Abstract
Background
Cardiac pulsation propels blood through the cerebrovascular network to maintain cerebral homeostasis. The cerebrovascular network is uniquely surrounded by paravascular cerebrospinal fluid (pCSF), which plays a crucial role in waste removal, and its flow is suspected to be driven by arterial pulsations. Despite its importance, the relationship between vascular and paravascular fluid dynamics throughout the cardiac cycle remains poorly understood in humans.
Methods
In this study, we developed a non-invasive neuroimaging approach to investigate the coupling between pulsatile vascular and pCSF dynamics within the subarachnoid space of the human brain. Resting-state functional MRI (fMRI) and dynamic diffusion-weighted imaging (dynDWI) were retrospectively cardiac-aligned to represent cerebral hemodynamics and pCSF motion, respectively. We measured the time between peaks (∆TTP) in \(\frac{d}{d\phi }fMRI\) and dynDWI waveforms and measured their coupling by calculating the waveforms correlation after peak alignment (correlation at aligned peaks). We compared the ∆TTP and correlation at aligned peaks between younger [mean age: 27.9 (3.3) years, n = 9] and older adults [mean age: 70.5 (6.6) years, n = 20], and assessed their reproducibility within subjects and across different imaging protocols.
Results
Hemodynamic changes consistently precede pCSF motion. ∆TTP was significantly shorter in younger adults compared to older adults (−0.015 vs. −0.069, p < 0.05). The correlation at aligned peaks were high and did not differ between younger and older adults (0.833 vs. 0.776, p = 0.153). The ∆TTP and correlation at aligned peaks were robust across fMRI protocols (∆TTP: −0.15 vs. −0.053, p = 0.239; correlation at aligned peaks: 0.813 vs. 0.812, p = 0.985) and demonstrated good to excellent within-subject reproducibility (∆TTP: intraclass correlation coefficient = 0.36; correlation at aligned peaks: intraclass correlation coefficient = 0.89).
Conclusion
This study proposes a non-invasive technique to evaluate vascular and paravascular fluid dynamics. Our findings reveal a consistent and robust cardiac pulsation-driven coupling between cerebral hemodynamics and pCSF dynamics in both younger and older adults.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Introduction
The proper flow of para-arterial cerebrospinal fluid (pCSF) is essential to maintain cerebral homeostasis by contributing to the delivery of nutrients and the removal of waste products throughout the brain [1, 2]. Despite its importance, the exact mechanisms behind pCSF flow are not completely understood. Accumulating evidence suggests that vessel wall motion is a major contributing factor toward the distribution of pCSF throughout the brain [3,4,5,6,7,8,9,10]. Accordingly, animal studies evaluating conditions that alter cerebral vascular health including aging [11], arterial ligation [6], hypertension [5], and cerebral amyloid angiopathy have reported evidence of impaired cerebrospinal fluid (CSF) clearance [12, 13]. Therefore, it is imperative to develop a comprehensive understanding of the coupling between cerebral hemodynamics and pCSF flow in humans, particularly given its potential implications in aging [11, 14] and neurodegenerative pathologies [12, 15].
Recent animal and human studies have unveiled a close association between hemodynamics and CSF dynamics. Notably in humans, strong correlations have been observed between cerebral blood volume (CBV) and CSF inflow and outflow at the fourth ventricle [3, 4, 16, 17]. However, these studies specifically measure CSF dynamics at the fourth ventricle and may not be reflective of the coupling between cerebral hemodynamics and CSF flow within the para-arterial space, which is more relevant to pCSF influx into the brain parenchyma and the corresponding waste clearance function. Animal studies utilizing two-photon microscopy have provided compelling evidence of the interplay between pulsation-driven arterial wall motion and pCSF flow [5, 9, 10]. In humans, the para-arterial space of the major cerebral arteries has been identified as a compartmentalized subarachnoid space [18], and its pulsatile dynamics have been studied using dynamic diffusion-weighted imaging (dynDWI) [19,20,21,22]. However, the interpretation of data in humans is currently limited to pCSF dynamics alone and lacks integration with cerebral hemodynamics. Additional evidence from contrast-enhanced MRI studies suggests that arterial pulsations contribute to the entry of CSF into the brain parenchyma through the paravascular spaces [23]. Nevertheless, these invasive scanning protocols do not directly provide evidence regarding the dependence of CSF clearance on vascular mechanics [24]. Overall, human studies have been limited in their ability to assess the coupling between cerebral hemodynamics and pCSF dynamics within subarachnoid spaces.
Exploring the potential interplay between cerebral hemodynamics and pCSF flow becomes crucial in understanding the impact of changes in vascular stiffening and cerebrovascular reactivity that occur with aging [25,26,27]. Studies in mice suggest that aging is associated with a decrease in both vessel wall pulsatility and CSF clearance [11, 14]. However, in humans, age-related changes in hemodynamics and CSF clearance have only been independently reported, mainly due to the challenge of simultaneously assessing both neurofluid systems. Consequently, the biomechanical processes underlying the two co-occurring events in aging remain unclear.
In this study, we hypothesized that arterial hemodynamics and para-arterial fluid dynamics exhibit a tight coupling during cardiac pulsation. To investigate this hypothesis, we used a non-invasive multi-modal imaging protocol to assess the coupling between cerebral hemodynamics and pCSF motion along the major cerebral arteries of the human brain. We measured cerebral hemodynamics using resting-state functional MRI (fMRI), interpreting alterations in signal within the major cerebral arteries as reflective of changes in hemodynamics [28,29,30,31]. To quantify pCSF motion, we employed a dynDWI technique specifically designed to measure CSF dynamics in the paravascular space [19]. The coupling between cerebral hemodynamics and pCSF was assessed across age groups (experiment 1), with differing fMRI protocols (experiment 2), and for its reproducibility (experiment 3). In our analysis, we observed consistent coupling between cerebral hemodynamics and pCSF dynamics throughout the cardiac cycle, establishing a clear dynamic relationship between these two neurofluid compartments.
Methods
Human participants
Data from thirty-five participants (25–87 years old, 13 Male/22 Female) were used to complete this study. The older participants (in experiment 1) were cognitively normal and did not have a history of significant cerebrovascular disease or malformations, chemotherapy or radiation therapy, Parkinson's disease, brain surgery, brain infection, or significant head injury with loss of consciousness exceeding 30 min. Cognitive status was assessed using the Uniform Data Set version 3 battery, a standardized cognitive assessment tool used in all National Institute of Aging Alzheimer Disease Centers [32]. The younger participants (and older participants in experiments 2 and 3) were graduate students and research assistants and were not tested for cognitive status or medical history. All participants provided written informed consent according to procedures approved by the Institutional Committee for the Protection of Human Participants at Indiana University.
Image acquisition
Participants were imaged with a 3 T MR scanner (Magnetom Prisma, Siemens Medical Solutions, Erlangen, Germany) using a 64-channel head-neck coil. Three MR sequences within the same scanning session were used to complete this study including a T1-weighted (T1W) anatomical, resting-state fMRI scan, and dynamic diffusion-weighted imaging (dynDWI). fMRI and dynDWI scans were completed sequentially in all experiments. T1W anatomical imaging data were collected using a 3D magnetization rapid gradient echo (MPRAGE) with variable resolutions, including 0.8 × 0.8 × 0.8 mm3, 1 × 1 × 1 mm3, and 1.1 × 1.1 × 1.2 mm3 voxels.
This study utilized two fMRI protocols with longer and shorter TRs to assess the reproducibility between protocols (experiment 2). The first fMRI protocol, fMRI_1, had the following acquisition parameters: repetition time (TR) = 1200 ms, echo time (TE) = 29.8 ms, flip angle (FA) = 65°, voxel size = 2.5 × 2.5 × 2.5 mm3, 200 volumes, multiband factor = 3, and acquisition time = 4 min. The second fMRI protocol, fMRI_2, had the following acquisition parameters: TR = 366 ms, TE = 29.8 ms, FA = 35°, voxel size = 2.5 × 2.5 × 2.5 mm3, 500 volumes, multiband factor = 8, and, and acquisition time = 3 min and 3 s. In this study, experiment 1 utilized fMRI_1, experiment 2 utilized fMRI_1 and fMRI_2, and experiment 3 utilized fMRI_2 (experiments are described in experimental design and statistical analysis).
dynDWI used a single-shot echo-planar imaging sequence with Stejskal-Tanner pulsed gradient spin echo with the following imaging parameters: 134 × 134 acquisition matrix (GRAPPA factor = 2, partial Fourier = 75%), field of view = 240 × 240 × 140 mm3, TR = 1999 ms, TE = 48.6 ms, diffusion time = 22.6 ms, and voxel size = 1.8 × 1.8 × 4 mm3. DWIs were collected at three cardinal diffusion encoding directions (x/y/z) using the b-value of 150 s/mm2, with each diffusion direction repeated 50 times. Ten b = 0 s/mm2 were collected to calculate the apparent diffusion coefficient (ADC). The total acquisition time was 5 min and 40 s. The dynDWI scanning protocol is discussed in-depth by Wen et al. [19].
Data-driven mask creation for arterial-rich and paravascular spaces
Creating an arterial-rich mask in the fMRI space
To identify arterial-rich regions in fMRI datasets, we used a data-driven artery segmentation approach that locates voxels with highly pulsatile patterns in fMRI, which is fully is outlined in Wright et al. [33] (preprint article). We started by applying a voxel-wise high-pass Butterworth filter with a cutoff frequency of 0.005 Hz to remove the signal’s DC offset. Subsequently, we randomly split the fMRI signals into two temporal subsets and retrospectively aligned each to the cardiac cycle resulting in a voxel-wise cardiac-aligned signal pair. These two cardiac-aligned fMRI signals were used to compute a voxel-wise correlation coefficient between each signal pair. This process was repeated 100 times, and then the mean voxel-wise correlation was calculated to create a correlation map. A higher correlation coefficient corresponded to a higher likelihood of arterial regions as these regions expressed increased pulsatility compared to signals from the rest of the brain.
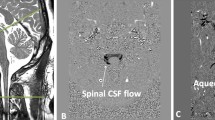
We then created a preliminary artery mask by selecting voxels with a high correlation coefficient using a data-driven cutoff, which was based on three standard deviations from the mean. To refine this preliminary region, we calculated the mean cardiac-aligned waveform within this preliminary region and calculated a final whole-brain voxel-wise correlation between the voxel’s cardiac-aligned waveform and the preliminary region’s mean waveform. The voxels with a high correlation coefficient (> 0.5) were considered cerebral vessels. Lastly, to restrict the mask to arterial-rich regions, the voxel must be located within the three major cerebral arteries (anterior cerebral artery, ACA; middle cerebral artery, MCA; posterior cerebral artery, PCA) defined by modifying a publicly available cerebral artery atlas, referred to as the UBA atlas [34]. The UBA atlases were transformed into the participant’s native anatomical space (T1W) (transformations were derived from non-linear registration of MNI space to T1W space, ANTS: antsRegistration) and then independently transformed into the fMRI space (transformations were derived from linear registration of T1W space to fMRI space, FSL: epi_reg). This process resulted in a data-driven mapping of the arterial-rich regions within the native fMRI space (Fig. 1a). The mean fMRI time-domain signal within the arterial-rich mask is displayed in Fig. 1b. A 3D projection of the arterial-rich mask is displayed in Supplemental Fig. 1.
Overall fMRI and dynDWI signal processing approach. a fMRI data-driven mapping of the arterial-rich vessel regions overlaid on T2-weighted MR imaging. b The mean fMRI time-domain signal defined by the arterial-rich region of interest. c The retrospectively aligned arterial fMRI signal into 50 cardiac phases. The cardiac-aligned signal was smoothed using a Savitzky-Golay filter. d The derivative of the cardiac-aligned fMRI signal was calculated and inverted. The inversion of the signal, results in a maximal \(\frac{d}{d\phi }fMRI\) coinciding with cardiac systole. e dynDWI data-driven mapping of pCSF regions overlaid on T2-weighted MR images. f The mean dynDWI time-domain signal defined by the pCSF region of interest in three directions (x/y/z) g The mean retrospectively aligned pCSF dynDWI signal into 50 cardiac phases (mean in all directions). Note: The definition of cardiac systole is approximated and not used in any data processing. AU arbitrary units, dynDWI dynamic diffusion-weighted-imaging, pCSF paravascular cerebrospinal fluid, ADC apparent diffusion coefficient
Creating a paravascular CSF (pCSF) mask in the dynDWI space
The paravascular space mask was created using a previously developed approach that leverages the pulsatile characteristics of the pCSF waveform [19]. In an iterative approach, the algorithm creates a pCSF mask by defining voxels as paravascular space if they exhibit large differences in systolic vs. diastolic ADC (> 200 × 10–6 mm2/s). The same approach was validated in a publication by Wen et al. [19]. The final pCSF mask was created by incorporating the UBA atlas (brought to dynDWI space using the same methodology as fMRI) to limit the pCSF space mask to the regions near the three major cerebral arteries (ACA, MCA, PCA) to match the locations of interest in the fMRI space (Fig. 1e). The mean dynDWI time-domain signal, in x/y/z directions, within the pCSF mask is displayed in Fig. 1f.
Signal post-processing, interpretation, and analysis
Retrospective cardiac alignment in fMRI and dynDWI
Images were inspected for motion and image artifacts resulting in the exclusion of four participants because of motion detected in fMRI scans. The scanner’s built-in wireless fingertip pulse oximeter was attached to the participant’s left index finger and recorded throughout the acquisition of the fMRI and dynDWI datasets. The first 10 volumes of fMRI data were discarded to allow for steady-state signal equilibration. Both the fMRI and dynDWI temporal image sets were retrospectively aligned to the cardiac cycle by matching the time of slice acquisition with the time of the acquired pulse oximetry [19, 35]. The resulting 4D datasets were aligned with the cardiac cycle in 50 equally spaced phases, where phases 0 and 1 correspond to consecutive peaks in the finger plethysmography signal.
Signal processing and coupling measurement
Following data-driven masking and cardiac alignment, the mean fMRI signal within the arterial-rich mask was computed. The mean signal was smoothed using a Savitzky-Golay filter (Fig. 1c) (Matlab: smooth, polynomial degree = 6, span = 0.15). The derivative of the fMRI signal was taken, and the signal was inverted so that the peak signal corresponds to the timing of cardiac systole (\(\frac{d}{d\phi }fMRI\), Fig. 1d). The \(\frac{d}{d\phi }fMRI\) waveform represents the change in hemodynamics with respect to the cardiac cycle phase (further described in signal interpretation). The mean dynDWI signal within the pCSF mask was computed and used for all future analyses (Fig. 1g).
To assess the coupling between \(\frac{d}{d\phi }fMRI\) and dynDWI, the time delay between the first peaks in \(\frac{d}{d\phi }fMRI\) and dynDWI was measured and recorded as delta time to peak (∆TTP). The ∆TTP was used as a common reference point that occurs in the \(\frac{d}{d\phi }fMRI\) and dynDWI waveforms to normalize the coupling measure between participants. Next, a normalized correlation was calculated between the shifted \(\frac{d}{d\phi }fMRI\) (by ∆TTP) and dynDWI using Eq. 1:
where x = \(\frac{d}{d\phi }fMRI\), y = dynDWI, and circshift (Matlab: circshift) circularly shifts the elements in x by \(\Delta TTP,\) the dynamic range of \(\frac{d}{d\phi }fMRI\) and dynDWI was set to be from −1 to 1 (min = −1 and max = 1) and their autocorrelations at zero lag were set to equal 1. Finally, \({\widehat{C}}_{x, y, normalized}(\Delta TTP)\) was reported as the “correlation at aligned peaks”.
Signal interpretation
dynDWI waveforms were measured in ADC, which scales with the pCSF movement providing a measure of CSF dynamics (Fig. 1g) [19]. Its specificity to pCSF rather than blood flow is achieved through the blood-nulling effect of the diffusion gradient, as extensively discussed in previous research [19]. For fMRI, the fundamental principles resulting in a change of signal in a large vessel must be carefully considered. Typically, fMRI signals are sensitive to changes in deoxygenated hemoglobin levels. However, since arterial spaces exhibit minimal changes in oxygen saturation, the fMRI signal changes are unlikely to be linked to this phenomenon in these experiments. Previous studies have shown that fMRI signals are influenced by cardiac-related physiology with a significant influence on signal modulation near major arterial regions [36,37,38,39]. Hermes et al. [37] and Rajna et al. [40] both studied fMRI signal fluctuations in arterial regions and found that the event of cardiac systole resulted in a decreased fMRI signal. In our analysis, we found a similar signal drop in arterial-rich fMRI signal during cardiac systole (Fig. 1c). Overall, the fMRI signal fluctuation is dependent on various hemodynamic parameters, including cerebral blood volume and cerebral blood flow velocities [28,29,30, 41, 42]. Furthermore, the \(\frac{d}{d\phi }fMRI\) signal captures when hemodynamic changes are most prominent in the brain throughout the cardiac cycle.
Experimental design and statistical analysis
Statistical analysis
All statistical analyses were completed using R (version 4.3.1). Data normality was assessed with the Shapiro–Wilks test. When data normality was present, the mean and standard deviation were used to summarize the data, and parametric statistical tests were used to assess differences (t-test, ANOVA). When data normality was not present (in at least one test cohort), the median and interquartile range were used to summarize the data, and non-parametric tests were used to assess differences (Wilcoxon test). All normality tests and summary statistics are summarized in Supplemental Table 1. Statistical significance was declared if p < 0.05.
Experiment 1
In experiment 1, using the fMRI_1 protocol (TR = 1200 ms), the \(\frac{d}{d\phi }fMRI\) and dynDWI coupling was compared between younger [n = 9; 25–35 years; mean age = 27.9 (3.3) years] and older adults [n = 20; 58–87 years; mean age = 70.5 (6.6) years].
Experiment 2
In experiment 2, a cohort of nine participants [25–62 years; mean age = 31.6 (11.9) years] was used to compare the \(\frac{d}{d\phi }fMRI\) and dynDWI coupling between the different fMRI protocols (fMRI_1 and fMRI_2).
Experiment 3
In experiment 3, using four repeats of the fMRI_2 (TR = 366 ms) and dynDWI scans, the within-participant reproducibility of the \(\frac{d}{d\phi }fMRI\) and dynDWI coupling was assessed [n = 7; 25–74 years; mean age = 39.4 (20.1) years] with an intraclass correlation coefficient test (two-way mixed effects, single rater, absolute-agreement).
Results
Pulsatile hemodynamics and pCSF dynamics were captured with fMRI and dynDWI
The automated segmentation and mean MR waveforms of a representative participant (26-year-old) are summarized in Fig. 2. Axial slices of T2-weighted MR images are overlaid with the data-driven segmentations of the arterial-rich vessel mask and the pCSF mask (Fig. 2a, b, respectively). Qualitatively the arterial-rich mask derived from the fMRI matches closely with the pCSF mask obtained from dynDWI. It was empirically observed that the derivative of the cardiac-aligned fMRI (\(\frac{d}{d\phi }fMRI\)) aligns closely with dynDWI signal, where the mean (solid line) and standard error of the mean (SEM, shaded) of \(\frac{d}{d\phi }fMRI\) and dynDWI are displayed in Fig. 2c (Note: The cardiac-aligned waveforms are concatenated to display two cardiac cycles for visualization purposes only). The difference in cardiac phase between the first peak \(\frac{d}{d\phi }fMRI\) and dynDWI was defined as \(\Delta\)TTP (Fig. 2c). The \(\frac{d}{d\phi }fMRI\) was circularly shifted by \(\Delta\)TTP, and the normalized correlation coefficient was reported as the correlation at aligned peaks (Fig. 2d). The \(\Delta\)TTP and the correlation at aligned peaks measured in this representative participant were −0.06 and 0.913, demonstrating that \(\frac{d}{d\phi }fMRI\) led the dynDWI signal and had a high coupling when aligning their first peaks (Fig. 2d).
A representative participant’s data-driven segmentations, cardiac-aligned waveforms, and the normalized correlation at aligned peaks between \(\frac{d}{d\phi }fMRI\) and dynDWI. a fMRI data-driven mapping of the arterial-rich vessel regions overlaid on T2-weighted MR imaging. b dynDWI data-driven mapping of pCSF regions overlaid on T2-weighted MR images. c The \(\frac{d}{d\phi }fMRI\) and dynDWI waveforms concatenated for two cardiac cycles (mean—solid line, SEM—shaded). d The \(\Delta\)TTP and normalized correlation at aligned peaks between the circularly shifted \(\frac{d}{d\phi }fMRI\) and dynDWI. pCSF paravascular cerebrospinal fluid, dynDWI dynamic diffusion-weighted-imaging, SEM standard error of the mean, ADC apparent diffusion coefficient, AU arbitrary units, TTP time to peak
Hemodynamic-pCSF coupling is substantial within younger and older participants (Experiment 1)
The mean (solid line) and standard deviation (shaded) of \(\frac{d}{d\phi }fMRI\) and dynDWI of younger [27.9 (3.3) years, n = 9] and older [70.5 (6.6) years, n = 20] adult cohorts are summarized in Fig. 3a, b. The \(\frac{d}{d\phi }fMRI\) waveform in the younger and older participants appears similar. However, the prominent peak may occur slightly earlier with a longer \(\Delta\)TTP in the older participants than in the younger participants. In the older participants’ dynDWI waveform, a second peak is present that is not as pronounced in the younger participants.
Close coupling between \(\frac{d}{d\phi }(fMRI)\) and dynDWI is present in both younger and older adult participants. The mean (solid line) and standard deviation (shaded) \(\frac{d}{d\phi }fMRI\) and dynDWI in a younger adults and b older adults. c The \(\Delta\)TTP between the first peaks of \(\frac{d}{d\phi }fMRI\) and dynDWI waveforms was shorter in younger compared to older participants. d The normalized correlation at aligned peaks between circularly shifted \(\frac{d}{d\phi }fMRI\) and dynDWI did not differ between younger and older participants. e. A proposed relationship between cerebral hemodynamics and pCSF motion: The arrival of the cardiac pressure wave leading to hemodynamic changes (Event 1) precedes an increase in pCSF motion (Event 2). Note: The cardiac-aligned waveforms are concatenated to display two cardiac cycles for visualization purposes only, the signals from 0 to 1 and 1 to 2 cardiac phases are identical. dynDWI dynamic diffusion-weighted-imaging, ADC apparent diffusion coefficient, AU arbitrary units, TTP time to peak, NS not significant (p > 0.05), *p < 0.05, pCSF paravascular cerebrospinal fluid
In both age groups, the dynamic changes occurring in the \(\frac{d}{d\phi }fMRI\) precede the dynamic changes in the dynDWI. The median (interquartile range) \(\Delta\)TTP was significantly shorter in younger, −0.040 (−0.059 to 0.015), compared to older adults, −0.069 (−0.079 to −0.059), adults (Fig. 1c, W = 142.5, p < 0.05, two-sided, unpaired Wilcoxon sign-ranked test). The median (interquartile range) correlation at aligned peaks in younger, 0.833 (0.776–0.867), and older, 0.776 (0.601–0.844), adults were not significantly different (Fig. 3d, W = 121, p = 0.153, two-sided, unpaired Wilcoxon sign-ranked test). The participant’s heart rate between fMRI and dynDWI scans were not significantly different (Supplemental Fig. 2, V = 137, p = 0.718, two-sided, paired Wilcoxon sign-ranked test).
The group averaged waveforms and high coupling between \(\frac{d}{d\phi }fMRI\) and dynDWI were used to infer a potential model linking cardiac-related hemodynamics and pCSF motion (Fig. 3e). The proposed model is described by two events: The arrival of the cardiac pressure wave leading to hemodynamic changes (Event 1, representative of \(\frac{d}{d\phi }fMRI\)), which precedes an observed increase in pCSF motion (Event 2, representative of dynDWI). This model may partially explain the close coupling observed between cardiac aligned \(\frac{d}{d\phi }fMRI\) and dynDWI.
Hemodynamic-pCSF coupling is consistent with varying fMRI acquisition parameters (Experiment 2)
Nine participants underwent two distinct fMRI scans with varied parameters, referred to as fMRI_1 and fMRI_2. The mean (solid line) and standard deviation (shaded) of \(\frac{d}{d\phi }fMRI\) between fMRI_1 (Fig. 4a) and fMRI_2 (Fig. 4b) appeared to agree closely. The \(\Delta\)TTP and normalized correlation at aligned peaks did not significantly differ between acquisitions (\(\Delta\)TTP: t = 1.272, p = 0. 239; correlation at aligned peaks: t = 0.020, p = 0.985; two-sided, paired student’s t-tests), suggesting the coupling was consistent between fMRI protocols. The mean and standard deviation of the ΔTTP were −0.015 (0.062) and −0.053 (0.028), for fMRI_1 and fMRI_2, respectively (Fig. 4c). The mean and standard deviation of the correlation at aligned peaks were 0.813 (0.085) and 0.812 (0.107) for fMRI_1 and fMRI_2, respectively (Fig. 4d). The participant’s heart rates were not significantly different between fMRI_1, fMRI_2, and dynDWI scans (Supplemental Fig. 3, F = 0.019, p = 0.982, repeated measures ANOVA).
The coupling between \(\frac{d}{d\phi }fMRI\) and dynDWI remains consistent with varying fMRI acquisition parameters in nine participants. The mean and standard deviation (shaded) of \(\frac{d}{d\phi }fMRI\) and dynDWI, for the a fMRI_1 and b fMRI_2 protocols. c The \(\Delta\)TTP did not differ between the first peaks of \(\frac{d}{d\phi }fMRI\) and dynDWI waveforms in fMRI_1 and fMRI_2 protocols. d The normalized correlation at aligned peaks between circularly shifted \(\frac{d}{d\phi }fMRI\) and dynDWI did not differ between fMRI_1 and fMRI_2 protocols. dynDWI dynamic diffusion-weighted-imaging, ADC apparent diffusion coefficient, AU arbitrary units, TTP time to peak, NS not significant (p > 0.05)
Hemodynamic-pCSF coupling is reproducible (Experiment 3)
Figure 5a–g illustrates the mean (solid line) and SEM (shaded) of five younger (26–35 years) and two older (62 & 74 years) participants for four repeated scan pairs collected for each participant. Across the participants with repeated scans, the intraclass correlation of \(\Delta\)TTP was 0.362 (95% CI: 0.014 to 0.798), indicating poor to good agreement. The intraclass correlation coefficient of the correlation at aligned peaks was 0.976 (95% CI: 0.928 to 0.995, p < 0.001), indicating excellent agreement. The group mean and standard deviation of \(\Delta\)TTP and the correlation at aligned peaks were −0.065 (0.016) and 0.750 (0.032) (Fig. 5h). The participant’s heart rate between fMRI and dynDWI scans were not significantly different (Supplemental Fig. 4, V = 71.5, p = 0.830, two-sided, paired Wilcoxon sign-ranked test).
Cardiac-aligned \(\frac{d}{d\phi }fMRI\) (fMRI_2) and dynDWI waveforms are reproducible within participants and demonstrate consistent ΔTTP and correlation at aligned peaks measures. a–g The mean and SEM (shaded) waveforms for each participant with four repeated scans. h Summary table of the mean (std) for each participant’s \(\Delta\)TTP, correlation at aligned peaks, fMRI scan HR, and dynDWI scan HR between repeated scans, as well as the participant’s age. Note: The cardiac-aligned waveforms are concatenated to display two cardiac cycles for visualization purposes. dynDWI dynamic diffusion-weighted-imaging, TTP time to peak, HR heart rate, ADC apparent diffusion coefficient, AU arbitrary units, BPM beats per minute
Discussion
In this study, we combined fMRI and dynDWI to investigate the coupling between cerebral hemodynamics and pCSF dynamics in humans. Our data unveiled a robust and reproducible coupling between these two essential neurofluid compartments along the major cerebral arteries. We observed high levels of coupling between cerebral hemodynamics and pCSF dynamics in both younger and older participants. Based on our imaging findings, we proposed a vascular-pCSF coupling model describing the arrival of the cardiac pressure wave leading to hemodynamic changes (captured with \(\frac{d}{d\phi }fMRI\)), which precedes an increase in pCSF motion (captured with dynDWI).
While previous human studies have primarily focused on identifying the coupling between cerebral hemodynamics and CSF flow specifically at the fourth ventricle [3, 4, 16, 17, 43], our study demonstrates the ability to assess this coupling at the paravascular space surrounding major cerebral arteries. The results reveal that both cerebral hemodynamics and pCSF dynamics exhibit prominent cardiac cycle-driven pulsatility that are strongly coupled. This coupling has been studied in an animal model by Mestre et al. [5] using two-photon imaging, where they observed a close coupling between vessel wall velocities and pCSF motion. Arterial pulsation-driven pCSF flow has been suggested as early as 1985 [44] and has gained attention due to the proposed glymphatic system hypothesis [12, 45, 46]. However, the majority of evidence has been collected through invasive animal studies [5,6,7, 11, 12, 47]. Our study provides non-invasive evidence in humans that illustrates the fluctuations in cerebral hemodynamics preceding the observed pCSF dynamics.
The current study provides evidence that the cardiac pulsations influencing cerebral hemodynamics similarly impact pCSF motion and result in a close coupling between these two neurofluids, which is consistently strong across age groups. This observation indicates that the interaction between hemodynamic and CSF flow dynamics is essential throughout life, including into old age. However, it is important to note that this does not imply that the CSF dynamics remain unchanged with age. In this study, ∆TTP was increased in older participants, suggesting that the response time of pCSF motion to changes in cerebral hemodynamics is extended in older individuals. The slower response of pCSF motion could result from two distinct age-related changes: (1) an increase in vascular stiffness [25, 26] and/or (2) an increase in paravacular space. An increase in vascular stiffness may lead to a less effective force driving the pCSF flow from the arterial wall motion and, therefore, a potentially slower response and reduction in pCSF motion. This reduction in pCSF motion has been observed in acute hypertension mice [5]. The increase in the paravascular space with aging results in an increase in CSF surrounding the vessel, which may reduce the efficiency of the transfer of energy from the arterial wall motion to the pCSF motion.
Additionally, the pCSF motion in older participants displayed a prominent widened peak waveform which was absent in the younger participants. This finding is consistent with the previous study by Wen et al., which demonstrated that the pCSF motion in humans is altered with aging [19]. The widened peak likely indicates a more chaotic pCSF motion, corresponding to a less effective CSF pumping. These pCSF alterations may be impacted by various age-related brain changes such as altered vessel wall mechanics, enlarged paravascular space, or a reduction in the downstream exchange between pCSF and interstitial fluid, as observed in mice [11]. This finding suggests that while the coupling persists with aging, the pCSF’s response to hemodynamics is altered, indicating additional factors may influence pCSF alterations in the aging brain. However, understanding the determinants impacting pCSF motion remains to be explored in future research and is out of the scope of this work.
Although there was no significant difference in correlation at aligned peaks between younger and older participants, the lowest correlations in the study were observed in the older participants. This suggests that a reduced correlation at aligned peaks may be observed in pathological conditions. Ischemic stroke can lead to reduced cerebral blood delivery [48], disrupted blood–brain barrier function [49], vascular wall breakdown [50], and increased paravascular spaces [51], all of which may impact cerebral hemodynamics and pCSF flow. Our proposed neuroimaging methodology may be sensitive to these changes. Additionally, our technique provides the opportunity to investigate vascular involvement in Alzheimer’s disease (AD) and other neurodegenerative diseases, including the potential vascular deficiency-related hypoperfusion that may trigger events contributing to AD [52]. Notably, previous MR studies observed a reversed cardiac pulse propagation in participants with AD [53], which our coupling method can further explore to assess its impact on pCSF flow.
The present study demonstrates that the coupling analysis is consistent between different fMRI protocols and reproducible within subjects. This study focused on the reproducibility of fMRI protocols because the reproducibility of dynDWI acquisition (i.e. impact of varying b-value 50–300 s/mm2 and imaging resolutions) has been previously demonstrated [19]. The differing fMRI acquisition parameters in the cardiac-aligned fMRI signal appeared to have a negligible impact on the measured coupling with dynDWI because similar results were observed in participants who underwent both fMRI protocols. Hermes et al. similarly reported robustness in cardiac-aligned fMRI waveforms when comparing protocols [37]. In future cardiac-related coupling studies, either fMRI acquisition is suitable for retrospective cardiac binning. However, faster imaging will adequately sample cardiac-related signal changes at resting heart rates (60–80 beats per minute), allowing spectral analyses to study cardiac, respiratory, and low-frequency signal components simultaneously.
This study has certain limitations that warrant future investigation. First, the total number of participants is limited, and additional studies with larger samples are needed to draw further conclusions. Another limitation pertains to the spatial resolution of the MRI studies conducted in this research. At the current spatial resolution fMRI evaluation of major arteries may suffer from signal contamination from the paravascular CSF. To address this, we performed an analysis demonstrating that our data-driven arterial mask effectively isolates arterial-rich regions, ensuring fMRI signals predominantly from arterial blood with minimal CSF contamination (Supplemental Fig. 5). Imaging at a higher resolution would further mitigate this issue and allow for the investigation of smaller pial arteries. Additionally, it is important to consider physiological components beyond the influence of only cardiac pulsation. Factors such as respiration and low-frequency oscillations have been shown to drive CSF flow in other regions of the brain [3, 4, 16, 54], and their impact on vascular-pCSF coupling within these specific regions should be explored in future investigations.
In conclusion, our proposed method provides a non-invasive and clinically feasible (combined acquisition time < 10 min and fully automated post-processing) approach to studying the crucial coupling between the cerebral vascular system and paravascular CSF. Our results reveal a consistent coupling between the cardiac pulsation-driven cerebral hemodynamics and pCSF dynamics within the subarachnoid space. Notably, this coupling remained consistently high with increasing age. Altogether, this work supports the feasibility of non-invasive evaluations of neurofluid dynamics coupling and may be expanded to assess various disease conditions.
Availability of data and materials
All relevant codes are available from the corresponding author upon request. Due to the ethics and privacy issues of clinical data, the original imaging data will not be made openly available to the public. The imaging data are not openly available as we do not have participant permission to share this data. A deidentified demo dataset will be provided upon reasonable request.
Abbreviations
- pCSF:
-
Para-arterial/paravascular cerebrospinal fluid
- fMRI:
-
Functional magnetic resonance imaging
- dynDWI:
-
Dynamic diffusion weighted imaging
- CSF:
-
Cerebrospinal fluid
- CBV:
-
Cerebral blood volume
- T1W:
-
T1-weighted
- MPRAGE:
-
Magnetization rapid gradient echo
- TR:
-
Repetition time
- TE:
-
Echo time
- FA:
-
Flip angle
- \(\Delta\) :
-
Diffusion time
- ADC:
-
Apparent diffusion coefficient
- MCCC:
-
Maximum circular correlation coefficient
- SEM:
-
Standard error of the mean
- AU:
-
Arbitrary units
References
Nedergaard M. Garbage truck of the brain. Science (1979). 2013. https://doi.org/10.1126/science.1240514.
Spector R, Robert Snodgrass S, Johanson CE. A balanced view of the cerebrospinal fluid composition and functions: focus on adult humans. Exp Neurol. 2015;273:57–68.
Yang H-C, Inglis B, Talavage TM, Nair VV, Yao J, Fitzgerald B, Schwichtenberg AJ, Tong Y. Coupling between cerebrovascular oscillations and CSF flow fluctuations during wakefulness: an fMRI study. J Cereb Blood Flow Metab. 2022;42:1091–103.
Vijayakrishnan Nair V, Kish BR, Inglis B, Yang H-C, Wright AM, Wu Y-C, Zhou X, Schwichtenberg AJ, Tong Y. Human CSF movement influenced by vascular low frequency oscillations and respiration. Front Physiol. 2022. https://doi.org/10.3389/fphys.2022.940140.
Mestre H, Tithof J, Du T, Song W, Peng W, Sweeney AM, Olveda G, Thomas JH, Nedergaard M, Kelley DH. Flow of cerebrospinal fluid is driven by arterial pulsations and is reduced in hypertension. Nat Commun. 2018. https://doi.org/10.1038/s41467-018-07318-3.
Iliff JJ, Wang M, Zeppenfeld DM, Venkataraman A, Plog BA, Liao Y, Deane R, Nedergaard M. Cerebral arterial pulsation drives paravascular CSF-interstitial fluid exchange in the murine brain. J Neurosci. 2013. https://doi.org/10.1523/JNEUROSCI.1592-13.2013.
Iliff JJ, Lee H, Yu M, Feng T, Logan J, Nedergaard M, Benveniste H. Brain-wide pathway for waste clearance captured by contrast-enhanced MRI. J Clin Invest. 2013;123:1299–309.
Bohr T, Hjorth PG, Holst SC, et al. The glymphatic system: Current understanding and modeling. iScience. 2022;25:104987.
Holstein-Rønsbo S, Gan Y, Giannetto MJ, et al. Glymphatic influx and clearance are accelerated by neurovascular coupling. Nat Neurosci. 2023;26(6):1042–53.
Boster KAS, Cai S, Ladrón-de-Guevara A, Sun J, Zheng X, Du T, Thomas JH, Nedergaard M, Karniadakis GE, Kelley DH. Artificial intelligence velocimetry reveals in vivo flow rates, pressure gradients, and shear stresses in murine perivascular flows. Proc Natl Acad Sci. 2023. https://doi.org/10.1073/pnas.2217744120.
Kress BT, Iliff JJ, Xia M, et al. Impairment of paravascular clearance pathways in the aging brain. Ann Neurol. 2014. https://doi.org/10.1002/ana.24271.
Iliff JJ, Wang M, Liao Y, et al. A paravascular pathway facilitates CSF flow through the brain parenchyma and the clearance of interstitial solutes, including amyloid β. Sci Transl Med. 2012. https://doi.org/10.1126/scitranslmed.3003748.
Kim SH, Ahn JH, Yang H, Lee P, Koh GY, Jeong Y. Cerebral amyloid angiopathy aggravates perivascular clearance impairment in an Alzheimer’s disease mouse model. Acta Neuropathol Commun. 2020. https://doi.org/10.1186/s40478-020-01042-0.
Benveniste H, Liu X, Koundal S, Sanggaard S, Lee H, Wardlaw J. The glymphatic system and waste clearance with brain aging: a review. Gerontology. 2019;65:106–19.
Rasmussen MK, Mestre H, Nedergaard M. The glymphatic pathway in neurological disorders. Lancet Neurol. 2018;17:1016–24.
Fultz NE, Bonmassar G, Setsompop K, Stickgold RA, Rosen BR, Polimeni JR. Lewis LD (2019) Coupled electrophysiological, hemodynamic, and cerebrospinal fluid oscillations in human sleep. Science. 1979. https://doi.org/10.1126/science.aax5440.
Nair VV, Diorio TC, Wen Q, Rayz VL, Tong Y. Using respiratory challenges to modulate CSF movement across different physiological pathways: an fMRI study. Imaging Neurosci. 2024;2:1–14.
Eide PK, Ringstad G. Functional analysis of the human perivascular subarachnoid space. Nat Commun. 2024;15:2001.
Wen Q, Tong Y, Zhou X, Dzemidzic M, Ho CY, Wu Y-C. Assessing pulsatile waveforms of paravascular cerebrospinal fluid dynamics within the glymphatic pathways using dynamic diffusion-weighted imaging (dDWI). Neuroimage. 2022;260:119464.
Wen Q, Wright A, Tong Y, Zhao Y, Risacher SL, Saykin AJ, Wu Y, Limaye K, Riley K. Paravascular fluid dynamics reveal arterial stiffness assessed using dynamic diffusion-weighted imaging. NMR Biomed. 2023. https://doi.org/10.1002/nbm.5048.
Han G, Jiao B, Zhang Y, Wang Z, Liang C, Li Y, Hsu Y-C, Bai R. Arterial pulsation dependence of perivascular cerebrospinal fluid flow measured by dynamic diffusion tensor imaging in the human brain. Neuroimage. 2024;297:120653.
Adam M., Wright Yu-Chien, Wu Li, Feng Qiuting, Wen. Diffusion magnetic resonance imaging of cerebrospinal fluid dynamics: Current techniques and future advancements. Abstract NMR in Biomedicine. 2024;37(9). https://doi.org/10.1002/nbm.v37.9, https://doi.org/10.1002/nbm.5162
Ringstad G, Valnes LM, Dale AM, Pripp AH, Vatnehol SAS, Emblem KE, Mardal KA, Eide PK. Brain-wide glymphatic enhancement and clearance in humans assessed with MRI. JCI Insight. 2018. https://doi.org/10.1172/JCI.INSIGHT.121537.
Patel M, Atyani A, Salameh JP, McInnes M, Chakraborty S. Safety of intrathecal administration of gadolinium-based contrast agents: a systematic review and meta-analysis. Radiology. 2020;297:75–83.
Reneman RS, Van Merode T, Hick P, Muytjens AMM, Hoeks APG. Age-related changes in carotid artery wall properties in men. Ultrasound Med Biol. 1986;12:465–71.
Zimmerman B, Rypma B, Gratton G, Fabiani M. Age-related changes in cerebrovascular health and their effects on neural function and cognition: a comprehensive review. Psychophysiology. 2021. https://doi.org/10.1111/psyp.13796.
Zhou Y, Cai J, Zhang W, Gong X, Yan S, Zhang K, Luo Z, Sun J, Jiang Q, Lou M. Impairment of the glymphatic pathway and putative meningeal lymphatic vessels in the aging human. Ann Neurol. 2020;87:357–69.
Von Schulthess GK, Higgins CB. Blood flow imaging with MR: Spin-phase phenomena. Radiology. 1985. https://doi.org/10.1148/radiology.157.3.2997836.
Buxton RB, Wong EC, Frank LR. Dynamics of blood flow and oxygenation changes during brain activation: The balloon model. Magn Reson Med. 1998;39:855–64.
Ances BM, Leontiev O, Perthen JE, Liang C, Lansing AE, Buxton RB. Regional differences in the coupling of cerebral blood flow and oxygen metabolism changes in response to activation: implications for BOLD-fMRI. Neuroimage. 2008;39:1510–21.
Magnuson M, Majeed W, Keilholz SD. Functional connectivity in blood oxygenation level-dependent and cerebral blood volume-weighted resting state functional magnetic resonance imaging in the rat brain. J Magn Reson Imaging. 2010;32:584–92.
Weintraub S, Besser L, Dodge HH, et al. Version 3 of the Alzheimer disease centers’ neuropsychological test battery in the Uniform Data Set (UDS). Alzheimer Dis Assoc Disord. 2018;32:10–7.
Wright AM, Xu T, Ingram J, Koo J, Zhao Y, Tong Y, Wen Q. Robust data-driven segmentation of pulsatile cerebral vessels using functional magnetic resonance imaging. bioRxiv 2024.07.17.603932; 2024.
Dunås T, Wåhlin A, Ambarki K, Zarrinkoob L, Malm J, Eklund A. A Stereotactic probabilistic Atlas for the major cerebral arteries. Neuroinformatics. 2017;15:101–10.
Tong Y, Hocke LM, De Frederick BB. Short repetition time multiband echo-planar imaging with simultaneous pulse recording allows dynamic imaging of the cardiac pulsation signal. Magn Reson Med. 2014;72:1268–76.
Dagli MS, Ingeholm JE, Haxby JV. Localization of cardiac-induced signal change in fMRI. Neuroimage. 1999. https://doi.org/10.1006/nimg.1998.0424.
Hermes D, Wu H, Kerr AB, Wandell BA. Measuring brain beats: Cardiac-aligned fast functional magnetic resonance imaging signals. Hum Brain Mapp. 2023;44:280–94.
Chang C, Cunningham JP, Glover GH. Influence of heart rate on the BOLD signal: The cardiac response function. Neuroimage. 2009;44:857–69.
Raitamaa L, Huotari N, Korhonen V, Helakari H, Koivula A, Kananen J, Kiviniemi V. Spectral analysis of physiological brain pulsations affecting the BOLD signal. Hum Brain Mapp. 2021;42:4298–313.
Rajna Z, Mattila H, Huotari N, et al. Cardiovascular brain impulses in Alzheimer’s disease. Brain. 2021. https://doi.org/10.1093/brain/awab144.
Weisskoff RM, Kiihne S. MRI susceptometry: image-based measurement of absolute susceptibility of MR contrast agents and human blood. Magn Reson Med. 1992;24:375–83.
Ogawa S, Lee TM, Kay AR, Tank DW. Brain magnetic resonance imaging with contrast dependent on blood oxygenation. Proc Natl Acad Sci U S A. 1990;87:9868.
Williams SD, Setzer B, Fultz NE, Valdiviezo Z, Tacugue N, Diamandis Z, Lewis LD. Neural activity induced by sensory stimulation can drive large-scale cerebrospinal fluid flow during wakefulness in humans. PLoS Biol. 2023;21:e3002035.
Rennels ML, Gregory TF, Blaumanis OR, Fujimoto K, Grady PA. Evidence for a ‘Paravascular’ fluid circulation in the mammalian central nervous system, provided by the rapid distribution of tracer protein throughout the brain from the subarachnoid space. Brain Res. 1985;326:47–63.
Rennels ML, Blaumanis OR, Grady PA. Rapid solute transport throughout the brain via paravascular fluid pathways. Adv Neurol. 1990;52:431–9.
Schley D, Carare-Nnadi R, Please CP, Perry VH, Weller RO. Mechanisms to explain the reverse perivascular transport of solutes out of the brain. J Theor Biol. 2006;238:962–74.
Peng W, Achariyar TM, Li B, et al. Suppression of glymphatic fluid transport in a mouse model of Alzheimer’s disease. Neurobiol Dis. 2016;93:215–25.
Lin TN, Sun SW, Cheung WM, Li F, Chang C. Dynamic changes in cerebral blood flow and angiogenesis after transient focal cerebral ischemia in rats: Evaluation with serial magnetic resonance imaging. Stroke. 2002;33:2985–91.
Lecrux C, Campinho P, Zhang J, Nian K, Harding IC, Herman IM, Ebong EE. Blood-brain barrier damage in ischemic stroke and its regulation by endothelial mechanotransduction. Front Physiol. 2020. https://doi.org/10.3389/fphys.2020.605398.
Del Zoppo GJ, Mabuchi T. Cerebral microvessel responses to focal ischemia. J Cereb Blood Flow Metabol. 2003. https://doi.org/10.1097/01.WCB.0000078322.96027.78.
Mestre H, Du T, Sweeney AM, et al. Cerebrospinal fluid influx drives acute ischemic tissue swelling. Science. 2020;367:1198–9.
Kelleher RJ, Soiza RL. Evidence of endothelial dysfunction in the development of Alzheimer’s disease: Is Alzheimer’s a vascular disorder? Am J Cardiovasc Dis. 2013;3:197.
Kiviniemi V, Wang X, Korhonen V, et al. Ultra-fast magnetic resonance encephalography of physiological brain activity-Glymphatic pulsation mechanisms? J Cereb Blood Flow Metab. 2016;36:1033–45.
Chen L, Beckett A, Verma A, Feinberg DA. Dynamics of respiratory and cardiac CSF motion revealed with real-time simultaneous multi-slice EPI velocity phase contrast imaging. Neuroimage. 2015;122:281–7.
Acknowledgements
The illustration in Fig. 3e was created with BioRender.com. The authors would like to acknowledge Jacob Ingram for his support in manually confirming all automated data measures.
Funding
The work was supported by F30AG084336, RF1AG083762, R21AG068962, R01AG053993, and P30AG072976.
Author information
Authors and Affiliations
Contributions
AMW, QW, and YT conceived the study and wrote the manuscript. AMW, QW, YT, H-CY, SLR, Y-CW, and AJS contributed to data collection. AMW performed data analysis. AMW, QW, and YT interpreted the results and developed the theory. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
All participants provided written informed consent according to procedures approved by the Institutional Committee for the Protection of Human Participants at Indiana University.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wright, A.M., Wu, YC., Yang, HC. et al. Coupled pulsatile vascular and paravascular fluid dynamics in the human brain. Fluids Barriers CNS 21, 71 (2024). https://doi.org/10.1186/s12987-024-00572-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12987-024-00572-2