Abstract
Background
Epidermal growth factor receptor (EGFR) fusions are rare but potentially actionable oncogenic drivers across multiple solid tumors. However, the distribution and molecular characteristics of EGFR fusions in Chinese patients with solid malignancies have not been explored.
Methods
Panel-based next-generation sequencing (NGS) data of 35,023 patients with various types of solid tumors was collected and analyzed from the Simcere Diagnostics (Nanjing, China) database. A 9563-patient cohort was derived from The Cancer Genome Atlas (TCGA) to explore the relationship between EGFR fusion status and overall survival (OS).
Results
In this study, prevalence of functional EGFR fusions was 0.303% (106/35,023) in total across solid tumors, which occur more commonly in gastroesophageal junction cancer (1/61, 1.613%), followed by medulloblastoma (1/66, 1.515%) and glioma (33/2409, 1.370%). Analysis showed a prevalence for fusion partners in different tumor types. The top 3 co-mutant genes with EGFR fusion were TP53 (mutation frequency, MF: 65%), BRCA2 (MF: 43%), and ALK (MF: 41%). Furthermore, patients in the EGFR fusion group had a significantly shorter OS than those in the non-EGFR fusion group (p < 0.0001) in the TCGA cohort, suggesting that EGFR fusion might be a high-risk factor for poor prognosis.
Conclusions
Our study is the first retrospective analysis of EGFR fusions in a large-scale solid tumor population, which may provide a reference for future EGFR-TKI clinical trials with EGFR fusions.
Similar content being viewed by others
Introduction
Epidermal growth factor receptor (EGFR) is a receptor tyrosine kinase that belongs to the ERBB protein family, which includes three other members, namely ErbB2/HER-2, ErbB3/HER-3 and ErbB4/HER-4 [1,2,3]. Overexpression of EGFR or activation through ligand-dependent and ligand-independent mechanisms are common driving mechanisms in cancer [3, 4]. Subsequently, a cascade of multiple events in the cytoplasm occurs, leading to cell proliferation, survival, and inhibition of apoptosis [1, 5]. The key role of EGFR in cell signaling pathways makes it a major therapeutic target for cancer [6,7,8]. EGFR tyrosine kinase inhibitors (TKIs), as single-target inhibitors of EGFR, have completely changed the treatment model for patients with non-small cell lung cancer (NSCLC) [9, 10]. To date, many different small molecules have been developed as potential EGFR TKIs, from first to fourth generation, some of which are used in the clinical treatment of cancer [10,11,12,13].
EGFR mutations occur more commonly in lung cancer and glioma. In NSCLC, the common mutation of EGFR is located in exons 18–21. Deletions in exon 19 and single amino acid substitutions in exon 21 to L858R, commonly referred to as “classic” EGFR mutations, together account for approximately 80-85% of EGFR mutations observed in NSCLC [14,15,16]. In glioma, the most common cause of abnormal activation of the EGFR pathway is EGFR amplification and mutation [17]. The most common EGFR mutation in glioma is EGFRvIII mutation caused by deletion of exons 2–7. The incidence of EGFRvIII mutations in primary glioblastoma is only about 20–30%. Both EGFR amplification and the occurrence of EGFRvIII mutations predict a poor survival prognosis in glioma patients [18].
As detection technology advances, some rare or atypical EGFR mutations have been identified. Gene fusions involving EGFR are rare in various cancers. One study reported the discovery of 5 EGFR fusions in analysis of 10,000 NSCLC case [19]. Kartik Konduri et al. first reported carcinogenic EGFR fusion in lung cancer, the most common being EGFR-RAD51 [20]. Subsequently, several cases of EGFR fusion were reported in lung cancer, colorectal cancer, glioblastoma, and other cancers, of which lung cancer was the most common. The patients with EGFR fusion can benefit from the corresponding EGFR-TKIs, including erlotinib, icotinib, and afatinib [21,22,23,24]. The mechanism of EGFR fusion in tumors remains unclear, and EGFR fusion could be a potential target.
In this study, we molecularly characterized 35,023 Chinese patients’ tumor samples across multiple solid tumors by next-generation sequencing (NGS) and further analyzed the data of 106 patients with EGFR fusions. The aim is to provide an outlook for patients with EGFR fusions in solid tumors and evidence for more effective treatment.
Methods
Patient information and sample collection
A total of 35,023 patients with 13 cancer types diagnosed between January 2019 and December 2022 were enrolled in this study. The cohort underwent comprehensive genomic profiling of the targeted panel by Simcere Diagnostics Co., Ltd. (Nanjing, China), of which 14,874 patients were detected using a 539-gene panel. The study was conducted in accordance with the Declaration of Helsinki. The written informed consent was waived for this retrospective analysis.
Next-generation sequencing detection
DNA extraction and library preparation
Formalin-fixed paraffin-embedded (FFPE) tumor slides and paired blood samples were collected. Two DNA extraction kits were used: a Tissue Sample DNA Extraction Kit (Kai Shuo) for genomic DNA (gDNA) extraction from tumor tissue and a MagMAXTM DNA Multi-Sample Ultra Kit (Thermo) for gDNA extraction from leukocytes. The extraction procedures were performed according to the manufacturer’s protocol. Library construction used the probe hybridization capture method. Briefly, 15–200 ng gDNA was fragmented into 200 ~ 350 bp fragments by fragmentation enzymes. After end repair, poly(A)-tailing and adaptor ligation, customized probes and a commercial kit were used for hybridization capture and library quantification, respectively.
Library sequencing and bioinformatics analysis
The qualified DNA libraries were sequenced on an Illumina NovaSeq6000 platform (Illumina, San Diego, CA) to generate 150 bp paired-end reads. Base calls from Illumina NovaSeq6000 were conducted to FASTQ files. The software Fastp (v.2.20.0) was used for adapter trimming and filtering of low-quality bases [25]. The BWA-MEM (v.0.7.17) algorithm was performed to align to the reference genome (UCSC hg19 GRCh37) [26]. Duplicate reads from PCR were excluded using Dedup with Error Correct. SNVs/InDels were called and annotated via VarDict (v.1.5.7) [27] and InterVar [28]. The variants were filtered against the common SNPs in the public database, including the 1000 Genome Project (Aug 2015) and Exome Aggregation Consortium (ExAC) Browser28 (v.0.3). Copy number variations (CNVs) and fusions were analyzed by CNVkit (dx1.1) [29] and factera (v1.4.4) [30], respectively.
TMB was defined as the number of somatic, coding, base substitution, and indel mutations per megabase of genome examined. The 539-cancer gene-targeted NGS panel TMB was counted by summing all base substitutions and indels in the coding region of targeted genes, excluding synonymous alterations, alterations of AF < 0.02, and alterations listed as known somatic alterations in COSMIC.
Data acquisition and analysis from TCGA
All clinical and genomic data of 9563 solid tumors across 21 tumor types were retrieved from cBioPortal (www.cbioportal.org). These data were collected from The Cancer Genome Atlas (TCGA) Pan-Cancer analysis project [31]. The CNV pipeline and pipelines for the detection of gene fusions are described on the GDC documentation website (https://docs.gdc.cancer.gov/Data/Introduction/).
Statistical analysis
All statistical analyses were performed using R V4.0.5 (https://www.r-project.org). Differences in TMB between subgroups stratified by EGFR fusion status were analyzed by the Wilcoxon test. The landscape of the co-occurring gene alteration events in a 539-gene panel was generated using ComplexHeatmap (R package). The circos plot showing the chromosome distribution of EGFR fusion was analyzed by RCircos (R package). The MutationMapper module (https://www.cbioportal.org/mutation_mapper) from cBioPortal was used to investigate the distribution of mutations at the protein domain. The Kaplan‒Meier curve analysis of OS was compared using the log-rank test. All reported P values were two-tailed, and P < 0.05 was considered statistically significant.
Results.
Clinical characteristics of patients with EGFR fusion
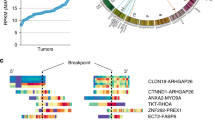
In total, 0.303% (106/35,023) of patients in our cohort harbored EGFR fusion. EGFR fusion is classified into variant type 1 and type 2 according to the direction of fusion and the principle of activation. Variant type 1 is EGFR-X, which may result in the loss of the EGFR autophosphorylation site in the C-terminal tail of the receptor [20]. Variant type 2 is the “X-EGFR” and has a common fusion activation mechanism in which the partner gene leads to the activation and continued expression of the EGFR protein. All 106 EGFR fusions included the full EGFR kinase domain, which is encoded by exons 18–24. As seen in Fig. 1A, we present a schematic with an EGFR partner gene occurred ratio higher than 4%. The activation and treatment effectiveness with TKI have been reported in both EGFR fusion variant types [22, 23]. Patient characteristics are summarized in Table 1. Patients’ age ranged from 5 to 82 years, with a median age of 60.5 years. Sixty-three (59.43%) patients were male, and 43 (40.57%) were female. A total of 51.89% had clinical-stage III/IV disease versus 5.66% with stage I-II disease, and other stages were unavailable (NA). Among the top two cancers, adenocarcinoma (31, 64.58%) predominates in lung cancer, and glioblastoma (16, 48.48%) predominates in glioma. Variant type 1 accounted for 53.77%, variant type 2 accounted for 37.74%, and contained both two variant types accounted for 8.49%.
EGFR rearrangements in solid tumors. (A) Schematic of all EGFR fusions of two variant types. The dark blue in the upper part represents variant type 1, and the red represents variant type 2. The partners are colored green, with the EGFR kinase domain colored yellow and other EGFR exons colored blue by individual boxes. (B) The frequencies of EGFR fusions in diverse tumor types. Blue represents variant type1, red represents variant type1 and green represents both two variant types
In total, EGFR fusions were detected in 0.303% (106/35,023) of the patients, with varying frequencies across diverse tumor types. Lung cancer exhibited a prevalence of 0.236% (48/20,345), glioma 1.370% (33/2,409), gastric cancer 0.676% (11/1,627), colorectal cancer 0.084% (3/3,582), breast cancer 0.743% (2/269), bone and soft tissue sarcoma 0.277% (2/721), and a single case of EGFR fusion was identified in gastroesophageal junction cancer, medulloblastoma, cervical cancer, pancreatic cancer, biliary tumors, liver cancer, and other types as shown in Fig. 1B.
Identification of EGFR fusion partners in patients with different cancer types
The partner genes of the two fusion types are shown in Table 2. SEPT14, ELDR, and LOC100996654 are the most frequent in variant type (1) ELDR and LOC100996654 are the most common in variant type (2) The fusion partner genes are also different in various cancer types. The partner genes in different cancer types are shown in Supplementary Fig. 1A-B. The chromosome distribution of the fusion partner genes of the two types is shown in Fig. 2A-B and the partner genes are scattered across the chromosomes. Breakpoint positions of different EGFR fusion variant types are shown in Fig. 2C. The breakpoints of EGFR in patients with lung cancer were concentrated in exons 15, 16, 27, and 28. In patients with glioma, breakpoints of EGFR were concentrated in exons 7, 8, 24, and 25. The breakpoints of EGFR in other cancer types are scattered.
Co-occurrence genetic mutants and TMB of patients harboring EGFR fusion
Among 106 patients with EGFR fusion, 55 patients’ tumors were examined by NGS using a 539-gene panel, and the co-mutation status of these patients is shown in Fig. 3A. Among the 55 patients with EGFR fusion, 31 (56%) patients had EGFR amplification, and 26 (47%) patients had EGFR single nucleotide variants (SNV, including missense mutation, in-frame deletion, in-frame insertion and multi-hit) mutations. Notably, there were 19 (35%) patients with sensitive mutations. The top 5 genes co-mutated with EGFR fusion were TP53 (mutation frequency, MF: 65%), BRCA2 (MF: 43%), ALK (MF: 41%), MUC16 (MF: 39%), and MYC (MF: 39%).
The median TMB among the 55 patients with EGFR fusion was 4.26 (0.71-139.01) Muts/Mb in solid tumors. The median TMB was 3.97 (0.71-139.01) in the 26 patients with lung cancer, 9.22 (1.42–63.83) in the nine patients with gastric cancer and 2.49 (0.71–4.26) in the eight patients with glioma. There was no significant difference in TMB between EGFR fusion and non-EGFR fusion groups, whether among pan-cancer (p = 0.054), lung cancer (p = 0.28), glioma (p = 0.27) or gastric cancer patients (p = 0.11, Supplementary Fig. 2).
Among 106 patients with EGFR fusion, there were 77 patients with EGFR CNV and 62 patients with EGFR SNV (including L858R, 19 exon deletion, L861Q, T790M, 20 exon insertion and so on; Fig. 3B). EGFR L858R, L861Q, T790M, and 20 exon insertion are mutation types that have only been detected in lung cancer patients in our cohort, while other types of cancer have not been detected. Furthermore, according to Fig. 3C and D, the EGFR mutation sites in lung cancer and glioma are different. In lung cancer, EGFR mutations mostly occurred in functional kinase domain, mainly tyrosine kinase catalytic (TyrKc). But in glioma, the most common EGFR mutation is EGFRvIII mutation, which may be caused by deletion of exons 2–7. These results are basically consistent with the results reported in other previous studies [14,15,16, 19].
The prognostic impact of EGFR fusions
We collected 9,563 tumor variant data in 30 cancer types from TCGA to calculate the proportion of EGFR fusions, and 27 (0.28%) samples were identified harboring EGFR fusions in six cancer types, including 22 patients with variant type 1 and 5 patients with variant type 2. The proportion of glioma was the highest (16/1094, 1.46%), followed by bladder urothelial carcinoma (3/410, 0.73%) and head and neck squamous cell carcinoma (3/522, 0.57%) (Table 3). Notably, there was no co-mutant EGFR-sensitive mutation, but 26 EGFR CNV co-mutations were found among the 27 patients with EGFR fusion.
We further explored the relationship between EGFR fusion and overall survival (OS) in the TCGA cohort. Patients with EGFR fusion had a significantly shorter OS than those without EGFR fusion in pan-cancer (p < 0.0001, Fig. 4A) and in glioma (p = 0.028, Fig. 4B), suggesting that EGFR fusion might be a high-risk factor for poor prognosis. Notably, there were 26 EGFR amplification co-mutations among the 27 EGFR fusion samples. Therefore, we specifically detected the relationship between EGFR fusion, only the EGFR amplification group and others (without EGFR fusion and EGFR amplification). The results showed that in pan-cancer, the EGFR fusion group had the worst OS, and the EGFR CNV group had a comparably worse OS than others group (p < 0.0001, Fig. 4C). In glioma, the EGFR fusion and the EGFR amplification groups had a comparably worse OS than others group (p < 0.0001, Fig. 4D).
Kaplan‒Meier survival curves of patients with pan-cancer TCGA. (A) Kaplan‒Meier survival curves of patients with pan-cancer in the EGFR fusion group and non-EGFR fusion group in TCGA in pan-cancer. (B) Kaplan‒Meier survival curves of patients with pan-cancer in the EGFR fusion group and non-EGFR fusion group in TCGA in glioma. (C) The Kaplan‒Meier survival curves of patients with pan-cancer in the EGFR fusion with CNV co-mutation group and only EGFR CNV group in TCGA. (D) The Kaplan‒Meier survival curves of patients with pan-cancer in the EGFR fusion with CNV co-mutation group and only EGFR CNV group in TCGA
Case presentation
A 71-year-old male patient was admitted to the hospital for 15 days with an irritant cough and two days with bloody sputum. A space-occupying lesion in the left upper lobe was discovered using computed tomography (CT) (Fig. 5A). Following the appropriate preoperative examination, a left total pneumonectomy was performed on November 2, 2020 (Fig. 5B). Percutaneous lung biopsy yielded a diagnosis of lung adenocarcinoma. Finally, the disease was diagnosed as stage IIIA (T1bN2M0). The patient then received postoperative adjuvant chemotherapy for six months, CT results were shown in Fig. 5C. Subsequently, FFPE samples were analyzed by NGS, and a novel EGFR-SEPT14 (11.36% abundance) fusion was detected (Fig. 5E-F). Several other mutations, such as TP53, CCNE1 and HRAS were also observed (Supplementary Table 1). The patient then received almonertinib mesilate tablets (500 mg daily) and remained relapse-free until July 8, 2023 (Fig. 5D).
Chest computed tomography revealed the different stages of the patient’s lung tumor and Next-generation sequencing findings of EGFR-SEPT14 fusion. (A) Computed tomography diagnosis of lung tissue. (B) CT scan after the surgery. (C) CT scan after adjuvant chemotherapy. (D) CT scan after almonertinib treatment for 26 months. (E) Sequencing reads of EGFR are shown by the Integrative Genomics Viewer. (F) Illustration of the EGFR-SEPT14 fusion
Discussion
Our study is the first extensive sample study analysis of EGFR fusion in solid tumors. We retrospectively analyzed EGFR fusion among 35,023 patients with solid tumors who underwent NGS detection and found that the overall incidence of EGFR fusion in solid tumors was 0.303%. A total of 106 patients were found harboring EGFR fusion, of which 57 patients harbored variant type 1, 40 harbored variant type 2, and 9 harbored both types 1 and 2. The incidence of EGFR fusion in various cancer types was quite different, ranging from 0.034 to 1.613%. The incidences of glioma, gastric cancer, and lung cancer with a large number of participants were 1.37% (33/2409), 0.676% (11/1627), and 0.236% (48/20,345), respectively. This is the first time that the frequency of the EGFR fusion has been disclosed in detail.
EGFR fusion represents a novel oncogenic driver across different cancer types. There could be two distinct activation mechanisms of EGFR fusion. The variant type 1 is EGFR-X, and the mechanism has been reported to be that the loss of the C-terminal tail autophosphorylation site leads to the loss of the Cbl binding site, which makes the EGFR fusion protein more stable and finally activates tumorigenic signaling and forms an oncogenic phenotype [20, 32]. This activation mechanism of variant type 1 has been reported, and several cases are successful against EGFR TKI, including in lung cancer and colorectal cancer [20, 23, 33, 34]. Variant type 2 is X-EGFR, and its activation mechanism is similar to that of ALK fusion, in which EGFR retains the complete kinase domain and the partner gene may contain promoters or CC domains. Preclinical carcinogenic effects of this fusion have also been reported [24], as well as cases of TKI sensitivity [22, 35]. Gene rearrangement plays an essential role in the occurrence and development of tumors, and rearrangement can be treated by TKI targeting with generally satisfactory effects. Both variant types 1 and 2 can activate EGFR and operate as effective TKI targets, making them novel therapeutic targets for patients with solid tumors.
The partner gene was further investigated, and the distribution of EGFR partner genes was found to be relatively scattered across the chromosome. Variant types 1 and 2 are evenly distributed on chromosomes. Unlike the case report, there is no RAD51 gene, which is more reported in other literature. In addition to some reported EGFR fusions, including EGFR-ZNF713, EGFR-TNS3, and EGFR-SEPT14, various rare and novel EGFR fusion partners were identified in this study. In addition, multiple EGFR fusions were identified in one individual patient. The distribution of EGFR partner genes in each cancer species is also scattered, which may introduce challenges for the widespread detection of partners. NGS can accurately identify fusion partners and breakpoints, which may become the most widely used procedure in clinical practice.
Previous reports of EGFR fusions were based mainly on the occurrence and cases of lung cancer. Our study is the first large-scale retrospective study of EGFR fusion occurrence in pan-solid tumors. The frequencies of EGFR fusions in pan cancer and some cancers are similar in Chinese and Western populations (TCGA), including glioma (1.37% vs. 1.46%) and lung cancer (0.236% vs. 0.20%). In addition, we analyzed the co-mutations of EGFR fusion, including the combination of EGFR-sensitive mutations and EGFR CNV, of which 30 (56%) patients had EGFR CNV and 19 (35%) patients had EGFR-sensitive mutations in our cohort. However, the combination of EGFR-sensitive mutations is different from that of TCGA, which may be related to ethnic differences, and Asians have a higher frequency of EGFR-sensitive mutations. Additionally, the TMB of EGFR fusion patients was further analyzed, which indicated that the TMB of EGFR fusion was higher than that of non-EGFR fusion but with no statistical significance, no matter in pan cancer or in the lung, gastric cancer, and glioma.
Patients with EGFR fusion had a significantly shorter OS than patients without EGFR fusion in TCGA, which suggests that EGFR fusion is probably a high-risk factor for poor prognosis. Emerging evidence supports that EGFR amplification predicts worse outcomes in patients with lung cancer [36]. Almost all patients with EGFR fusion had EGFR amplification (96.30%, 26/27) in TCGA. We explored the relationship between EGFR fusion and only EGFR CNV in the TCGA cohort. Patients in the EGFR fusion group had a significantly shorter OS than those in the group only harboring EGFR amplification, which means that patients with EGFR fusion have a worse prognosis.
A few of cases have been reported in which patients with EGFR fusions responded to corresponding EGFR-TKIs, including erlotinib, icotinib, and afatinib [21,22,23,24]. Guoqing Zhang et al. described a patient with NSCLC with EGFR fusion and EGFR amplification who achieved a significant antitumor response from treatment with gefitinib combined with cetuximab [37]. Our case described success in treating with EGFR-TKIs, which provides a reference for future EGFR-TKI clinical trials with EGFR fusions.
However, some limitations are worth noting. There was no RNA-based NGS or protein level validation to confirm the functional activation of EGFR fusion. Some cancer types had limited numbers of patients, and clinical follow-up data were unavailable from our cohort. The distribution of partner genes is not concentrated, and future detection of EGFR fusion needs to be carried out by NGS.
In summary, this is the first retrospective analysis of EGFR fusions in a large-scale solid tumor population. Our study may provide a reference for designing future EGFR TKI clinical trials with EGFR fusions.
Data availability
No datasets were generated or analysed during the current study.
References
Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2(2):127–37.
Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer. 2005;5(5):341–54.
Scaltriti M, Baselga J. The epidermal growth factor receptor pathway: a model for targeted therapy. Clin Cancer Res. 2006;12(18):5268–72.
Butti R, et al. Receptor tyrosine kinases (RTKs) in breast cancer: signaling, therapeutic implications and challenges. Mol Cancer. 2018;17(1):34.
Ullrich A, et al. Human epidermal growth factor receptor cDNA sequence and aberrant expression of the amplified gene in A431 epidermoid carcinoma cells. Nature. 1984;309(5967):418–25.
Tan L, et al. Development of dual inhibitors targeting epidermal growth factor receptor in Cancer Therapy. J Med Chem. 2022;65(7):5149–83.
Bhatia P, et al. Novel quinazoline-based EGFR kinase inhibitors: a review focussing on SAR and molecular docking studies (2015–2019). Eur J Med Chem. 2020;204:112640.
Abourehab MAS et al. Globally approved EGFR inhibitors: insights into their syntheses, Target Kinases, Biological activities, receptor interactions, and metabolism. Molecules, 2021. 26(21).
Soltan OM, et al. Molecular hybrids: a five-year survey on structures of multiple targeted hybrids of protein kinase inhibitors for cancer therapy. Eur J Med Chem. 2021;225:113768.
Douillard JY, et al. First-line gefitinib in caucasian EGFR mutation-positive NSCLC patients: a phase-IV, open-label, single-arm study. Br J Cancer. 2014;110(1):55–62.
Camidge DR, et al. Exploratory Analysis of Brigatinib Activity in patients with anaplastic lymphoma kinase-positive non-small-cell Lung Cancer and Brain metastases in two clinical trials. J Clin Oncol. 2018;36(26):2693–701.
Pirker R, et al. Cetuximab plus chemotherapy in patients with advanced non-small-cell lung cancer (FLEX): an open-label randomised phase III trial. Lancet. 2009;373(9674):1525–31.
Herbst RS. Review of epidermal growth factor receptor biology. Int J Radiat Oncol Biol Phys. 2004;59(2 Suppl):21–6.
Lynch TJ, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–39.
Paez JG, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497–500.
Gazdar AF. Activating and resistance mutations of EGFR in non-small-cell lung cancer: role in clinical response to EGFR tyrosine kinase inhibitors. Oncogene. 2009;28(Suppl 1):S24–31.
Wong AJ, et al. Structural alterations of the epidermal growth factor receptor gene in human gliomas. Proc Natl Acad Sci U S A. 1992;89(7):2965–9.
Jiang T, et al. CGCG clinical practice guidelines for the management of adult diffuse gliomas. Cancer Lett. 2016;375(2):263–73.
Zehir A, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23(6):703–13.
Konduri K, et al. EGFR fusions as Novel therapeutic targets in Lung Cancer. Cancer Discov. 2016;6(6):601–11.
Zhu YC, et al. Identification of a Novel Icotinib-Sensitive EGFR-SEPTIN14 Fusion variant in Lung Adenocarcinoma by Next-Generation sequencing. J Thorac Oncol. 2019;14(8):e181–3.
Wang X, et al. A novel KIF5B-EGFR Fusion variant in non-small-cell lung Cancer and response to Afatinib: a Case Report. Onco Targets Ther. 2021;14:3739–44.
Li Y, et al. A Rare EGFR-SEPT14 Fusion in a patient with colorectal adenocarcinoma responding to Erlotinib. Oncologist. 2020;25(3):203–7.
Komuro A, et al. Identification of a novel fusion gene HMGA2-EGFR in glioblastoma. Int J Cancer. 2018;142(8):1627–39.
Chen S, et al. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):i884–90.
Hwang KB, et al. Comparative analysis of whole-genome sequencing pipelines to minimize false negative findings. Sci Rep. 2019;9(1):3219.
Lai Z, et al. VarDict: a novel and versatile variant caller for next-generation sequencing in cancer research. Nucleic Acids Res. 2016;44(11):e108.
Li Q, Wang K. InterVar: clinical interpretation of genetic variants by the 2015 ACMG-AMP guidelines. Am J Hum Genet. 2017;100(2):267–80.
Talevich E, et al. CNVkit: genome-wide Copy Number Detection and visualization from targeted DNA sequencing. PLoS Comput Biol. 2016;12(4):e1004873.
Newman AM, et al. FACTERA: a practical method for the discovery of genomic rearrangements at breakpoint resolution. Bioinformatics. 2014;30(23):3390–3.
Weinstein JN, et al. The Cancer Genome Atlas Pan-cancer analysis project. Nat Genet. 2013;45(10):1113–20.
Paik PK. Something Old, something New, something borrowed, something fused: Novel EGFR rearrangements in lung adenocarcinomas. Cancer Discov. 2016;6(6):574–5.
Zhu YC, et al. EGFR-RAD51 fusion variant in lung adenocarcinoma and response to erlotinib: a case report. Lung Cancer. 2018;115:131–4.
Raez LE, et al. EGFR-RAD51 Fusion: a Targetable Partnership originated from the Tumor Evolution? J Thorac Oncol. 2018;13(3):e33–4.
Xu H, Shao C. KIF5B-EGFR Fusion: a Novel EGFR Mutation in Lung Adenocarcinoma. Onco Targets Ther. 2020;13:8317–21.
Chang H, et al. Epidermal growth factor receptor gene amplification predicts worse outcome in patients with surgically resected Nonadenocarcinoma Lung Cancer. Clin Lung Cancer. 2019;20(1):7–e121.
Zhang G, et al. Gefitinib Combined with Cetuximab for the treatment of Lung Adenocarcinoma harboring the EGFR-Intergenic Region (SEC61G) Fusion and EGFR amplification. Oncologist. 2021;26(11):e1898–902.
Acknowledgements
The authors thank Ms. Xiaofeng Zhu, Mr. Chuang Qi, Mr. Wanglong Deng, Mr. Dongsheng Chen, Mr. Ran Ding and Ms. Qin Shuai from Simceredx for the kindly assistance.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
H.Z., J.W., and T.W. planed the study. X.L., D.Z., X.W., and Q.Z. analyzed data and interpreted the results. Y.Q. and N.L. wrote the manuscript. H.Z., J.W., and T.W. participated in manuscript revision and editing. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
No ethical certificate is required.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Zhang, H., Wang, J., Li, X. et al. Distribution of EGFR fusions in 35,023 Chinese patients with solid tumors-the frequency, fusion partners and clinical outcome. World J Surg Onc 22, 194 (2024). https://doi.org/10.1186/s12957-024-03463-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12957-024-03463-w