Abstract
Gephyrin is thought to play a critical role in clustering glycine receptors at synapses within the central nervous system (CNS). The main in vivo evidence for this comes from Gephyrin (Gphn)-null mice, where glycine receptors are depleted from synaptic regions. However, these mice die at birth, possibly due to impaired molybdenum cofactor (MoCo) synthesis, an essential role Gephyrin assumes throughout an animal. This complicates the interpretation of synaptic phenotypes in Gphn-null mice and raises the question whether the synaptic and enzymatic functions of Gephyrin can be investigated separately. Here, we generated a gephyrinb zebrafish mutant, vo84, that almost entirely lacks Gephyrin staining in the spinal cord. gephyrinbvo84 mutants exhibit normal gross morphology at both larval and adult stages. In contrast to Gphn-null mice, gephyrinbvo84 mutants exhibit normal motor activity and MoCo-dependent enzyme activity. Instead, gephyrinbvo84 mutants display impaired rheotaxis and increased mortality in late development. To investigate what may mediate these defects in gephyrinbvo84 mutants, we examined the cell density of neurons and myelin in the spinal cord and found no obvious changes. Surprisingly, in gephyrinbvo84 mutants, glycine receptors are still present in the synaptic regions. However, their abundance is reduced, potentially contributing to the observed defects. These findings challenge the notion that Gephyrin is absolutely required to cluster glycine receptors at synapses and reveals a new role of Gephyrin in regulating glycine receptor abundance and rheotaxis. They also establish a powerful new model for studying the mechanisms underlying synaptic, rather than enzymatic, functions of Gephyrin.
Similar content being viewed by others
Background
Gephyrin is a multifunctional 93-kDa protein with distinct roles in various tissues [1]. In the central nervous system (CNS), Gephyrin plays a crucial role in inhibitory synaptic transmission [2]. Meanwhile, Gephyrin is also essential for molybdenum cofactor (MoCo) synthesis [3], acting downstream of MoCo synthesis 1 (MoCS1) and MoCS2. MoCo-dependent enzymes, such as sulfite oxidase and xanthine oxidase, are critical for proper cellular metabolism. MoCo deficiency in humans leads to death in early infancy and severe neurological defects including seizures, feeding difficulties, cerebral atrophy, and white matter lesions [3, 4]. Mocs1 and Mocs2 mice die within the first postnatal week and exhibit smaller body size and neuronal apoptosis [5,6,7]. Similarly, Gephyrin (Gphn)-null mice die perinatally and exhibit a range of neurological defects. Gphn-null mice lack glycine receptors (GlyRs) at synapses [8, 9], supporting a synaptic role of Gephyrin to cluster GlyRs. However, given the severe CNS defects caused by MoCo deficiency, it has been challenging to fully separate the synaptic role of Gephyrin from its MoCo enzymatic role.
An intrabody-based approach targeting Gephyrin for degradation in zebrafish led to a 64% reduction of Gephyrin levels in a sparse population of motoneurons [10]. However, GlyRs in these fish were not examined, and it is unclear whether a 64% reduction is sufficient to abolish Gephyrin’s function. In vitro assays have been more successful in dissecting the synaptic function of Gephyrin, as disruption of Gephyrin in cultured neurons leads to abolished GlyR clustering [9, 11, 12]. Nonetheless, the in vivo relevance of these defects remains unclear. Hence, there is a need for an in vivo approach that can effectively abolish Gephyrin in the CNS without severely affecting other tissues, thereby facilitating a deeper understanding of its role in synapse organization within the CNS.
In zebrafish, gephyrin is duplicated such that gephyrina (gphna) and gephyrinb (gphnb) orthologs are encoded in the genome. mRNA expression analysis via in situ hybridization indicates that gphnb is specific to the CNS, while gphna is more widely expressed [13, 14]. Previous work in zebrafish using morpholinos found that disrupting both gphna and gphnb is necessary to abolish Gephyrin levels in the spinal cord as assessed by antibody staining. This disruption abolished GlyR clustering and led to abnormal coiling behaviors [13]. However, the known off-target effects of morpholinos [15] and the incomplete gene disruption through transient morpholino injection raise questions about whether stable mutants would recapitulate these phenotypes.
Here, we generated a new gphnb mutant in zebrafish (gphnbvo84) with nearly absent Gephyrin staining in the spinal cord. Despite increased mortality in late development, many gphnbvo84 mutants developed normally and survived to adulthood. They also exhibited normal activity of xanthine oxidase. Therefore, these mutants establish an excellent in vivo model where the synaptic function of Gephyrin in the CNS can be studied independently of its enzymatic function. These otherwise normal-appearing mutants displayed impaired rheotaxis (the ability to swim against currents). To our surprise, GlyRs, although at reduced levels, still localized to synaptic regions. These results suggest new roles of Gephyrin and refine our understanding of its synaptic clustering function.
Methods
Experimental model and subject details
Animal care and use
We used the following zebrafish lines: AB, nacre, and Tg(mbp:GFP-CAAX) [16]. We generated gphnbvo84 mutants and transgenic Tg(olig1:mScarlet) for these studies. The genotypes of fish and the number of sections used for each experiment are indicated in the figure legends. Animals in the fish facility were maintained at 28° C with a 14 h/10 h light/dark cycle. Larvae and juveniles were nurtured with rotifer suspension and dry food (Gemma 75 and 150, respectively). Adults were fed with a combination of brine shrimp and dry food (Gemma 300). Male and female fish at 3 to 12 months-old were mated; larval fish at 3 dpf to 5 dpf were used for experiments.
Generation of gphnb vo84/vo84 mutants
The sgRNA sequence for gphnb exon 7 (GGAGAGCCGGCAGGATGAACTGG) was previously selected and exhibited high cutting efficiency [17]. We injected the sgRNA together with 1 ng Cas9 protein. We sequenced the F1 offsprings from fin clips and found 11 different gphnb alleles. We chose the vo84 allele (cagtt deletion within the sgRNA targeting loci) because it causes a frameshift and premature stop codon in an early exon. For genotyping, we used amplicon amplification near the target locus and enzyme digestion. Briefly, the following primers were used to amplify the region near the target locus: Forward TGTCCAGCCCCCTCTGAAACATCC; Reverse TGGAGGGGACGGCAGGTCTTC. The amplicon (243 bp) from wildtype is recognized by BsrI and digested into 106 bp and 137 bp, while the amplicon from gphnbvo84 is not recognized by BsrI (Fig. 1B). These bands can be readily distinguished on 3% gels (Fig. 1B). gphnbvo84/vo84 mutant animals are from gphnbvo84/+ in-crosses; WT in this case refers to the siblings. MZ gphnbvo84/vo84 mutant animals are from gphnbvo84/vo84 in-crosses; WT in this case refers to AB strain.
Disruption of gphna
The sgRNA sequences for gphna exon 2 (GGTCATGAACGAGATCTTTGAGG) and exon 4 (GTGGGACGGGTTTTGCGCCTCGG) were previously selected and exhibited high cutting efficiency [17]. We injected WT or gphnbvo84/vo84 embryos with gphna sgRNA and Cas9 protein at the single-cell stage to disrupt gphna.
Generation of Tg(olig1:mScarlet) animals
p5E_olig1 was created using the olig1:Kalta4 plasmid [18], pME_mScarlet was generated using the pME_myrmScarlet plasmid [17], and p3E_polyA was from the Tol2 kit [19]. We injected one-cell stage embryos with 1 nl of an injection solution containing 20 ng µl−1 plasmid DNA, 25 ng µl−1Tol2 transposase mRNA, 0.02% phenol red and 0.2 M KCl. F1 progenies were screened for germline transmission by fluorescence.
Tissue sectioning and immunofluorescence
Zebrafish larvae were anesthetized with 0.16 mg/mL tricaine at 5 dpf and then fixed in 4% PFA-0.1% Triton at 4°C overnight with gentle rocking. Heads and posterior tail tips were first removed for better penetration of fixative. The tissue was then washed in PBS-0.1% Triton, passed through 10%, 20%, and 30% sucrose in PBS, and embedded in OCT and frozen at 20 °C. The OCT blocks were cryosectioned into 20 µm transverse slices on Leica Surgipath X-tra slides (3800200). Slides for immunostaining were brought to room temperature for 45 min and rehydrated in PBS-0.1% Triton for one hour. 3% BSA, 5% NGS, 0.2% Triton, and 0.02% sodium azide in 1X PBS was used as blocking buffer. The following primary antibodies were used: mouse Gephyrin mAb7a IgG1 (Synaptic Systems, 1:1000) [17], rabbit Synapsin1/2 (Synaptic Systems, 1:1000) [20], DAPI (Invitrogen, 1:10,000), mouse HuC mIgG2b (Invitrogen, 1:1000) [21], rabbit GlyR IgG1 (mAb4a, Synaptic Systems, 1:500) [22, 23], mouse SV2 mIgG1 (DSHB, 1:1000) [24]. The following secondary antibodies were used at 1:1000: goat anti-rabbit IgG (H + L) Alexa Fluor 488, goat anti-rabbit IgG (H + L) Alexa Fluor Plus 594, goat anti-mouse IgG Alexa Fluor 647. Slides used for GlyR staining were additionally fixed with 95:5 methanol:acetic acid for 10 min at -20°C prior to rehydration. Stained samples were treated with ProLong Glass (Invitrogen P36980) at least one day prior to imaging.
Mounting larvae for in vivo imaging
For live imaging experiments, larvae at indicated stages were anesthetized with 0.16 mg/mL (600 µM) tricaine in embryo medium. The fish were mounted in 0.8% low-melt agarose (Sigma A9414) on a coverslip.
Confocal microscopy
All fluorescent imaging was performed on an upright Zeiss LSM 980 confocal with Airyscan 2 in 4Y fast mode. For fixed tissues, a 63x/1.4 NA oil objective was used, and for in vivo imaging, a 20x/1.0 NA water objective was used. All images were taken in Zeiss software. The following lasers were used: 488 nm for Alexa488 and GFP, 561 nm for Alexa594, 405 nm for DAPI, and 639 nm for Alexa647. For fixed tissue, z spacing was set to 0.13 µm. For in vivo imaging, z spacing was set to 1.0 µm.
Xanthine oxidase activity
Larvae were anesthetized with 0.16 mg/mL tricaine in embryo medium and their tails were collected individually for genotyping. The rest of larval bodies were collected individually in tubes, flash frozen, and stored at -80°C. Subsequently, 15 larvae of the same genotype were pooled together and homogenized with a handheld tissue homogenizer (VWR, 47747–370). The assessment of xanthine oxidase activity was conducted using a commercial kit (Sigma, MAK078-1KT). Fluorescence readings were obtained using a plate reader (CLARIOstarPlus, BMG LABTECH).
Swimming behavior assays
Coiling behavior recording
At 1 dpf, individual embryos from a gphnvo84/+ in-cross were placed under the scope and recorded in brightfield for 1 min at 40 Hz. The whole embryos were then used for genotyping.
Head touch response
At 5 dpf, fish were anesthetized using 0.16 mg/mL (600 µM) tricaine. Subsequently, the fish were transferred to individual wells within a 12-well plate filled with fresh embryo medium. Plates were placed on a hotplate set to 28°C, and the fish were allowed to recover from the effects of the tricaine. Each fish was gently prodded on the head using a probe.
Forced group swimming test
Dishes containing 25 mL of embryo medium and 50 5-dpf larvae were placed on a dark background on a 28°C hotplate and allowed to acclimatize. Subsequently, each dish was manually swirled ten times and then recorded for one minute using a Samsung Galaxy S20 phone. The first still was taken after the 11th swirl of embryo medium in the dish, allowing time for the hand to move out of frame after the ten swirls. Subsequent stills were taken every two seconds for eight total timepoints. The Cell Counter plugin in ImageJ was then used to mark the positions of the heads of all fish in each still, as well as the center of the dish. Matlab was used to measure the distance of each fish from the center and quantify the number within each concentric regions of increasing distance from the center at each timepoint.
Forced individual swimming test
Dishes containing 25 mL of embryo medium and one 5 dpf larvae were placed in a customized chamber and allowed to acclimatize. The chamber was equipped with 940 nm infrared strip lights and an acA1920-155um camera (Basler 106,879) with a 6 mm C VIS–NIR lens (Edmund Optics 39–939). The images were acquired with pylon Viewer 64-bit (Basler). During the imaging session, each dish was manually swirled three times, kept within a marked area, and recorded for 25 s at a frame rate of 25Hz. The images were processed in ImageJ (1.54f) and the fish was traced with TrackMate (v7.11.1). Fish were subsequently genotyped.
Animal survival
Three tanks of 50 fish each of WT and gphnbvo84/vo84 were placed on our system at 5 dpf. Fish were maintained in accordance with standard protocols of our fish facility. Briefly, the tanks are initially half full with no water flow, and the larvae are fed a diet of Gemma 75 and rotifers until 21 dpf. At 21 dpf, water flow is introduced, and fish are switched to Gemma 150 and rotifers. At three months, fish are switched to the adult diet of Gemma 300 and brine shrimp.
Quantification and statistical analysis
Fluorescent signal analysis
After Airyscan processing of confocal images in Zen, all images were analyzed using ImageJ. For fluorescent intensity analyses, maximum projections were generated. For Gephyrin, Synapsin, and GlyR fluorescence, ROIs were drawn delineating the neuropil and soma regions of each projection. The neuropil region was defined by the presence of Synapsin, and the soma region was defined by the lack of Synapsin and presence of DAPI. Integrated density was measured, restricted to the respective ROIs with a standard intensity threshold set for each round of stains. For GFP signal analysis in Tg(mbp:GFP-CAAX), a rectangular ROI of 100-µm length was created around the dorsal region of the spinal cord (body segments 7 or 15). The GFP signal was measured without a threshold.
Cell density analysis in the spinal cord sections
We restricted our analysis to a 2 µm section from the center of the z-stack. The Cell Counter plugin in ImageJ was used to mark all cells, identified by nuclear DAPI staining. We manually went through the 2 µm stack to ensure all cells were accounted for. We noted that sometimes HuC fluorescence appeared to colocalize with part of one nucleus but was actually associated with a different nucleus situated above or below. Therefore, HuC + cells were only scored if the HuC fluorescence was visible surrounding the nucleus in all directions to ensure HuC fluorescence is not from any overlapping cells.
3D reconstruction in Imaris
OPCs within a defined 300 µm length of the spinal cord between body segments 7 and 15 in Tg(olig1:mScarlet) were analyzed in Imaris. The Surface function was used to identify individual cells (diameter guide 10 µm, surface grain size 0.148 µm, threshold > 700, and quality > 150).
The Spots function was used to identify Gephyrin, GlyR, and SV2 puncta. XY diameter was set to 0.300 µm and Z diameter was set to 0.600 µm. Max intensity threshold was set to at least 1300 for Gephyrin, 2800 for GlyR, and 2200 for SV2. For colocalization analysis, a new set of spots was generated for distance between GlyR and SV2 puncta less than or equal to 0.5 µm, similar to previous work [17]. This set of spots represents colocalization locations between GlyR and SV2. The number of colocalization spots was divided by the total number of GlyR puncta to calculate colocalization ratio.
Statistics
All data are shown as mean ± SEM, with individual data points shown as circles. The datapoint that corresponds to the raw data image is indicated in pink. We conducted Shapiro–Wilk’s test and Levene’s test to verify data normality and homoscedasticity, respectively, before performing two-sided t-test for unpaired groups, one-way ANOVA test for more than 3 groups, and two-way ANOVA test for more than 1 variable. These analyses were performed in GraphPad Prism. The significance and statistical analyses employed were indicated in figure legends. Animal and cell number used for each experiment, t and df for t-tests, and F[DFn, DFd] for one-way or two-way ANOVA were indicated in figure legends. In some cases, data collection was not performed blindly due to the experimental conditions, but analyses were performed blindly. Sample size was chosen based on previous studies [17, 25]. Sex is not yet determined in zebrafish at the ages employed in our studies and so cannot be considered as a biological variable.
Results
Gephyrin is largely absent in gphnb vo84/vo84 spinal cord
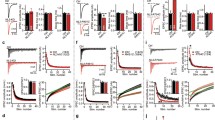
Gephyrin contains three domains: the N-terminal G-domain and the C-terminal C- and E-domains (Fig. 1A). G- and E-domains are required for Gephyrin to form multimers, which are proposed to anchor inhibitory receptors at synapses [26, 27]. We generated a gphnb mutant in zebrafish with CRISPR/Cas9-mediated genome editing and designated this allele as vo84. gphnbvo84 represents a 5-bp deletion in exon 7 in a region encoding the G-domain, which is predicted to cause a frameshift and premature stop codon (Fig. 1A). We developed a genotyping assay that can reliably identify the homozygous mutants (Fig. 1A, B; see Methods). In the spinal cord, Gephyrin is concentrated in the neuropil regions [8, 13] (Fig. 1C, D). Immunofluorescence (IF) analysis revealed that Gephyrin signal in the neuropil of gphnbvo84/vo84 mutants was reduced by 89.89 ± 7.61% compared to wild-type (WT) neuropil at 5 days post-fertilization (dpf) (Fig. 1D, E). To validate our synaptic staining, we co-stained Synapsin and normalized Gephyrin signal to Synapsin signal. We found that Synapsin signals in the mutant were unchanged, and normalized Gephyrin was reduced by 92.92 ± 1.56% (Fig. 1F, G). Some gphnbvo84/vo84 mutants survive to adulthood, so we utilized maternal-zygotic (MZ) gphnbvo84/vo84 mutants to assess maternal Gephyrin contributions. We found no significant differences in the residual Gephyrin IF signal between MZ and zygotic gphnbvo84/vo84 mutants (Fig. 1D-G), indicating lack of maternal contribution. A previous zebrafish study using morpholinos found that knockdown of gphna or gphnb alone did not reduce Gephyrin, but knockdown of both gphna and gphnb reduced Gephyrin in the spinal cord, suggesting that gphna contributes to Gephyrin abundance [13]. Therefore, we sought to test if gphna disruption can further reduce the Gephyrin protein abundance in gphnbvo84/vo84. We injected two high cutting-efficiency gphna sgRNAs [17] with Cas9 protein into embryos, such that gphna would be knocked down (“crispants”). We observed neither reduction of Gephyrin IF in gphnacrispant larvae compared to WT nor additional reduction of Gephyrin IF in gphnbvo84/vo84, gphnacrispant larvae compared to gphnbvo84/vo84 mutants at 5 dpf (Fig. 1D-G), indicating a minimal or no role of gphna in Gephyrin abundance in the spinal cord. Together, these results suggest that the 10% remaining Gephyrin signal in these mutants likely comes from background staining. Therefore, gphnbvo84 is likely a null loss-of-function allele, sufficient to abolish Gephyrin abundance in the developing spinal cord of zebrafish as assessed by IF.
Generation and validation of gphnbvo84 mutants. A Schematic of Gephyrin protein showing its three domains and the 5 bp deletion in gphnbvo84 causing a frameshift and premature stop. The BsrI recognition site used for genotyping was indicated in orange. B Representative gel images for genotyping gphnbvo84. A BsrI recognition site is disrupted by gphnbvo84 mutation in the middle of the amplicon. C Schematic of neuropil (magenta) and soma (black) regions in transverse spinal cord section of zebrafish at 5 dpf. D Representative transverse sections in the spinal cord at 5 dpf with Gephyrin (green) and Synapsin (magenta) staining from the following conditions: wildtype (WT); gphnacrispant, gphnbvo84/vo84; maternal zygotic (MZ) gphnbvo84/vo84; and gphnbvo84/vo84, gphnacrispant. E–G Gephyrin signal (E), Synapsin signal (F) and Gephyrin normalized to Synapsin (G) from the spinal sections. nWT = 19 fish, 41 sections, 3 technical replicates; ngphna = 12 fish, 20 sections, 1 technical replicate; ngphnb = 18 fish, 42 sections, 3 technical replicates; nMZ gphnb = 7 fish, 13 sections, 1 technical replicate; and n = 8 fish, 15 sections, 1 technical replicate. E, F[4, 126] = 45.29; F, F[4, 116] = 3.303; G, F[4, 129] = 56.29. All data are represented as mean ± SEM; ns, not significant; *, p < 0.05; ****, p < 0.0001; the data points corresponding to the representative images are noted in pink; ns, not significant; (E–G) one-way ANOVA with Tukey’s post-hoc test; scale bars 10 μm
Gphnb vo84/vo84 mutants exhibit normal gross morphology and xanthine oxidase activity
Gphn-null mice die within 1 day of birth [8], at least in part due to failures in MoCo synthesis outside the CNS. In contrast, gphnbvo84/vo84 developed normally at embryonic stages and exhibited normal gross morphology during larval stages (Fig. 2A, B). We observed a higher mortality of gphnbvo84/vo84 during late development, particularly among juveniles from 3 to 12 weeks post-fertilization (wpf) (Fig. 2C). While 74.66 ± 2.67% of WT fish survived to 12 wpf, only 48.66 ± 2.91% of gphnbvo84/vo84 mutants did (Fig. 2C). gphnbvo84/vo84 adults exhibited normal morphology (Fig. 2D) and no increased mortality (Fig. 2C). This preservation of normal development could be attributed to the presence of gphna, which can maintain MoCo synthesis throughout the organism. To test this, we measured the activity of a major MoCo-dependent enzyme, xanthine oxidase, previously shown to exhibit significantly reduced activity in Gphn-null mice [8]. We found no difference in enzyme activity between WT and gphnbvo84/vo84 larvae (Fig. 2E, F). Together, despite the absence of Gephyrin the spinal cord, gphnbvo84/vo84 mutants exhibit normal gross morphology and are capable of synthesizing MoCo.
gphnbvo8/vo84 mutants exhibit normal gross morphology and xanthine oxidase activity. A Representative images of WT and gphnbvo84/vo84 zebrafish larvae at 5 dpf. B The body length of larvae at 5 dpf. nWT = 13 animals, ngphnb = 12 animals, 2 technical replicates. t[0.7040], df[23]. C Fish survival tracked from larvae to adults. 3 groups of 50 fish were tested per condition. Fwpf[2.289, 9.156] = 144.2, Fgenotype[1, 4] = 20.58, Finteraction[15, 60] = 16.14, Ftank[4, 60] = 67.54. D Representative images of 1.5 years old adult zebrafish of the indicated genotypes and sexes. E Xanthine oxidase activity measured by fluorometric assays over time in WT and gphnbvo84/vo84. Each trace represents the total activity of 15 animals combined at 5 dpf. F Quantification of the xanthine oxidase activity at the end of recording from (E). Two sets of homogenates, each consisting of 15 animals were tested for each condition. t[0.7709], df[2]. All data are represented as mean ± SEM; ns, not significant; *, p < 0.05; (B,F) unpaired t-tests; (C) two-way repeat measure ANOVA with Sidak’s multiple comparison test
gphnb vo84/vo84 mutants exhibit swimming deficits against currents
Gphnbvo84/vo84 mutants offer an opportunity to study the role of Gephyrin at synapses independent of its role in MoCo synthesis. We first tested if the absence of Gephyrin in the spinal cord led to any behavioral defects. Previous studies using morpholinos to disrupt both gphna and gphnb reported defective coiling behavior (side-to-side alternating muscle contractions) at 24 h post-fertilization (hpf) and a failed touch response at 48 hpf [13]. In contrast, gphnbvo84/vo84 larvae exhibited normal spontaneous coiling behavior at 24 hpf (nWT = 28, ngphnb = 13; Videos 1–2) and were capable of swimming away in response to head touch at 5 dpf (nWT = ngphnb = 24).
In handling larvae, we noticed that gphnbvo84/vo84 mutants tended to cluster near the dish center after a swirl, indicating potential defects in rheotaxis (ability to swim against currents). To test this, we developed a forced group swimming assay, in which we swirled a dish containing 50 larvae, creating a current towards the center. WT larvae typically swam against the current and distributed relatively evenly in the dish (Fig. 3A, B and Video 3). However, gphnbvo84/vo84 larvae clustered closer to the center immediately after the swirl (Fig. 3A, B and Video 4). To further analyze this behavior, we customized a dark chamber and tracked individual zebrafish with infrared light to eliminate any influence from visible light (Fig. 3C and Videos 5). In this forced individual swimming assay, gphnbvo84/vo84 mutants also localized closer to the dish center after the swirl (Fig. 3D and Video 6). The swimming velocity of individual gphnbvo84/vo84 fish was slower than that of WT fish initially (Fig. 3E), likely contributing to their centered distribution after swirls. Interestingly, these mutants exhibited a burst of speed a few seconds later when the current subsided (Fig. 3E), suggesting that the current suppressed the swimming ability of gphnbvo84/vo84 animals. Together, these data suggest that synaptic Gephyrin in the spinal cord is dispensable for coiling behavior and touch response but plays an important role in rheotaxis.
gphnbvo84/vo84 mutants exhibit swimming deficits against currents. A Representative snapshots of WT and MZ gphnbvo84/vo84 larvae in a forced group swimming assay at 0 and 14 s after the dish swirl. Inset shows the distribution of fish (green dots) with reference concentric regions. B The number of fish within concentric regions ranging from the center to the edge of a dish at 0s and 14s after the dish swirl. 5 dishes each containing 50 fish were tested per condition. Fconcentric[7, 72] = 56.910s, 79.5314s; Fgenotype[1, 72] = 0.034190s, 0.108114s; Finteraction[7, 72] = 1.6700s, 1.71314s. C Representative snapshots of an individual larva in a forced swimming assay with infrared light. The larva (red circle) was traced over time (blue). Inset shows the larva at higher magnification. D Distance of individual larvae of WT and gphnbvo84/vo84 from the dish center after the dish swirl. nWT = 21 animals, ngphnb = 11 animals, 1 technical replicate. E Swimming velocity of individual zebrafish larva after the dish swirl. nWT = 21 animals, ngphnb = 11 animals, 1 technical replicate. All data are represented as mean ± SEM; *, p < 0.05; (B,E) two-way ANOVA with Sidak’s multiple comparison test
Cell densities and myelin are unchanged in the spinal cord of gphnb vo84/vo84 mutants
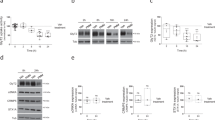
We wondered if the rheotaxis defects observed in gphnbvo48/vo84 mutants could be attributed to cellular changes in the spinal cord. Examination of spinal cord sections from gphnbvo84/vo84 animals revealed similar area of neuropil and soma regions compared to WT (Fig. 4A-C). Subsequently, we assessed cell density in the spinal cord using the nuclear marker DAPI and the neuronal marker HuC (Fig. 4A, B). Our analyses indicated that overall cell densities and percentage of neurons (HuC+) and non-neurons (HuC-) were comparable between WT and gphnbvo84/vo84 animals (Fig. 4D, E). Given the role of myelin in animal movement [28],the recent association of Gephyrin with myelination [17], and white matter lesions in MoCo deficiency [3, 4], we next examined oligodendrocyte development and myelination. Our findings revealed similar numbers of oligodendrocyte precursor cells (OPCs) in WT and gphnbvo84/vo84 spinal cords (Fig. 5A, B). Myelination in gphnbvo84/vo84, evaluated using the transgenic reporter Tg(mbp:GFP-CAAX), also appeared normal (Fig. 5C, D). These results suggest that overall cell density and myelination are not affected in the spinal cord of gphnbvo84/vo84mutants.
Cell densities are unchanged in gphnbvo84/vo84 mutant spinal cord. A Representative single-plane images of transverse larval spinal cord sections of WT and gphnbvo84/vo8 with HuC (green) and DAPI (blue) staining at 5 dpf. B Zoom-in image from (A) showing HuC+ (magenta arrowheads) and HuC- (yellow arrowheads) cells. C-E Quantifications of (C) transverse spinal cord area, (D) cell density, and (E) cell percentage from (A, B). C nWT = 22 fish, 31 sections; ngphnb = 20 fish, 32 sections; 6 technical replicates; tneuropil[1.483], tsoma[1.021], ttotal[0.1881], df[61]. (D,E) n = 7 fish, 10 sections per condition; 1 technical replicate; tHuC+density[1.619], tHuC-density[0.5591], t%[0.4604], df[18]. All data are represented as mean ± SEM; ns, not significant; (C-E) unpaired t-tests
Oligodendrocyte precursors and myelin are unchanged in gphnbvo84/vo84 mutant spinal cord. A Representative in vivo images from Tg(olig1:mScarlet) and 3D reconstruction showing individual OPCs in the spinal cord of WT and gphnbvo84/vo84 at 5 dpf. B OPC number in 300-µm-length spinal cord from (A). nWT = 11 animals, ngphnb = 11 animals, 1 technical replicate, t[0.5192] df[20],. C Representative in vivo images of myelin in the dorsal spinal cord from Tg(mbp:GFP-CAAX) at body segment 7 (S7) and 15 (S15) of WT and gphnbvo84/vo84 at 5 dpf. D GFP signal intensity from (C). nWT = 8 animals, ngphnb = 11 animals, 1 technical replicate, F[3,, 33] = 0.6057. All data are represented as mean ± SEM; ns, not significant; (B) unpaired t-tests; (D) one-way ANOVA with Tukey’s post-hoc test
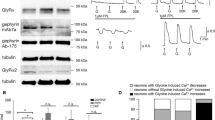
GlyRs are reduced, but remain proximal to synaptic regions in the spinal cord of gphnb vo84/vo84 animals
In Gphn-null mice, GlyRs are diffuse, absent from synapses, and do not colocalize with presynaptic markers [8]. To test if gphnbvo84/vo84 mutants possessed similar defects, we examined the abundance and localization of GlyRs in the spinal cord at 5 dpf with antibody staining. Interestingly, GlyRs still localized to the neuropil regions of gphnbvo84/vo84 spinal cord (Fig. 6A). We did observe a 44.15 ± 15.06% reduction in GlyR levels in the neuropil of gphnbvo84/vo84 animals compared to WT (Fig. 6A, B). Minimal GlyR signal was observed in the soma region, showing no significant differences between WT and gphnbvo84/vo84 zebrafish (Fig. 6A, B). We further identified each individual GlyR puncta using Imaris and observed a 31.70 ± 14.15% reduction in puncta number in the spinal cord of gphnbvo84/vo84 mutants compared to WT (Fig. 6C, D). To further investigate if the GlyRs localize to synapses, we examined GlyRs’ colocalization with presynaptic marker SV2. Similar to previous Gphn-null mice [8], SV2 abundance was not affected in the spinal cord (Fig. 6E, F). Surprisingly, we found similar levels of colocalization of SV2 and GlyRs in both WT and gphnbvo84/vo84 animals (0.5 μm distance threshold; WT, 26.18 ± 2.68%; gphnbvo84/vo84, 26.20 ± 2.32%) (Fig. 6G, H), indicating that the remaining GlyRs can localize to synaptic regions in the absence of Gephyrin. Together, these results suggest that while GlyRs are reduced in the neuropil of gphnbvo84/vo84, they remain in close proximity to synaptic regions.
GlyRs are reduced, but remain proximal to synaptic regions in the spinal cord of gphnbvo84/vo84 animals. A Representative transverse images of Gephyrin and GlyR staining from WT and gphnbvo84/vo84 at 5 dpf. From left, maximum z-projection, single plane, and zoom-in images. B GlyR signal normalized to WT from (A). nWT = 14 fish, 38 sections, ngphnb = 15 fish, 29 sections, 3 technical replicates, tneuropil[2.931] df[65], tsoma[0.2904] df[19]. C 3D reconstruction of GlyR from (A) with Imaris. D GlyR puncta number in 3D reconstruction from (C). nWT = 14 fish, 39 sections, ngphnb = 15 fish, 35 sections, 3 technical replicates, t[2.239], df[72]. E Representative transverse images of SV2 and GlyR staining from WT and gphnbvo84/vo84 at 5 dpf (maximum z-projection). F SV2 signal normalized to WT from (E). nWT = 7 fish, 19 sections, ngphnb = 8 fish, 27 sections, 2 technical replicates, t[1.602], df[44]. G 3D reconstruction of SV2 and GlyR from (E) with Imaris with zoom-in images on the right. In the zoom-in images, colocalizing puncta are in solid color and non-colocalizing puncta are transparent. H The percentage of GlyR that colocalize with SV2 from (G). nWT = 7 fish, 27 sections, ngphnb = 8 fish, 25 sections, 2 technical replicates, t[0.004848], df[50]. All data are represented as mean ± SEM; ns, not significant; *, p < 0.05; **, p < 0.01; (B,D,F,H) unpaired t-tests
Discussion
Postsynaptic organization plays a pivotal role in synaptic function and plasticity at inhibitory synapses [1]. Gephyrin acts as the postsynaptic scaffold, interacting with a myriad of proteins to enable inhibitory synaptic transmission. Among these interactions, Gephyrin’s association with GlyRs is best known, as exemplified in foundational work co-purifying Gephyrin and GlyRs from spinal cords using strychnine affinity columns [29, 30]. Currently, Gephyrin is thought to directly regulate GlyR clustering at postsynaptic sites, as best shown by the absence of GlyRs at synapses in Gphn-null mice [8]. However, assessing Gephyrin’s specific synaptic function has been challenging due to its dual role as a critical enzyme for MoCo synthesis. Indeed, the Gphn-null mice die perinatally, complicating the interpretation of the synaptic phenotypes.
In zebrafish, Gephyrin is duplicated and expressed in various tissues. Here, we report a new mutant allele, gphnbvo84, that led to nearly complete absence of Gephyrin protein in the spinal cord. With maternal depletion and additional gphna disruption, we did not observe further Gephyrin reduction, establishing gphnb as the source for Gephyrin in the spinal cord. Interestingly, these animals appear grossly normal at larval and adult stages and exhibit normal activity of xanthine oxidase and neuronal densities. Unlike Gphn-null mice or zebrafish with morpholinos targeting both gphna and gphnb [8, 13], gphnbvo84/vo84 mutants show overall normal movement and response to touch. These data suggest that the lack of synaptic Gephyrin in the spinal cord does not cause perinatal lethality or severely impaired movement. Instead, the reduced MoCo enzymatic activity of Gephyrin in other cell types and tissues possibly mediates these defects, consistent with the early lethality and neuronal death in Mocs1 and Mocs2 mice [5,6,7]. Transcriptomic data from mice reveal that Gphn is also expressed in OPCs, oligodendrocytes, and astrocytes [31, 32]. These CNS glia have recently been linked to movement disorders, such as Parkinson’s disease [33, 34]. It would be intriguing to explore how Gephyrin in glia or other cell types contribute to movement and response to touch in future studies.
We observed rheotaxis defects in gphnbvo84/vo84 larvae. Rheotaxis relies on lateral line neurons to detect currents [35,36,37]. These sensory inputs are then processed in the hindbrain, involving inhibitory neurons and resulting in distinct motor outputs between the left and right sides of the body [38]. Our observations of reduced GlyRs in gphnbvo84/vo84 larvae likely impaired the function of inhibitory transmission within this circuit, disrupting the correct sequence of muscle contraction and relaxation between the two sides of the body. Alternatively, it is possible that a “suppressive” signal is produced in the brain in response to currents in the mutants and interferes with proper rheotaxis. We suspect that such defects in rheotaxis may contribute to the increased mortality in late development of the mutants. In surviving adults, we did not observe obvious swimming defects in gphnbvo84/vo84 mutants. One possibility is that the increased mortality in late development of the mutants might lead to selective survival of healthier animals, potentially masking behavior defects in adults. Therefore, future directions rescuing the late development mortality (e.g., reducing water flow) might provide more insight into the role of Gephyrin in adult behaviors. In addition, it would be interesting to test if other types of directional movement are also affected in these mutants.
Mutations in GPHN and GLRA1 (Glycine receptor alpha 1) have been linked to hyperekplexia in humans, a disorder that manifests from childhood and is characterized by exaggerated startle responses, stiffness, and an inability to move [39, 40]. Interestingly, in our swimming assay, we observed that gphnbvo84 larvae initially exhibited impairment in response to currents (within 2.5 s), followed by a burst of activity thereafter (5–10 s). We speculate that such distinct phases of activity also occur in hyperekplexia. It is worth noting that reduced GlyRs can affect not only neurons but also glia [41, 42]; yet the role of glia in hyperekplexia remains unclear. To better characterize this disease and dissect its underlying mechanisms, zebrafish could provide the necessary tools to combine in vivo imaging and behavior assays to assess the underlying circuits from early developmental stages.
Surprisingly, in gphnbvo84/vo84 animals, the remaining GlyRs still localize to the synaptic regions and exhibit a similar colocalization ratio with SV2. We noted that only a portion of GlyRs colocalize with SV2 in WT, indicating SV2 may not label all inhibitory presynaptic terminals. Nonetheless, these results challenge the previous notion that the depletion of Gephyrin at synapses would inevitably result in diffuse and non-synaptic GlyR localization. While electron microscopy would offer the resolution needed to precisely define the synaptic location of GlyRs, our results suggest that GlyRs are transported to axons and are capable of forming clusters near synapses even in the absence of Gephyrin. Overall, this viable gphnbvo84 allele provides an excellent tool for studying the synaptic function of Gephyrin without the confounding factors of major defects in animal health.
Conclusions
We identified a viable zebrafish gphnb mutant that shows nearly no defects that were previously associated with MoCo deficiency. Instead, this mutant exhibits impaired rheotaxis. Moreover, GlyRs still cluster to some degree in the synaptic regions of gphnb mutant, contrasting the diffuse GlyRs observed when both gphna and gphnb were disrupted in previous studies [13]. These observations underscore the distinct roles of Gephyrin in synaptic function and MoCo synthesis.
Availability of data and materials
All data used during the current study are available from the corresponding author on reasonable request.
Abbreviations
- CNS:
-
Central nervous system
- MoCo:
-
Molybdenum cofactor
- GlyR:
-
Glycine receptor
- gphnb :
-
gephyrinb
- gphna :
-
gephyrina
- WT:
-
Wild-type/wildtype
- hpf:
-
Hours post-fertilization
- dpf:
-
Days post-fertilization
- wpf:
-
Weeks post-fertilization
- MZ:
-
Maternal-zygotic
- OPC:
-
Oligodendrocyte precursor cell
- Mbp :
-
Myelin basic protein
References
Tyagarajan SK, Fritschy JM. Gephyrin: a master regulator of neuronal function? Nat Rev Neurosci. 2014;15:141–56.
Jung H, Kim S, Ko J, Um JW. Intracellular signaling mechanisms that shape postsynaptic GABAergic synapses. Curr Opin Neurobiol. 2023;81:102728.
Atwal PS, Scaglia F. Molybdenum cofactor deficiency. Mol Genet Metab. 2016;117:1–4.
Bayram E, Topcu Y, Karakaya P, Yis U, Cakmakci H, Ichida K, et al. Molybdenum cofactor deficiency: Review of 12 cases (MoCD and review). Eur J Paediatr Neurol. 2013;17:1–6.
Reiss J, Bonin M, Schwegler H, Sass JO, Garattini E, Wagner S, et al. The pathogenesis of molybdenum cofactor deficiency, its delay by maternal clearance, and its expression pattern in microarray analysis. Mol Genet Metab. 2005;85:12–20.
Lee HJ, Adham IM, Schwarz G, Kneussel M, Sass JO, Engel W, et al. Molybdenum cofactor-deficient mice resemble the phenotype of human patients. Hum Mol Genet. 2002;11:3309–17.
Jakubiczka-Smorag J, Santamaria-Araujo JA, Metz I, Kumar A, Hakroush S, Brueck W, et al. Mouse model for molybdenum cofactor deficiency type B recapitulates the phenotype observed in molybdenum cofactor deficient patients. Hum Genet. 2016;135:813–26.
Feng G, Tintrup H, Kirsch J, Nichol MC, Kuhse J, Betz H, et al. Dual-requirement for gephyrin in glycine receptor clustering and molybdoenzyme activity. Science (80- ). 1998;282:1321–4.
Fischer F, Kneussel M, Tintrup H, Haverkamp S, Rauen T, Betz H, et al. Reduced synaptic clustering of GABA and glycine receptors in the retina of the gephyrin null mutant mouse. J Comp Neurol. 2000;427:634–48.
Gross GG, Straub C, Perez-Sanchez J, Dempsey WP, Junge JA, Roberts RW, et al. An E3-ligase-based method for ablating inhibitory synapses. Nat Methods. 2016;13:673–8.
Lévi S, Logan SM, Tovar KR, Craig AM. Gephyrin is critical for glycine receptor clustering but not for the formation of functional GABAergic synapses in hippocampal neurons. J Neurosci. 2004;24:207–17.
Kirsch J, Wolters I, Triller A, Betz H. Gephyrin antisense oligonucleotides prevent glycine receptor clustering in spinal neurons. Nature. 1993;366:745–8.
Ogino K, Ramsden SL, Keib N, Schwarz G, Harvey RJ, Hirata H. Duplicated gephyrin genes showing distinct tissue distribution and alternative splicing patterns mediate molybdenum cofactor biosynthesis, glycine receptor clustering, and escape behavior in zebrafish. J Biol Chem. 2011;286:806–17.
Lange M, Granados A, VijayKumar S, Bragantini J, Ancheta S, Santhosh S, et al. Zebrahub – Multimodal Zebrafish Developmental Atlas Reveals the State Transition Dynamics of Late Vertebrate Pluripotent Axial Progenitors. bioRxiv. 2023;2023.03.06.531398.
Kok FO, Shin M, Ni CW, Gupta A, Grosse AS, vanImpel A, et al. Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Dev Cell. 2015;32:97–108.
Almeida RG, Czopka T, Ffrench-Constant C, Lyons DA. Individual axons regulate the myelinating potential of single oligodendrocytes in vivo. Development. 2011;138:4443–50.
Li J, Miramontes TG, Czopka T, Monk KR. Synaptic input and Ca2+ activity in zebrafish oligodendrocyte precursor cells contribute to myelin sheath formation. Nat Neurosci. 2024;27:219–31.
Marisca R, Hoche T, Agirre E, Hoodless LJ, Barkey W, Auer F, et al. Functionally distinct subgroups of oligodendrocyte precursor cells integrate neural activity and execute myelin formation. Nat Neurosci. 2020;23:363–74.
Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, et al. The Tol2kit: A multisite gateway-based construction Kit for Tol2 transposon transgenesis constructs. Dev Dyn. 2007;236:3088–99.
Easley-Neal C, Fierro J, Buchanan JA, Washbourne P. Late Recruitment of Synapsin to Nascent Synapses Is Regulated by Cdk5. Cell Rep. 2013;3:1199–212.
Kay JN, Finger-Baier KC, Roeser T, Staub W, Baier H. Retinal ganglion cell genesis requires lakritz, a zebrafish atonal homolog. Neuron. 2001;30:725–36.
Ogino K, Yamada K, Nishioka T, Oda Y, Kaibuchi K, Hirata H. Phosphorylation of gephyrin in zebrafish mauthner cells governs glycine receptor clustering and behavioral desensitization to sound. J Neurosci. 2019;39:8988–97.
McDearmid JR, Liao M, Drapeau P. Glycine receptors regulate interneuron differentiation during spinal network development. Proc Natl Acad Sci U S A. 2006;103:9679–84.
Emond MR, Jontes JD. Inhibition of protocadherin-α function results in neuronal death in the developing zebrafish. Dev Biol. 2008;321:175–87.
Mensch S, Baraban M, Almeida R, Czopka T, Ausborn J, El Manira A, et al. Synaptic vesicle release regulates myelin sheath number of individual oligodendrocytes in vivo. Nat Neurosci. 2015;18:628–30.
Herweg J, Schwarz G. Splice-specific glycine receptor binding, folding, and phosphorylation of the scaffolding protein gephyrin. J Biol Chem. 2012;287:12645–56.
Sander B, Tria G, Shkumatov AV, Kim EY, Grossmann JG, Tessmer I, et al. Structural characterization of gephyrin by AFM and SAXS reveals a mixture of compact and extended states. Acta Crystallogr Sect D Biol Crystallogr. 2013;69:2050–60.
Chernoff GF. Shiverer: an autosomal recessive mutant mouse with myelin deficiency. J Hered. 1981;72:128.
Pfeiffer P, Graham D, Betz H. Purification by affinity chromatography of the glycine receptor of rat spinal cord. J Biol Chem. 1982;257:9389–93.
Graham D, Pfeiffer F, Simler R, Betz H. Purification and characterization of the glycine receptor of pig spinal cord. Biochemistry. 1985;24:990–4.
Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34:11929–47.
Hrvatin S, Hochbaum DR, Nagy MA, Cicconet M, Robertson K, Cheadle L, et al. Single-cell analysis of experience-dependent transcriptomic states in the mouse visual cortex. Nat Neurosci. 2018;21:120–9.
Xu J, Farsad HL, Hou Y, Barclay K, Lopez BA, Yamada S, et al. Human striatal glia differentially contribute to AD- and PD-specific neurodegeneration. Nat Aging. 2023;3:346–65.
Dehestani M, Kozareva V, Blauwendraat C, Fraenkel E, Gasser T, Bansal V. Transcriptomic changes in oligodendrocytes and precursor cells predicts clinical outcomes of Parkinson’s disease. bioRxiv. 2023;2023.05.11.540329.
Oteiza P, Odstrcil I, Lauder G, Portugues R, Engert F. A novel mechanism for mechanosensory-based rheotaxis in larval zebrafish. Nature. 2017;547:445–8.
Suli A, Watson GM, Rubel EW, Raible DW. Rheotaxis in larval zebrafish is mediated by lateral line mechanosensory hair cells. PLoS ONE. 2012;7:e29727.
Newton KC, Kacev D, Nilsson SRO, Saettele AL, Golden SA, Sheets L. Lateral line ablation by ototoxic compounds results in distinct rheotaxis profiles in larval zebrafish. Commun Biol. 2023;6:84.
Valera G, Markov DA, Bijari K, Randlett O, Asgharsharghi A, Baudoin JP, et al. A neuronal blueprint for directional mechanosensation in larval zebrafish. Curr Biol. 2021;31:1463–1475.e6.
Saini AG, Pandey S. Hyperekplexia and other startle syndromes. J Neurol Sci. 2020;416:117051.
Harvey RJ, Topf M, Harvey K, Rees MI. The genetics of hyperekplexia: more than startle! Trends Genet. 2008;24:439–47.
Morais TP, Coelho D, Vaz SH, Sebastião AM, Valente CA. Glycine receptor activation impairs ATP-induced calcium transients in cultured cortical astrocytes. Front Mol Neurosci. 2018;10:444.
Belachew S, Malgrange B, Rigo JM, Register B, Leprince P, Hans G, et al. Glycine triggers an intracellular calcium influx in oligodendrocyte progenitor cells which is mediated by the activation of both the ionotropic glycine receptor and Na+-dependent transporters. Eur J Neurosci. 2000;12:1924–30.
Acknowledgements
We thank members of the Monk lab for discussions and critical reading of the manuscript. We are indebted to Austin Forbes and Tia Perry for fish care. We also thank Taylor Jay and the Freeman lab for instrument use and technical help with the plate reader.
Funding
This work was supported by a Warren Alpert Distinguished Scholar Award (JL), and NIH/NINDS grant 1R21NS120650 (KRM).
Author information
Authors and Affiliations
Contributions
E.J.B. and J.L. conceived the project with input from K.R.M. E.J.B. carried out experiments and data analyses with input from J.L. J.L. generated the gphnb mutant and the transgenic Tg(olig1:mScarlet) line and performed analysis on OPCs. E.J.B. and J.L. wrote the manuscript with input from K.R.M. All authors edited the manuscript and approved of the final version.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All zebrafish experiments and procedures were performed in compliance with institutional ethical regulations for animal testing and research at Oregon Health and Science University (OHSU). Experiments were approved by the Institutional Animal Care and Use Committee of OHSU.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Video 1. Spontaneous coiling behavior of WT larva at 1 dpf.
Additional file 2: Video 2. Spontaneous coiling behavior of gphnbvo84/vo84 larva at 1 dpf.
Additional file 3: Video 3. WT larvae in forced group swimming assay at 5 dpf.
Additional file 4: Video 4. MZ gphnbvo84/vo84larvae in forced group swimming assay at 5 dpf.
Additional file 5: Video 5. WT larva in individual forced swimming assay at 5 dpf with path tracked.
Additional file 6: Video 6. gphnbvo84/vo84larva in individual forced swimming assay at 5 dpf with path tracked.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Brennan, E.J., Monk, K.R. & Li, J. A zebrafish gephyrinb mutant distinguishes synaptic and enzymatic functions of Gephyrin. Neural Dev 19, 14 (2024). https://doi.org/10.1186/s13064-024-00191-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13064-024-00191-5