsequent experiment, PKH67-stained EVs isolated from the plasma of pregnant mice were adoptively transferred into recipient mice via tail vein injection. The presence of the green fluorescent PKH67 dye in the liver of recipient mice indicated in vivo uptake of EVs (Fig. 1E).
EVs isolated from the plasma of pregnant women and internalization of EVs by liver cells. (A) Representative transmission electron microscopy images of isolated EVs. EVs displayed a cup-shaped morphology. (B) The distribution of EVs size by NTA analysis. (C) The expressions of EVs markers (CD81, CD63, and TSG101), and cellular markers (Calnexin) were assessed by western blotting. Appearance of PKH67-stained EVs in THLE-2 cells (D), and liver of recipient mice (E) after 24 h administration of EVs. DAPI stained the nucleus blue, and PKH67 stained EVs green (bar, 50 μm)
The effects of EVs on hepatic glucose metabolism and energy metabolism
To evaluate the potential bioactivity of EVs on the glucose metabolism of liver cells, we conducted experiments on glucogenesis and mitochondrial respiration in THLE-2 cells and primary mouse hepatocytes incubated with plasma EVs from GDM and normal pregnant women. We found that GDM-EVs significantly augmented glucose production and upregulated the gene expression of key gluconeogenic enzymes, including glucose-6-phosphatase catalytic subunit (G6PC) and phosphoenolpyruvate carboxykinase 1 (PCK1), in hepatocytes (Fig. 2A-B and D-E). Additionally, we observed decreased insulin-associated phosphorylation of AKT after GDM-EVs treatment (Fig. 2C and F), suggesting that EVs from GDM plasma can impair glucose metabolism. In order to determine the effects of EVs on hepatic energy metabolism, we measured mitochondrial respiration (OCR) in THLE-2 cells treated with EVs from GDM and normal pregnant women. Administration of GDM-EVs did not affect basal respiration compared to Normal-EVs (p > 0.05), while GDM-EVs induced a significant reduction in maximal respiration, ATP production, and spare respiratory capacity (Fig. 2G).
Effect of EVs from GDM plasma on hepatic glucose metabolism and energy metabolism. (A) Glucose production and (B) the mRNA levels of G6PC and PCK1 in THLE-2 cells after 24 h incubation with plasma EVs from GDM and normal pregnant women. (C) AKT phosphorylation levels in THLE-2 cells after treatment with GDM-EVs or Normal-EVs. (D) Glucose production and (E) the mRNA levels of G6pc and Pck1 in primary mouse hepatocytes after 24 h incubation with plasma EVs. (F) AKT phosphorylation levels in primary mouse hepatocytes after treatment with GDM-EVs or Normal-EVs. (G) Normalized OCR measurements of cells incubated with GDM-EVs and Normal-EVs. After recording three baseline OCR measurements, oligomycin (Oligo), Carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP), and Rotenone/Antimycin A (Rot/AA) were sequentially added, and OCR measurements were taken thrice after each treatment. After EVs were separately injected into mice via tail vein, OGTT(H) and ITT (I) were assessed in each group at GD 11.5. HOMA-IR (J), and the hepatic mRNA levels of G6PC and PCK1(K) in each group at GD 18.5. Results are means ± SEM (n = 3–7). *p < 0.05, **p < 0.01, ***p < 0.001
To delve deeper into the impact of GDM-EVs on insulin sensitivity, we conducted in vivo experiments by inducing GDM mice with a HFD, or injecting mice on a normal chow diet with GDM-EVs (derived from HFD mice), Normal-EVs (derived from LFD mice), or PBS (as vehicle control). According to the OGTT, ITT and mice characteristics results, the HFD induced insulin resistance, and the mice developed GDM (Supplementary Figure S1A-F, Table S4). Normal chow diet-fed pregnant mice were used as recipients and intravenously injected with GDM-EVs, Normal-EVs, and PBS. Although all recipient mice exhibited no difference in body weight after EVs injection, GDM-EVs injection had a tendency to increase body weights at GD 18.5 (p = 0.07) (Supplementary Table S5). GDM-EVs treatment led to impaired glucose tolerance at GD 11.5 and GD 16.5, compared to either mice treated with Normal-EVs or to control animals treated with vehicle (Fig. 2H, Supplementary Figure S2A-B). In addition, treatment with GDM-EVs led to increased insulin resistance, as measured by ITT and HOMA-IR (Fig. 2I-J, and Supplementary Figure S2C-F). Consistent with these results, insulin signaling was reduced in the liver, as evidenced by decreased insulin-stimulated phosphorylation of AKT (Supplementary Figure S2G). Similarly, GDM-EVs also increased the mRNA levels of G6pc and Pck1 in the livers (Fig. 2K). Together, these findings suggest that EVs from GDM plasma impair insulin sensitivity in insulin-targeted hepatic tissues.
GDM-EVs induced distinct metabolome alteration in liver cells
In an effort to elucidate global metabolic alterations, we conducted untargeted metabolomics to analyze the cell metabolic profiles in the GDM-EVs and Normal-EVs groups. Using our in-house mass spectrometry library and the NIST commercial database, we identified a total of 125 metabolites in cells after incubation with GDM-EVs and Normal-EVs. Subsequently, we performed a partial least square-discriminant analysis (PLS-DA) and found a clear separation between the two groups of cells (Fig. 3A). Additionally, through univariate analysis, we discovered fifteen metabolites that exhibited significantly different abundancies (p < 0.05) between the GDM-EVs and Normal-EVs groups (Fig. 3B, Supplementary Table S6). These metabolites were predominantly amino acids and fatty acids. Notably, the most important metabolic feature in the GDM-EVs group was increased oxidized glutathione (p < 0.05 and q < 0.05). Moreover, we observed a decrease in the level of 2-oxoglutaric acid, an intermediate of the Tricarboxylic Acid (TCA) cycle, in the GDM-EVs group. To gain insight into the biological functions of the identified metabolites, we used the KEGG database and the SMPD metabolic framework to annotate the metabolic pathways. Our analysis revealed that fourteen metabolic pathways were significantly differentially enriched (p < 0.05) in the intracellular metabolites of the GDM-EVs and Normal-EVs groups. These pathways included alanine metabolism, glyoxylate and dicarboxylate metabolism, glutathione metabolism, lysine degradation, purine metabolism, primary bile acid biosynthesis, phosphonate and phosphinate metabolism, methionine metabolism, arginine and proline metabolism, porphyrin metabolism, glycine and serine metabolism, thiamine metabolism, the mTOR signaling pathway, and glutamate metabolism (Fig. 3C). Subsequently, we annotated these fourteen metabolic pathways, and found that differential metabolites (glycine, alanine, cysteine, putrescine, 2-oxoglutaric acid and oxidized glutathione) were most involved in the two metabolic pathways: glutamate metabolism and glutathione metabolism (Fig. 3D). Based on these findings, we hypothesized that there was an increased oxidative stress in the cells. To test this hypothesis, we performed a ROS activity assay and observed the accumulation of ROS in cells treated with GDM-EVs (Fig. 3E).
Cellular metabolic profiles after administration of GDM-EVs and Normal-EVs to THLE-2 cells. (A) Partial least square discriminant analysis (PLS-DA) of metabolite patterns in THLE-2 cells incubated with plasma EVs from GDM and normal pregnant women. (B) Heatmap of metabolites with differential (p < 0.05) levels between the GDM-EVs group and the Normal-EVs group. (C) Activities of metabolic pathways in THLE-2 cells incubated with plasma EVs from GDM and normal pregnant women. Black dots represent metabolic activities in the Normal-EVs group that were adjusted to 0. Red dots represent metabolic activities in the GDM-EVs group. The metabolic activities were visualized using a log2 scale. The dot size indicates the number of metabolites of the pathway, and the dot color indicates the p value. (D) A Circos plot displaying the connectivity between significantly enriched pathways and metabolites. (E) GDM-EVs induced oxidative stress in THLE-2 cells. Results are means ± SEM (n = 5–6). *p < 0.05, **p < 0.01
To investigate more thoroughly the impact of GDM-EVs on cell metabolic reprogramming, we conducted isotope enrichment analysis by supplying GDM-EVs- and Normal-EVs-treated liver cells with 30% 13C-labelled glucose as the carbon source. This analysis allows us to track the rate of biochemical conversion from the labelled glucose to the metabolites. Our findings revealed a significant reduction in 13C-labelling of metabolites in the GDM-EVs group (Fig. 4). In particular, TCA cycle intermediates (e.g. citric acid, succinic acid, fumaric acid and malic acid) exhibited a lower rate of biochemical conversion, while 2-oxoglutaric acid also had a tendency to lower isotope enrichment (p = 0.07). However, most of the relevant metabolites involved in glutamate and glutathione metabolism did not show a significant difference between the groups, except for glutamic acid and proline. These results suggest that GDM-EVs may attenuate mitochondrial respiration by reducing the flux through the TCA cycle.
13C-labelled glucose metabolic flux and intracellular TCA cycle metabolism in EV-treated THLE-2 cells. TCA cycle metabolism showed a lower rate of biochemical conversion after the administration of GDM-EVs. The histograms show a percentage difference of 13C-labelling metabolites between the GDM-EVs and the Normal-EVs groups. M+ is the main molecular ion of an identified metabolite, + 1 is 1 m/z higher than the M+. Results are means ± SEM (n = 5). *p < 0.05, **p < 0.01
EVs miRNA expression changes in GDM
Although EVs carry diverse cargo, a significant portion of their biological effects have been attributed to miRNAs. Specifically, EVs miRNAs have exhibited significant regulatory roles in glucose metabolism and insulin sensitivity in individuals with diabetes and GDM [33]. To investigate the miRNAs associated with GDM-EVs, we obtained the GSE114860 miRNA expression dataset of circulating GDM EVs from the GEO database. From this dataset, we identified over 500 miRNAs and determined a set of miRNAs differentially expressed in GDM-EVs compared to Normal-EVs. Figure 5A displays a hierarchical clustering of the 27 significant differentially EVs miRNAs (p < 0.05). Subsequently, we selected 15 differentially expressed miRNAs based on a Volcano plot using a threshold of 1.5-fold change in expression and p < 0.05 to identify significantly regulated miRNAs. Among these, four miRNAs were upregulated, while eleven miRNAs were downregulated in the GDM group (Fig. 5B). miR-1299 exhibited the most significant upregulation in expression in GDM-EVs compared to Normal-EVs. To validate this finding, we performed qPCR and confirmed that miR-1299 was expressed approximately two-fold more in GDM-EVs than in Normal-EVs in both human and mouse samples (Fig. 5C-D).
Significant changes in the miRNA profile of circulating EVs in GDM. (A) The differential expression level of miRNAs between GDM-EVs and Normal-EVs. (B) Volcano plot showing four upregulated (red) and eleven downregulated (blue) miRNAs (log2 fold-change > 1.5). qPCR was used to determine the differential expression of miR-1299 in human (C) and murine (D) GDM-EVs compared to Normal-EVs. Results are means ± SEM (n = 3–5). *p < 0.05. (E) A miRNA-target gene topology network for miR-1299. The grey nodes represent genes, and the red nodes represent miRNAs. Nineteen putative target genes of miR-1299 were highlighted by enlarged blue circles with labelled gene names
The significant miRNAs were analyzed using miRNet 2.0 to construct an miRNA-target gene network. This network revealed that these miRNAs were linked to 5798 genes which were visually analyzed as a topology network (Supplementary Figure S3A). Module analysis reduced the network complexity while keeping the functions and connections of miR-1299 (Fig. 5E). To further explore the molecular mechanism of miR-1299 in GDM, 21 putative target genes were identified by miRNA target prediction algorithms and combining the results from the TargeScan, miRDB, RNA22, and TarBase databases (Supplementary Figure S3B). Among the 21 putative target genes, 19 genes were also found in the miRNA-target gene network and were listed in the topology network of miR-1299 (Fig. 5E). Furthermore, a review of the literature, indicated that signal transducer and activator of transcription 3 (STAT3) plays a role in influencing insulin sensitivity and energy metabolism. FAM3 metabolism regulating signaling molecule A (FAM3A), a major downstream effector of STAT3, has also been shown to impact energy metabolism [34]. Our western blot analysis showed that incubating liver cells with GDM-EVs led to a significant decrease in the abundance of STAT3, P-STAT3, and FAM3A (Supplementary Figure S3C).
Elevated miR-1299 levels in GDM-EVs induced insulin resistance through the STAT3/FAM3A pathway
To probe more deeply into the mechanisms underlying the induction of insulin resistance by GDM-EVs, we determined the effects of miR-1299 and STAT3/FAM3A on insulin sensitivity. We observed a significant increase in miR-1299 abundance within liver cells after incubation with GDM-EVs (Fig. 6A). After the administration of Colivelin (activator of STAT3), there was a reduction in the promotion of cellular glucose output and gluconeogenic gene by GDM-EVs (Fig. 6B-C). Meanwhile, the abundance of STAT3, P-STAT3, FAM3A, and P-AKT were downregulated after treatment with GDM-EVs (Fig. 6D-E). The administration of Colivelin significantly attenuated the suppressive effect of GDM-EVs on the expression of STAT3, P-STAT3, FAM3A, and P-AKT (Fig. 6D-E). In vivo, we observed a reduction in the hepatic expression of STAT3, P-STAT3, and FAM3A following the administration of GDM-EVs to mice (Fig. 6F-G).
GDM-EVs induced insulin resistance through STAT3/FAM3A. (A) After 24 h exposure of THLE-2 cells to GDM-EVs or Normal-EVs, the levels of miR-1299 in cells were measured by qPCR analysis. The effects of Colivelin on recovery of glucose output (B) and gluconeogenic gene (C) in liver cells with treatment of GDM-EVs. (D) The effects of Colivelin on recovery of STAT3, FAM3A, P-STAT3, and P-AKT expression levels in cells exposed to GDM-EVs. (E) Statistical analysis of western blot results. Representative immunofluorescent staining of STAT3, FAM3A (F), and P-STAT3 (G) localization in the liver from mice injected GDM-EVs or Normal-EVs. (bar, 50 μm). Results are means ± SEM (n = 3–6). *p < 0.05, **p < 0.01, ***p < 0.001
To assess the effect of miR-1299 on insulin resistance, THLE-2 cells were transfected with miR-1299-mimics. The overexpression of miR-1299 significantly increased gluconeogenesis in liver cells (Fig. 7A-B). Concomitantly, the expressions of STAT3, P-STAT3, FAM3A, and P-AKT were also decreased in liver cells with miR-1299 overexpression (Fig. 7E). Conversely, when liver cells were transfected with miR-1299 inhibitor the opposite trend was observed (Fig. 7C-E). Bioinformatics algorithms predicted that the 3ʹUTR of STAT3 WT contained complementary base pairing to miR-1299. Subsequently, a dual-luciferase reporter assay showed that THLE-2 cells co-transfected with STAT3 WT and miR-1299 displayed decreased luciferase activity. However, this inhibition was blocked by mutation of the STAT3 binding domains (Fig. 7F). Overall, these results suggest that elevated miR-1299 in GDM-EVs targeted STAT3 and inhibited the expression of STAT3/FAM3A, which might contribute to insulin resistance in GDM.
MiR-1299 impairs insulin sensitivity through STAT3/FAM3A. (A-D) Glucose output and gluconeogenic gene in THLE-2 cells after transfection with miR-1299 mimics or inhibitor. (E) Expression of STAT3, FAM3A, P-STAT3, and P-AKT after overexpressing or inhibiting of miR-1299. (F) MiR-1299 directly targets STAT3. The binding site of the miRNA on its target was predicted by TargetScan. Results are means ± SEM (n = 3–6). *p < 0.05, **p < 0.01, ***p < 0.001. WT = wild type, MUT = mutation
Discussion
There is increasing evidence that insulin resistance is influenced by inter-organ communication, with EVs playing a role in this crosstalk [33]. Circulating EVs comprise a heterogeneous population of vesicles released by various tissues. EVs not only act as a waste disposal system by packaging and secreting unwanted components from different organs [35], but their cargos (e.g. miRNAs, oxidized proteins, and metabolites) also contribute to global metabolic reprogramming [35, 36]. Recent studies have indicated that the number of circulating EVs are elevated during pregnancy, and GDM patients have higher levels of EVs than women with normal pregnancies [37]. Considering the liver receives blood from both the portal vein and the arterial system, the elevated levels of circulating EVs in GDM may directly impact the liver and influence insulin sensitivity. Therefore, the aim of this study was to investigate how circulating EVs from GDM patients affect peripheral insulin resistance and glucose intolerance in the liver through the action of miRNA. We showed that there was increased glucogenesis, impaired energy metabolism, and upregulation of miR-1299 in hepatocytes exposed to circulating EVs derived from GDM patients. Notably, miR-1299 was found to be the most overexpressed miRNA in EVs from GDM blood samples. The elevated levels of miR-1299, derived from circulating EVs in GDM, induced insulin resistance in liver cells by repressing STAT3 and its downstream gene, FAM3A, through an miR-1299-mediated mechanism. In an in vivo mouse study, pregnant non-GDM mice injected with GDM-EVs developed systemic insulin resistance and glucose intolerance. Therefore, our investigation of EVs as paracrine or endocrine vectors may provide new insight on hepatic insulin resistance in GDM patients.
Hepatic uptake of circulating EVs
Circulating EVs were harvested from GDM plasma by differential centrifugation. We confirmed these vesicles as EVs with a diameter of about 100 nanometers by transmission electron microscopy, Nanoparticle Tracking Analysis, and measurement of the EVs markers CD63, CD81, and TSG101. The present study used two methods to demonstrate the direct transfer of circulating EVs into target cells. Firstly, liver cells were treated with PKH67 labeled EVs and the uptake of the fluorescent label into cells was directly observed ex vivo. The results of our in vivo experiments further supported this finding, as mice treated with PKH67-labeled EVs retained the EVs in the liver. Importantly, the liver is a crucial site for the development of insulin signaling abnormalities that affect glucose and lipid metabolism [38]. Previous studies have investigated the biodistribution of EVs and consistently reported their accumulation primarily in the liver [39, 40]. Wilklander et al. found that regardless of dose, time duration, route of injection, or tissue source, EVs tend to accumulate predominantly in the liver [40]. Moreover, EVs resemble liposomes in terms of size, shape, and structure but have more complex bilayers. In line with earlier findings on the biodistribution of synthetic liposomes, the accumulation of nanoparticles in the liver may be attributed to taken up by patrolling macrophages, for example, by Kupffer cells in liver [41]. Nevertheless, several nanoparticle studies have revealed that particle size affects the distribution and clearance of nanoparticles. As an example, small liposomes (≤ 100 nm) are less prone to opsonization and penetrate the fenestrations in the hepatic sinusoidal endothelium (about 150 nm) [42,43,44]. In line with earlier findings on the biodistribution of synthetic liposomes, our results showed the circulating EVs size distribution was 101.8 ± 28.2 nm. Most of EVs could penetrate these fenestrations, thus increasing the hepatocyte uptake of the total accumulation of EVs. Considering the fluorescent labeling of EVs, the physiology of the liver, and the structural characteristics of EVs, this information supports the notion that hepatocytes can uptake circulating EVs.
GDM-EVs induce insulin resistance through mitochondrial dysfunction and oxidative stress
Impairments in mitochondrial oxidative function, characterized by reduced numbers of mitochondria per cell, reduced ATP generation, and increased ROS production, could potentially affect insulin sensitivity within hepatocytes [45]. In addition, EVs have also been shown to inactivate the energetic networks of hepatic cells, leading to insulin resistance by increasing mitochondrial ROS and reducing AKT phosphorylation [15, 35, 46]. Our study showed that treatment with circulating GDM EVs led to increased glucogenesis, and reduced AKT phosphorylation in hepatocytes (Fig. 2). Furthermore, our results demonstrated that GDM-EVs attenuated mitochondrial respiration and energy production, as evidenced by a reduced OCR. These findings are supported by our metabolome profiling, which revealed a lower level of 2-oxoglutaric acid (a TCA cycle intermediate) and a reduced flux through the TCA cycle induced by GDM-EVs (Figs. 3 and 4). Thus, our data provide evidence that circulating EVs in GDM may impair mitochondrial function. Most of the cellular energy is produced by mitochondria via the TCA cycle and oxidative phosphorylation and is accompanied by the generation of ROS. Mitochondrial dysfunction often leads to impaired oxidative capacity and excessive ROS production. Indeed, oxidative stress is a well-recognized risk factor for insulin resistance and may increase the demand for glutathione synthesis. Glutathione exists in reduced (GSH) and oxidized (GSSG) forms, which are necessary for maintaining the redox balance of the cell. The de novo synthesis of GSH in the cell is limited by the availability of its constituent amino acids, and in particular by the availability of the sulfur amino acid precursor, cysteine [47]. Our hepatic metabolome results revealed higher levels of GSSG, lower levels of cysteine, and increased activity of glutathione-related metabolic pathways in the GDM-EVs group (Fig. 3). Meanwhile, excessive ROS accumulated in the liver cells after being treated with GDM-EVs, potentially indicating the pro-oxidant risk of GDM-EVs. Thus, GDM-EVs may elicit hepatic insulin resistance by triggering mitochondrial malfunction and oxidative stress, initiating a vicious cycle of imbalanced antioxidant status, ROS generation, and further damage to mitochondria.
GDM-EVs contain a high level of miR-1299
Although the EVs’ cargo consists of proteins, lipids, miRNAs, and other RNA species, many of the biological effects of EVs are often attributed to miRNAs [33]. For instance, miRNAs have exhibited critical roles in regulating glucose metabolism and insulin action in diabetes and GDM [48, 49]. In this study, we analyzed a sequencing dataset of circulating GDM EVs and identified 15 differentially expressed miRNAs. Among these, miR-1299 stood out as the most significantly increased miRNA in circulating GDM EVs relative to normal circulating EVs. Interestingly, elevated levels of miR-1299 have also been previously found in the blood of patients with type 2 diabetes mellitus and prediabetes [50,51,52]. Furthermore, miRNAs in EVs seem to mirror the metabolic status of the cells they originate from and can be protected from external ribonucleases [20]. Thus, we assumed that miRNAs, carried by circulating GDM EVs, taken up and released into liver cells might induce hepatic insulin resistance. It is worth noting that the upregulation of miR-1299 expression in blood EVs has only been reported in patients with polycystic ovary syndrome (PCOS), which is characterized by insulin resistance [53]. Additionally, a modest number of studies have shown that other miRNAs derived from circulating GDM EVs regulate glucose homeostasis and insulin sensitivity [21, 23]. Despite previous attempts to study miRNAs other than miR-1299 in circulating GDM EVs, the effect of circulating EVs-associated miRNAs on hepatic insulin resistance in GDM and the underlying mechanisms remain largely unknown.
miR-1299/STAT3/FAM3A axis may be involved in the mechanism of hepatic insulin resistance induction in GDM
To gain a better understanding of the metabolic effects associated with miR-1299, we performed a bioinformatic analysis to predict the target genes of miR-1299 using miRNA target prediction databases. This analysis showed that miR-1299 could bind to the 3ʹUTR of signal transducer and activator of transcription 3 (STAT3). STAT3 is a transcription factor expressed in hepatic tissue and plays a crucial role in regulating insulin sensitivity [54, 55]. STAT3 also regulates cell metabolism by promoting mitochondrial oxidative phosphorylation [56,57,58]. Furthermore, recent studies have identified FAM3A a direct downstream target of STAT3 and a vital regulator of glucose metabolism in the liver [34, 59]. It has been reported that FAM3A expression was reduced in the livers of obese diabetic mice, and the suppression of the FAM3A/ATP/AKT pathway caused insulin resistance, hyperglycemia, and fatty liver [60]. To investigate the involvement of the miR-1299/STAT3/FAM3A axis in insulin resistance mediated by circulating GDM EVs, we conducted further experiments. Our results demonstrated that GDM-EVs significantly reduced the expression of STAT3 and FAM3A in liver cells (Fig. 6). Moreover, the administration of a STAT3 activator significantly improved glucogenesis and the expressions of STAT3/FAM3A proteins, which were downregulated by circulating GDM EVs (Fig. 6). Luciferase reporter plasmids containing predictive STAT3 target gene sequences showed that STAT3 was indeed a direct target of miR-1299 (Fig. 7). Meanwhile, miR-1299 overexpression increased glucogenesis and downregulated STAT3, FAM3A, and P-AKT expression. Consistent with these results, GDM-EVs altered hepatic insulin signaling and the expression of STAT3 and FAM3A in vivo (Figs. 2 and 6). Previous studies have reported that EVs-trafficked miRNA downregulate STAT3 and impair insulin signaling in type 2 diabetes [61]. Nair et al. also highlighted that the STAT3 pathway was the most prominent biological process targeted by GDM-EVs-associated miRNAs [22]. Furthermore, hepatic STAT3 signaling was essential to maintain normal glucose homeostasis [62]. As a major STAT3 downstream effector, FAM3A is mainly present in mitochondria and can restore the reduction of mitochondrial respiration caused by STAT3 deficiency [34]. Evidence is mounting that mitochondrial dysfunction may be intimately linked to the development of hepatic insulin resistance [63]. In this way, circulating GDM EVs may contribute to hepatic insulin resistance through mitochondrial dysfunction via miR-1299/STAT3/FAM3A axis.
Limitations
One limitation of the current study is the unidentified source of the EVs in plasma. As EVs are released from various cells, those in blood circulation are often heterogeneous vesicles of unknown origin. Therefore, the standard isolation process does not differentiate vesicles based on their origin, presenting a challenge in determining the specific role and occurrence of miR-1299-containing EVs. In future studies, this issue will need to be investigated in more depth, to elucidate the cellular origins and the complex mechanisms that regulate the formation and behavior of these EVs. Another limitation of this study is that miR-1299 is unlikely to be the sole factor responsible for the differences in insulin sensitivity induced by EVs. It would be reasonable to assume that GDM EVs contain other miRNAs with important functions. The coordinated action of multiple miRNAs within EVs may also contribute to insulin-resistant phenotypes, and further research will be needed to demonstrate the full set of EV miRNAs that cause metabolic effects. While miR-1299 and its interaction with the STAT3/FAM3A axis are important, they are likely part of a larger system. Other regulatory molecules and signaling pathways could also play significant roles in modulating hepatic glucose metabolism. Further research is needed to elucidate the molecular interactions that contribute to this complex physiological process. In addition, the phenomenon and regulatory mechanisms involving miRNA and its target molecules may exhibit tissue-specific variations. Therefore, additional studies across muscle and other tissues are warranted to fully understand the universality and tissue-specificity of miRNA regulation.
Conclusion
In summary, our results demonstrate that circulating EVs influence hepatic insulin sensitivity through miRNA. Treatment with circulating GDM EVs leads to in vivo and in vitro insulin resistance. In particular, elevated miR-1299 levels in circulating GDM EVs altered STAT3/FAM3A signaling and induced hepatic insulin resistance through mitochondrial malfunction and oxidative stress (Fig. 8). Thus, we provide new evidence that EVs regulate glucose metabolism and may provide novel therapeutic options for GDM.
Schematic representation of the mechanisms of GDM EVs-derived miR-1299 induction of hepatic insulin resistance. In women with GDM, the levels of circulating EVs were elevated and miR-1299 was highly expressed in these EVs. Circulating GDM EVs were absorbed by hepatocytes, leading to an increase in the level of miR-1299 within these cells. Then, miR-1299 binding to the 3ʹUTR of STAT3 mRNA resulted in STAT3 translational arrest which consequently reduced the expression of FAM3A. The decreased levels of STAT3 and FAM3A can potentially affect mitochondrial function by slowing down TCA cycle flux and mitochondrial respiration, while increasing oxidative stress. Therefore, elevated miR-1299 in circulating GDM EVs is proposed to induce hepatic insulin resistance through impaired mitochondrial function via inhibiting the STAT3/FAM3A axis
Data availability
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- EVs:
-
Extracellular vesicles
- FAM3A:
-
FAM3 metabolism regulating signaling molecule A
- G6PC:
-
Glucose-6-phosphatase catalytic subunit
- GDM:
-
Gestational diabetes mellitus
- GSH:
-
Glutathione
- GSSG:
-
Oxidized glutathione
- HFD:
-
High-fat diet
- HOMA-IR:
-
Homeostatic Model Assessment for Insulin Resistance
- ITT:
-
Insulin tolerance test
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- LFD:
-
Low-fat diet
- NTA:
-
Nanoparticle tracking analysis
- OCR:
-
Oxygen consumption rate
- OGTT:
-
Oral glucose tolerance test
- PBS:
-
Phosphate Buffered Saline
- PCK1:
-
Phosphoenolpyruvate carboxykinase 1
- ROS:
-
Reactive oxygen species
- SEM:
-
Standard error of mean
- SMPD:
-
Small Molecule Pathway Database
- STAT3:
-
Signal transducer and activator of transcription 3
- TCA:
-
Tricarboxylic Acid
- TEM:
-
Transmission electron microscopy
- UTR:
-
Untranslated regions
References
American Diabetes Association. 2. Classification and diagnosis of diabetes: standards of medical care in diabetes-2021. Diabetes Care. 2021;44:S15–33.
McIntyre HD, Catalano P, Zhang C, Desoye G, Mathiesen ER, Damm P. Gestational diabetes mellitus. Nat Rev Dis Primers. 2019;5:47.
Czech MP. Insulin action and resistance in obesity and type 2 diabetes. Nat Med. 2017;23:804–14.
Zhao Y, Zhou X, Zhao X, Yu X, Wang A, Chen X, Qi H, Han TL, Zhang H, Baker PN. Metformin administration during pregnancy attenuated the long-term maternal metabolic and cognitive impairments in a mouse model of gestational diabetes. Aging. 2020;12:14019–36.
Wang A, Luo B, Chen Z, Xia Y, Chen C, Qi H, Baker PN, Saffery R, Han TL, Zhang H. Rodents on a high-fat diet born to mothers with gestational diabetes exhibit sex-specific lipidomic changes in reproductive organs. Acta Biochim Biophys Sin (Shanghai). 2022;54:736–47.
Barbour LA, McCurdy CE, Hernandez TL, Kirwan JP, Catalano PM, Friedman JE. Cellular mechanisms for insulin resistance in normal pregnancy and gestational diabetes. Diabetes Care. 2007;30(Suppl 2):S112–119.
Fasshauer M, Bluher M, Stumvoll M. Adipokines in gestational diabetes. Lancet Diabetes Endocrinol. 2014;2:488–99.
Mori MA, Ludwig RG, Garcia-Martin R, Brandao BB, Kahn CR. Extracellular miRNAs: from biomarkers to mediators of physiology and disease. Cell Metab. 2019;30:656–73.
Hoshino A, Kim HS, Bojmar L, Gyan KE, Cioffi M, Hernandez J, Zambirinis CP, Rodrigues G, Molina H, Heissel S, et al. Extracellular vesicle and particle biomarkers define multiple human cancers. Cell. 2020;182:1044–e10611018.
Mathieu M, Martin-Jaular L, Lavieu G, Thery C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat Cell Biol. 2019;21:9–17.
Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–9.
Salomon C, Torres MJ, Kobayashi M, Scholz-Romero K, Sobrevia L, Dobierzewska A, Illanes SE, Mitchell MD, Rice GE. A gestational profile of placental exosomes in maternal plasma and their effects on endothelial cell migration. PLoS ONE. 2014;9:e98667.
Salomon C, Scholz-Romero K, Sarker S, Sweeney E, Kobayashi M, Correa P, Longo S, Duncombe G, Mitchell MD, Rice GE, Illanes SE. Gestational diabetes mellitus is associated with changes in the concentration and bioactivity of placenta-derived exosomes in maternal circulation across gestation. Diabetes. 2016;65:598–609.
Freeman DW, Noren Hooten N, Eitan E, Green J, Mode NA, Bodogai M, Zhang Y, Lehrmann E, Zonderman AB, Biragyn A, et al. Altered extracellular vesicle concentration, cargo, and function in diabetes. Diabetes. 2018;67:2377–88.
Ying W, Riopel M, Bandyopadhyay G, Dong Y, Birmingham A, Seo JB, Ofrecio JM, Wollam J, Hernandez-Carretero A, Fu W, et al. Adipose tissue macrophage-derived exosomal miRNAs can modulate in vivo and in vitro insulin sensitivity. Cell. 2017;171:372–e384312.
Castano C, Mirasierra M, Vallejo M, Novials A, Parrizas M. Delivery of muscle-derived exosomal miRNAs induced by HIIT improves insulin sensitivity through down-regulation of hepatic FoxO1 in mice. Proc Natl Acad Sci U S A. 2020;117:30335–43.
Anfossi S, Babayan A, Pantel K, Calin GA. Clinical utility of circulating non-coding RNAs - an update. Nat Rev Clin Oncol. 2018;15:541–63.
Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–34.
Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14.
Cheng L, Sharples RA, Scicluna BJ, Hill AF. Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. J Extracell Vesicles. 2014;3:23743.
Gillet V, Ouellet A, Stepanov Y, Rodosthenous RS, Croft EK, Brennan K, Abdelouahab N, Baccarelli A, Takser L. miRNA profiles in extracellular vesicles from serum early in pregnancies complicated by gestational diabetes mellitus. J Clin Endocrinol Metab. 2019;104:5157–69.
Nair S, Guanzon D, Jayabalan N, Lai A, Scholz-Romero K, Kalita de Croft P, Ormazabal V, Palma C, Diaz E, McCarthy EA, et al. Extracellular vesicle-associated miRNAs are an adaptive response to gestational diabetes mellitus. J Transl Med. 2021;19:360.
Ye Z, Wang S, Huang X, Chen P, Deng L, Li S, Lin S, Wang Z, Liu B. Plasma exosomal miRNAs associated with metabolism as early predictor of gestational diabetes mellitus. Diabetes. 2022;71:2272–83.
Michael MD, Kulkarni RN, Postic C, Previs SF, Shulman GI, Magnuson MA, Kahn CR. Loss of insulin signaling in hepatocytes leads to severe insulin resistance and progressive hepatic dysfunction. Mol Cell. 2000;6:87–97.
Thomou T, Mori MA, Dreyfuss JM, Konishi M, Sakaguchi M, Wolfrum C, Rao TN, Winnay JN, Garcia-Martin R, Grinspoon SK, et al. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature. 2017;542:450–5.
International Association of D, Pregnancy Study Groups, Consensus P, Metzger BE, Gabbe SG, Persson B, Buchanan TA, Catalano PA, Damm P, Dyer AR, Leiva A, et al. International association of diabetes and pregnancy study groups recommendations on the diagnosis and classification of hyperglycemia in pregnancy. Diabetes Care. 2010;33:676–82.
McDonald MK, Capasso KE, Ajit SK. Purification and microRNA profiling of exosomes derived from blood and culture media. J Vis Exp. 2013;76:e50294.
Cabral F, Miller CM, Kudrna KM, Hass BE, Daubendiek JG, Kellar BM, Harris EN. Purification of hepatocytes and sinusoidal endothelial cells from mouse liver perfusion. J Vis Exp. 2018;132:56993.
Zhang Y, Zhou B, Zhang F, Wu J, Hu Y, Liu Y, Zhai Q. Amyloid-β induces hepatic insulin resistance by activating JAK2/STAT3/SOCS-1 signaling pathway. Diabetes. 2012;61:1434–43.
Mithieux G. Transcription factor p63, a member of the p53 family of tumour suppressors, regulates hepatic glucose metabolism. Gut. 2023;72:415–6.
Chen H, Wang SH, Chen C, Yu XY, Zhu JN, Mansell T, Novakovic B, Saffery R, Baker PN, Han TL, Zhang H. A novel role of FoxO3a in the migration and invasion of trophoblast cells: from metabolic remodeling to transcriptional reprogramming. Mol Med. 2022;28:92.
Han TL, Cannon RD, Gallo SM, Villas-Boas SG. A metabolomic study of the effect of Candida albicans glutamate dehydrogenase deletion on growth and morphogenesis. NPJ Biofilms Microbiomes. 2019;5:13.
Isaac R, Reis FCG, Ying W, Olefsky JM. Exosomes as mediators of intercellular crosstalk in metabolism. Cell Metab. 2021;33:1744–62.
Sala D, Cunningham TJ, Stec MJ, Etxaniz U, Nicoletti C, Dall’Agnese A, Puri PL, Duester G, Latella L, Sacco A. The Stat3-Fam3a axis promotes muscle stem cell myogenic lineage progression by inducing mitochondrial respiration. Nat Commun. 2019;10:1796.
Yin Y, Chen H, Wang Y, Zhang L, Wang X. Roles of extracellular vesicles in the aging microenvironment and age-related diseases. J Extracell Vesicles. 2021;10:e12154.
Crewe C, Funcke JB, Li S, Joffin N, Gliniak CM, Ghaben AL, An YA, Sadek HA, Gordillo R, Akgul Y, et al. Extracellular vesicle-based interorgan transport of mitochondria from energetically stressed adipocytes. Cell Metab. 2021;33:1853–e18681811.
James-Allan LB, Devaskar SU. Extracellular vesicles and their role in gestational diabetes mellitus. Placenta. 2021;113:15–22.
Haeusler RA, Accili D. The double life of Irs. Cell Metab. 2008;8:7–9.
Abello J, Nguyen TDT, Marasini R, Aryal S, Weiss ML. Biodistribution of gadolinium- and near infrared-labeled human umbilical cord mesenchymal stromal cell-derived exosomes in tumor bearing mice. Theranostics. 2019;9:2325–45.
Wiklander OP, Nordin JZ, O’Loughlin A, Gustafsson Y, Corso G, Mager I, Vader P, Lee Y, Sork H, Seow Y, et al. Extracellular vesicle in vivo biodistribution is determined by cell source, route of administration and targeting. J Extracell Vesicles. 2015;4:26316.
Moghimi SM, Hunter AC, Murray JC. Long-circulating and target-specific nanoparticles: theory to practice. Pharmacol Rev. 2001;53:283–318.
Braet F, Wisse E. Structural and functional aspects of liver sinusoidal endothelial cell fenestrae: a review. Comp Hepatol. 2002;1:1.
Garnett MC, Kallinteri P. Nanomedicines and nanotoxicology: some physiological principles. Occup Med (Lond). 2006;56:307–11.
Sarin H. Physiologic upper limits of pore size of different blood capillary types and another perspective on the dual pore theory of microvascular permeability. J Angiogenes Res. 2010;2:14.
Patti ME, Corvera S. The role of mitochondria in the pathogenesis of type 2 diabetes. Endocr Rev. 2010;31:364–95.
Kranendonk ME, Visseren FL, van Herwaarden JA, Nolte-‘t Hoen EN, de Jager W, Wauben MH, Kalkhoven E. Effect of extracellular vesicles of human adipose tissue on insulin signaling in liver and muscle cells. Obes (Silver Spring). 2014;22:2216–23.
Lu SC. Regulation of glutathione synthesis. Mol Aspects Med. 2009;30:42–59.
Ortega FJ, Mercader JM, Moreno-Navarrete JM, Rovira O, Guerra E, Esteve E, Xifra G, Martinez C, Ricart W, Rieusset J, et al. Profiling of circulating microRNAs reveals common microRNAs linked to type 2 diabetes that change with insulin sensitization. Diabetes Care. 2014;37:1375–83.
Rahimi G, Jafari N, Khodabakhsh M, Shirzad Z, Dogaheh HP. Upregulation of microRNA processing enzymes Drosha and Dicer in gestational diabetes mellitus. Gynecol Endocrinol. 2015;31:156–9.
Massaro JD, Polli CD, Costa ESM, Alves CC, Passos GA, Sakamoto-Hojo ET, Rodrigues de Holanda Miranda W, Bispo Cezar NJ, Rassi DM, Crispim F, et al. Post-transcriptional markers associated with clinical complications in type 1 and type 2 diabetes mellitus. Mol Cell Endocrinol. 2019;490:1–14.
Weale CJ, Matshazi DM, Davids SFG, Raghubeer S, Erasmus RT, Kengne AP, Davison GM, Matsha TE. Expression profiles of circulating microRNAs in South African type 2 diabetic individuals on treatment. Front Genet. 2021;12:702410.
Weale CJ, Matshazi DM, Davids SFG, Raghubeer S, Erasmus RT, Kengne AP, Davison GM, Matsha TE. MicroRNAs-1299, -126-3p and – 30e-3p as potential diagnostic biomarkers for prediabetes. Diagnostics (Basel). 2021;11:949.
Zhang F, Li SP, Zhang T, Yu B, Zhang J, Ding HG, Ye FJ, Yuan H, Ma YY, Pan HT, He Y. High throughput microRNAs sequencing profile of serum exosomes in women with and without polycystic ovarian syndrome. PeerJ. 2021;9:e10998.
Buettner C, Pocai A, Muse ED, Etgen AM, Myers MG Jr., Rossetti L. Critical role of STAT3 in leptin’s metabolic actions. Cell Metab. 2006;4:49–60.
Yu H, Lee H, Herrmann A, Buettner R, Jove R. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat Rev Cancer. 2014;14:736–46.
Wegrzyn J, Potla R, Chwae YJ, Sepuri NB, Zhang Q, Koeck T, Derecka M, Szczepanek K, Szelag M, Gornicka A, et al. Function of mitochondrial Stat3 in cellular respiration. Science. 2009;323:793–7.
Gough DJ, Corlett A, Schlessinger K, Wegrzyn J, Larner AC, Levy DE. Mitochondrial STAT3 supports ras-dependent oncogenic transformation. Science. 2009;324:1713–6.
Camporeale A, Demaria M, Monteleone E, Giorgi C, Wieckowski MR, Pinton P, Poli V. STAT3 activities and energy metabolism: dangerous liaisons. Cancers (Basel). 2014;6:1579–96.
Wang C, Chi Y, Li J, Miao Y, Li S, Su W, Jia S, Chen Z, Du S, Zhang X, et al. FAM3A activates PI3K p110alpha/Akt signaling to ameliorate hepatic gluconeogenesis and lipogenesis. Hepatology. 2014;59:1779–90.
Yang W, Wang J, Chen Z, Chen J, Meng Y, Chen L, Chang Y, Geng B, Sun L, Dou L, et al. NFE2 induces mir-423-5p to promote gluconeogenesis and hyperglycemia by repressing the hepatic FAM3A-ATP-Akt pathway. Diabetes. 2017;66:1819–32.
Katayama M, Wiklander OPB, Fritz T, Caidahl K, El-Andaloussi S, Zierath JR, Krook A. Circulating exosomal miR-20b-5p is elevated in type 2 diabetes and could impair insulin action in human skeletal muscle. Diabetes. 2019;68:515–26.
Inoue H, Ogawa W, Ozaki M, Haga S, Matsumoto M, Furukawa K, Hashimoto N, Kido Y, Mori T, Sakaue H, et al. Role of STAT-3 in regulation of hepatic gluconeogenic genes and carbohydrate metabolism in vivo. Nat Med. 2004;10:168–74.
Tilg H, Moschen AR, Roden M. NAFLD and diabetes mellitus. Nat Rev Gastroenterol Hepatol. 2017;14:32–42.
Acknowledgements
The authors thank all the study participants and all collaborators for data collection. The authors are grateful for the support provided by The Chongqing Key Laboratory of Translational Medicine in Major Metabolic Diseases. We also would like to thank Figdraw (www.figdraw.com).
Funding
This work was supported by the National Natural Science Foundation of China (No. 81971406, 81871185), The 111 Project (Yuwaizhuan (2016)32), Chongqing Science & Technology Commission (cstc2021jcyj-msxmX0213), Chongqing Municipal Education Commission (KJZD-K202100407), Chongqing Health Commission and Chongqing Science & Technology Commission (2021MSXM121, 2020MSXM101).
Author information
Authors and Affiliations
Contributions
X-B Z, T-L H, and H Z conceived the idea and designed the study. X-Y C and T-L H analyzed data and drafted the manuscript. X-Y C, X-Y T, and M W recruited participants, collected clinical data, and prepared samples. X-Y C performed EVs preparation, GC-MS analysis, bioinformatics analysis, luciferase reporter assay and other related experiments. X-Y C, X-Y Y, X-Y T and M W contributed to the animal-related experiments. R D C, R S, H-B Q, P N B, X-B Z, T-L H, and H Z critically reviewed the manuscript. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Ethical approval was granted by the Ethics committee of Chongqing Medical University (No. 2020 − 567 and No.2020 − 568).
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Chen, X., Tao, X., Wang, M. et al. Circulating extracellular vesicle-derived miR-1299 disrupts hepatic glucose homeostasis by targeting the STAT3/FAM3A axis in gestational diabetes mellitus. J Nanobiotechnol 22, 509 (2024). https://doi.org/10.1186/s12951-024-02766-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12951-024-02766-0