Abstract
Background
Biodiesel is a mixture of fatty acid short-chain alkyl esters of different fatty acid carbon chain lengths. However, while fatty acid methyl or ethyl esters are useful biodiesel produced commercially, fatty acid esters with branched-chain alcohol moieties have superior fuel properties. Crucially, this includes improved cold flow characteristics, as one of the major problems associated with biodiesel use is poor low-temperature flow properties. Hence, microbial production as a renewable, nontoxic and scalable method to produce fatty acid esters with branched-chain alcohol moieties from biomass is critical.
Results
We engineered Saccharomyces cerevisiae to produce fatty acid short- and branched-chain alkyl esters, including ethyl, isobutyl, isoamyl and active amyl esters using endogenously synthesized fatty acids and alcohols. Two wax ester synthase genes (ws2 and Maqu_0168 from Marinobacter sp.) were cloned and expressed. Both enzymes were found to catalyze the formation of fatty acid esters, with different alcohol preferences. To boost the ability of S. cerevisiae to produce the aforementioned esters, negative regulators of the INO1 gene in phospholipid metabolism, Rpd3 and Opi1, were deleted to increase flux towards fatty acyl-CoAs. In addition, five isobutanol pathway enzymes (Ilv2, Ilv5, Ilv3, Aro10, and Adh7) targeted into the mitochondria were overexpressed to enhance production of alcohol precursors. By combining these engineering strategies with high-cell-density fermentation, over 230 mg/L fatty acid short- and branched-chain alkyl esters were produced, which is the highest titer reported in yeast to date.
Conclusions
In this work, we engineered the metabolism of S. cerevisiae to produce biodiesels in the form of fatty acid short- and branched-chain alkyl esters, including ethyl, isobutyl, isoamyl and active amyl esters. To our knowledge, this is the first report of the production of fatty acid isobutyl and active amyl esters in S. cerevisiae. Our findings will be useful for engineering S. cerevisiae strains toward high-level and sustainable biodiesel production.
Similar content being viewed by others
Background
Biodiesel is a mixture of fatty acid short-chain alkyl esters of different fatty acid carbon chain lengths. FAMEs (fatty acid methyl esters) and FAEEs (fatty acid ethyl esters) are used as commercial biodiesel and obtained via transesterification of vegetable oils with an alcohol (methanol or ethanol) with the aid of a catalyst. However, while FAMEs or FAEEs are useful alternative diesel fuels currently being used in the market, fatty acid esters with branched-chain alcohol moieties have better fuel properties [1, 2]. Crucially, this includes improved cold flow characteristics where cloud points and pour points are reduced, as one of the major problems associated with biodiesel use is poor low-temperature flow properties [3].
Baker’s yeast Saccharomyces cerevisiae, which is used for industrial scale bioethanol production, brewing and winemaking, is an important host for biotechnological applications [4]. The ability to grow robustly and the abundance of genetic tools available for its manipulation make yeast an ideal host microbe for engineering biofuel production. Various research groups have engineered yeast for production of FAEEs by heterologous expression of an acyl-CoA: alcohol acyltransferase or wax ester synthase [5–11]. Five wax ester synthases, from Acinetobacter baylyi ADP1, Marinobacter hydrocarbonoclasticus DSM 8798, Rhodococcus opacus PD630, Mus musculus C57BL/6 and Psychrobacter arcticus 273-4 were expressed in yeast where wax ester synthase from M. hydrocarbonoclasticus DSM 8798 (ws2) was found to produce the most FAEEs (6.3 mg/L) [5]. Multiple metabolic engineering strategies were applied towards boosting the production of FAEEs in yeast. Abolishing protein kinase Snf1-dependent regulation of acetyl-CoA carboxylase Acc1 through Ser659 and Ser1157 mutation increased FAEEs titers to 15.8 mg/L [6]. Elimination of non-essential fatty acid utilization pathways through deletion of acyl-CoA: sterol acyltransferases (encoded by ARE1 and ARE2), diacylglycerol acyltransferases (encoded by DGA1 and LRO1) and fatty acyl-CoA oxidase (encoded by POX1) led to 17.2 mg/L FAEEs [7]. Integration of 6 copies of the ws2 expression cassette into the genome increased production of FAEEs to 34 mg/L, while further boosting of the fatty acyl-CoAs availability by acyl-CoA binding protein (encoded by ACB1) overexpression and NADPH supply by overexpression of bacterial NADP+-dependent glyceraldehyde-3-phosphate dehydrogenase (gapN) led to FAEEs production of 48 mg/L [8].
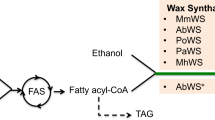
On the other hand, only trace amounts of fatty acid isoamyl esters (FAIEs) and FAEEs were produced in an engineered yeast strain expressing wax ester synthase from A. baylyi ADP1 and with ARE1, ARE2, DGA1 and LRO1 disrupted [12, 13]. In addition, the metabolic engineering of yeast to produce and accumulate fatty acid isobutyl esters (FABEs) and fatty acid active amyl esters (FAAEs) has not been reported. Here, we engineered yeast to produce fatty acid short- and branched-chain esters (FASBEs), including ethyl, isobutyl, active amyl and isoamyl esters, using endogenously synthesized fatty acids and alcohols (Fig. 1). First, two wax ester synthase genes (ws2 and Maqu_0168 from Marinobacter sp.) were cloned and expressed. Second, negative regulators of the INO1 gene in phospholipid metabolism, Rpd3 and Opi1, were deleted. INO1 gene encodes for inositol-3-phosphate synthase that makes inositol phosphates and inositol-containing phospholipids. As synthesis of phospholipids requires fatty acyl-CoAs as precursors, the removal of INO1 negative regulators may boost flux towards fatty acyl-CoAs-derived phospholipids and the abovementioned esters [14].The deletion of RPD3 and OPI1 was shown previously to enable simultaneous increase of phospholipids and desired product 1-hexadecanol [15]. Third, isobutanol pathway enzymes (acetolactate synthase Ilv2, ketoacid reductoisomerase Ilv5, dihydroxyacid dehydratase Ilv3, α-ketoacid decarboxylase Aro10, and alcohol dehydrogenase Adh7) targeted into the mitochondria were overexpressed to boost production of alcohol precursors. Ilv2, Ilv5 and Ilv3 are naturally located in the mitochondria, whereas Aro10 and Adh7 were re-targeted to the mitochondria using N-terminal fusion with mitochondria localization signal from subunit IV of the yeast cytochrome c oxidase (encoded by COX4) [16, 17]. Compartmentalization of this pathway into the mitochondria enabled high-level production of branched-chain alcohols. Finally, by combining these engineering strategies with high-cell-density fermentation, over 230 mg/L FASBEs were produced, which represents the highest titer reported in yeast to date.
Metabolic engineering strategy to produce FASBEs. FASBEs can be produced by expressing a wax ester synthase (ws2 or Maqu_0168). Isobutanol pathway enzymes (Ilv2, Ilv5, Ilv3, Aro10, Adh7) were overexpressed in the mitochondria to accumulate more isobutanol and FABEs. At the same time, production of isoamyl alcohol and active amyl alcohols were also increased, resulting in increased FAIEs and FAAEs production. To boost FASBEs production, negative regulators of INO1 (Opi1, Rpd3) were deleted. Genes overexpressed are shown in green. Red crosses gene deletions
Results and discussion
Substrate preference of the wax ester synthases
To produce FASBEs in S. cerevisiae, a wax ester synthase gene needs to be expressed. The ws2 enzyme, which was selected as the best wax ester synthase for FAEEs production in yeast [5], and the Maqu_0168 enzyme, which was shown in vitro to have high wax synthase activities [18], were, respectively, cloned into pESC-URA plasmid for expression in yeast. It was found that the yeast strain expressing ws2 produced mostly FAEEs (18.2 mg/L), with a small proportion of FABEs (1.85 mg/L) and FAIEs (2.21 mg/L) produced (Fig. 2). On the other hand, the yeast strain expressing Maqu_0168 resulted in a large proportion of FAIEs (96.2 mg/L), less FAEEs (12.2 mg/L), and a small amount of FABEs (3.82 mg/L). Further, as shown from the gas chromatography mass spectrometry (GC/MS) peaks (Fig. 2), the majority of the products were C16 and C18 fatty acid derived.
To determine whether having more alcohols present in the culture medium can further boost FASBEs product levels, 0.1 % ethanol, isobutanol or isoamyl alcohol was added exogenously. Additional file 1: Figure S1A shows that for the yeast strain expressing ws2, additional ethanol did not result in increased FAEEs titers, whereas additional isobutanol and isoamyl alcohol did result in ninefold increase in FABEs and FAIEs product titers, respectively. Therefore, while ethanol is not rate limiting in the yeast strains expressing ws2 for FAEEs production, isobutanol and isoamyl alcohol levels are rate limiting for FABEs and FAIEs production. Additional file 1: Figure S1B shows that for the yeast strain expressing Maqu_0168, neither exogenous ethanol nor isoamyl alcohol resulted in increased FAEEs or FAIEs titers, respectively, whereas additional isobutanol increased FABEs levels by eightfold. Hence, both ethanol and isoamyl alcohol are not rate limiting in the yeast strain expressing Maqu_0168, but isobutanol is rate limiting for FABEs production. In addition, this result indicates that Maqu_0168 has higher affinity for isoamyl alcohol compared to ws2.
Deletion of INO1 negative regulators to boost FASBEs production
Towards engineering yeast strains that produce high levels of fatty acid esters, we deleted two genes that are INO1 negative regulators, namely RPD3 and OPI1. Deletion of both genes was shown previously to boost phospholipid production in yeast, and simultaneously boost fatty acyl-CoA derived fatty alcohol titers [15]. Figure 3 shows the production of FAEEs, FABEs, FAIEs and total FASBEs in the yeast strains and mutants expressing ws2 over a period of 168 h. Fermentation of yeast was followed up to 168 h to enable yeast cells to biosynthesize and accumulate more products. Here, we use values obtained at 168 h to discuss the production levels of FABSEs. While production of FABEs and FAIEs is low in the wild-type strain (2.0 mg/L for FABEs and 2.1 mg/L for FAIEs), FAEEs production was higher at 14.0 mg/L. When OPI1 was deleted, the production of FAEEs increased 2.7-fold to 38.0 mg/L. However, deletion of RPD3 reduced fatty acid esters production, as opposed to the case for fatty alcohol production [15]. By carrying out a phospholipid assay, we found that indeed under our test conditions, OPI1 deletion increased phospholipid levels whereas RPD3 deletion decreased phospholipid levels (Additional file 1: Figure S3). This is possibly due to a difference in growth conditions, where glucose was used as carbon source in the previous report, while here, we have used galactose as inducer of protein expression and carbon source for growth. In the OPI1 knockout strain, a total FASBEs titer of 43.9 mg/L was obtained.
Production of a FAEEs, b FABEs, c FAIEs and d Total FASBEs, in BY4742 and knockout strains of INO1 negative regulators (rpd3Δ and opi1Δ) expressing wax ester synthase ws2. All strains were cultured in minimal medium lacking uracil with 0.2 % glucose and 1.8 % galactose. Values are the mean of biological triplicates ±SD at 0, 48, 96, and 168 h. Figures with adjusted y-axis scales for (b) and (c) are shown in Additional file 1: Figure S2
Figure 4 shows the production of FAEEs, FABEs, FAIEs, and total FASBEs in the yeast strains and mutants expressing Maqu_0168 over a period of 168 h. Here, the production of FAEEs and FABEs is low in the wild-type strain (11.3 mg/L for FAEEs and 3.4 mg/L for FABEs) compared to the production of FAIEs (95.6 mg/L). The total FASBEs produced was 110.3 mg/L. Also, it was found that deletion of OPI1 and RPD3 both resulted in reduced FASBEs production levels. As the production of fatty acid esters requires two substrates, fatty acyl-CoA and alcohol, a possible explanation is that deletion of OPI1 caused a reduction in isoamyl alcohol levels. This was confirmed by quantifying alcohol levels, where BY4742 opi1Δ (45.0 mg/L) had lower isoamyl alcohol levels compared to BY4742 (64.3 mg/L) (Table 1).
Production of a FAEEs, b FABEs, c FAIEs and d Total FASBEs, in BY4742 and knockout strains of INO1 negative regulators (rpd3Δ and opi1Δ) expressing wax ester synthase Maqu_0168. All strains were cultured in minimal medium lacking uracil with 0.2 % glucose and 1.8 % galactose. Values are the mean of biological triplicates ±SD at 0, 48, 96, and 168 h. Figures with adjusted y-axis scales for (a) and (b) are shown in Additional file 1: Figure S4
Both Figs. 3 and 4 show that fatty acid ester titers were comparable at 48, 96 and 168 h. This suggests that production of FASBEs occurs mainly during cell growth stage, and is greatly reduced or stopped in stationary phase (Additional file 1: Figure S5). In addition, FAEEs titers were higher at 48 h and decreased after that, suggesting that FAEEs may have diffused out of the cell, or possibly degraded. However, extraction of products in the medium did not yield any FAEEs, indicating that most FAEEs were intracellular, while extracellular FAEEs had likely evaporated. Similarly, no FABEs or FAIEs were detected in the medium.
Boosting FABEs production
As seen in Figs. 3 and 4, the production of FABEs is minimal. Hence, to boost FABEs production, genes that enable the production and accumulation of isobutanol need to be expressed in yeast. Here, we obtained a yeast strain (BY4742-IB) that has five isobutanol pathway enzymes (Ilv2, Ilv5, Ilv3, Aro10 and Adh7) overexpressed in the mitochondria of yeast to supply the required isobutanol [17]. This strain was constructed through integration of expression cassettes into δ-sites of retrotransposon elements of yeast chromosomes. Isobutanol produced is naturally transported out from the mitochondria into the cytoplasm where synthesis of FASBEs takes place. The increased isobutanol production was confirmed where an 18-fold increase to 176.6 mg/L isobutanol was obtained compared to BY4742 (9.61 mg/L) (Table 1).
We then expressed ws2 and Maqu_0168 wax ester synthases in BY4742-IB and its OPI1 knockout mutant (Fig. 5). A RPD3 knockout mutant was not used because RPD3 deletion did not increase FASBEs production in BY4742. BY4742-IB expressing ws2 produced 49.3 mg/L FABEs, whereas BY4742-IB expressing Maqu_0168 produced 26.6 mg/L FABEs. Also, the deletion of OPI1 increased the amount of FABEs produced for both ws2 and Maqu_0168 expressing strains to 69.8 and 30.2 mg/L, respectively.
Production of a FAEEs, b FABEs, c FAIEs and FAAEs, and d Total FASBEs, in BY4742-IB and BY4742-IB opi1Δ expressing wax ester synthase ws2 or Maqu_0168. All strains were cultured in minimal medium lacking uracil and valine with 0.2 % glucose and 1.8 % galactose. Values are the mean of biological triplicates ±SD at 0, 48, 96, and 168 h
At the same time, in BY4742-IB strains, FAAEs were also accumulated when active amyl alcohols levels were produced (Table 1). The production of active amyl alcohol is a result of overlapping synthesis pathway with the isobutanol producing pathway [16]. FAAEs were not detected in wild-type BY4742 strain due to the absence of active amyl alcohol. In BY4742-IB opi1Δ expressing ws2, 4.25 mg/L FAAEs were produced, whereas 3.01 mg/L FAAEs were obtained when Maqu_0168 was expressed. FAIEs were the major fatty acid amyl esters produced, where production levels of FAIE for BY4742-IB opi1Δ expressing ws2 and Maqu_0168 were 9.6 mg/L and 68.7 mg/L, respectively. For FAEEs, 32.4 mg/L was produced in BY4742-IB opi1Δ expressing ws2, whereas 8.3 mg/L was produced in BY4742-IB opi1Δ expressing Maqu_0168. The total FASBEs produced increased for both ws2 (83.2 mg/L to 116.1 mg/L) and Maqu_0168 (102.0 mg/L to 110.2 mg/L) expressing OPI1 mutant strains.
In addition, isoamyl alcohol concentration was found to be boosted more than 35 % for both BY4742-IB opi1Δ (87.2 mg/L) and BY4742-IB (90.1 mg/L) compared to BY4742 strain (64.3 mg/L) (Table 1). Indeed, the expression of the isobutanol pathway enzymes can also boost isoamyl alcohol levels, also as a result of overlapping synthesis pathway with the isobutanol producing pathway.
High-cell-density fermentation for FASBEs production
We next characterized four best strains for production of FASBEs using high-cell-density fermentation. High-cell-density fermentation would allow rapid and more efficient conversion of sugars into desired products [15]. Cells were inoculated with initial OD600 of 9 (Additional file 1: Figure S6). Figure 6 and Additional file 1: Figure S7 shows that yeast strain BY4742 opi1Δ expressing ws2 produced 33.4 mg/L FAEEs, 16.2 mg/L FABEs and 15.9 mg/L FAIEs, giving a total of 65.5 mg/L FASBEs (15.9 mg/gDCW). Yeast strain BY4742 expressing Maqu_0168 produced 16.9 mg/L FAEEs, 15.8 mg/L FABEs and 128.1 mg/L FAIEs, giving a total of 160.8 mg/L FASBEs (30.2 mg/gDCW). Yeast strain BY4742-IB opi1Δ expressing ws2 produced 16.9 mg/L FAEEs, 127.9 mg/L FABEs, 31.4 mg/L FAIEs and 12.9 mg/L FAAEs, giving a total of 189.1 mg/L FASBEs (42.2 mg/gDCW). Yeast strain BY4742-IB opi1Δ expressing Maqu_0168 produced 9.43 mg/L FAEEs, 63.8 mg/L FABEs, 151.5 mg/L FAIEs and 9.48 mg/L FAAEs, giving a total of 234.2 mg/L FASBEs (50.9 mg/gDCW). The majority of FASBEs had fatty acid components of carbon chain lengths of C16 and C18, while esters with shorter chain length fatty acids were minor products. Strains expressing ws2 gave a higher proportion of longer chain FASBEs (fatty acid components of carbon chain lengths of C16 and C18, ~92 %), whereas for strains expressing Maqu_0168, the longer chain FASBEs proportion was ~80 %. In addition, strains expressing ws2 resulted in a product profile that had higher proportion of FASBEs with saturated fatty acid components (~84 %) compared to strains expressing Maqu_0168 which gave ~77 % FASBEs with saturated fatty acid components. All FASBEs with unsaturated fatty acid components were monounsaturated.
High-cell-density fermentation for FASBEs production of different fatty acid chain lengths. Four strains were chosen for the fermentation: a BY4741 opi1Δ expressing ws2, b BY4742 expressing Maqu_0168, c BY4742-IB opi1Δ expressing ws2 and d BY4742-IB opi1Δ expressing Maqu_0168. All strains were cultured in minimal medium lacking appropriate amino acid and/or nucleotide with 2 % galactose at an initial OD600 of ~9. Values are the mean of biological triplicates ±SD at 48 h
Conclusions
Concerns about limited supply of nonrenewable fossil fuels and the environmental impact of their usage continue to encourage discovery and development of renewable advanced biofuels, such as alcohols, alkanes, fatty acid esters and isoprenoids [19–23]. Recently, Liu and coworkers engineered Escherichia coli to produce fatty acid esters with short- and branched-chain alcohol moieties from glycerol [22, 23]. Introduction of 2-keto acid pathway and metabolic engineering of the fatty acid pathway together with expression of a wax ester synthase enzyme from Acinetobacter baylyi enabled production of a range of fatty acid esters, including ethyl, propyl, isobutyl, butyl and isoamyl esters [22]. Further introduction of branched fatty acid biosynthetic pathway resulted in branched fatty acid branched-chain esters [23].
Here, we have engineered yeast S. cerevisiae to produce and accumulate FASBEs that can be used as biodiesel, including FAEEs, FABEs, FAIEs and FAAEs. However, challenges remain before large-scale bioproduction can be considered. First, even though we have produced up to 230 mg/L FASBEs, which is the highest reported fatty acid ester production titer in yeast to date, further manipulation of the cells through increase of intermediate supply and use of gene regulatory tools are required to maximize the production potential of yeast [24–27]. Second, the products were found to be accumulated intracellularly. Strategies for product recovery need to be developed to enable cell reuse [28, 29].
In this work, it was found that wax synthases ws2 and Maqu_0168 resulted in different product profiles where the former preferentially produced FAEEs, FABEs and FAAEs and the latter produced mostly FAIEs. In addition, more FABEs and FAIEs were produced compared to FAEEs, even though ethanol levels were much higher than the other alcohols (Table 1). This can be attributed to higher specific activity of wax synthases for longer chain alcohols [5, 18]. Hence, the identification and use of wax synthases according to substrate preference and desired products are crucial for improved biodiesel production.
In summary, we engineered yeast to produce FASBEs using endogenously synthesized fatty acids and alcohols. Two wax ester synthase genes (ws2 and Maqu_0168) were found to catalyze the formation of FASBEs, with different alcohol preferences. To boost the ability of yeast to produce the FASBEs, gene deletions (OPI1 and RPD3) were carried out to increase flux towards fatty acyl-CoAs. In addition, isobutanol pathway enzymes (Ilv2, Ilv5, Ilv3, Aro10 and Adh7) targeted into the mitochondria were overexpressed, producing branched alcohols for esterification. By combining these engineering strategies, and through high-cell-density fermentation, over 230 mg/L FASBEs were produced, which is the highest titer reported in yeast to date.
Methods
Yeast strains and media
The yeast strains used in this study were derived from S. cerevisiae strain BY4742 (MATα; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0) (ATCC, Manassas, VA, USA) (Table 2). Isobutanol producing strain M12 derived from BY4742 was a gift from Dr. Jifeng Yuan (National University of Singapore). Plasmids used are listed in Table 3. DNA primers used for PCR are listed in Additional file 1: Table S1. E. coli strain Top10 was used for transformation and amplification of plasmids, grown at 37 °C in Luria–Bertani broth supplemented with 100 μg/mL ampicillin. Yeast extract and peptone were obtained from BD (Franklin Lakes, NJ, USA), whereas other chemicals were purchased from Sigma Aldrich (St. Louis, MO, USA), unless otherwise stated. Yeast strains were grown in minimal medium (yeast nitrogen base 6.7 g/L, yeast synthetic drop-out medium supplements—Ura- 1.92 g/L, d-glucose 20 g/L) at 30 °C with 225 rpm shaking.
Cloning procedures
Restriction enzymes used were procured from New England Biolabs (Ipswich, MA, USA) and digestions were conducted according to the recommended protocols. PCR amplifications were performed with iProof High Fidelity DNA Polymerase (Bio-Rad, Hercules, CA, USA) at suggested conditions. Ligations were carried out with T4 DNA Ligase (New England Biolabs) at 16 °C. Gel extractions were performed using QIAquick gel extraction kit (Qiagen, Venlo, The Netherlands). Minipreps of plasmids from E. coli were carried out using QIAprep Spin Miniprep kit. Standard chemical transformation methods for E. coli were applied for transformation of ligation mixtures. The LiAc/PEG technique was used for transformation of yeast cells.
Plasmids and yeast knockout strains construction
To construct plasmids expressing wax ester synthase proteins, genes ws2 and Maqu_0168 were codon optimized and synthesized using GeneArt (Life Technologies, Carlsbad, CA, USA) (Additional file 1: Table S2). PCR was carried out using primers ws-f-EcoRI and ws-r-NotI for ws2 and Maqu-f-EcoRI and Maqu-r-NotI for Maqu_0168. The PCR products were then gel extracted and digested using EcoRI and NotI, and ligated into pESC-URA, also digested with the same enzymes. Correct clones were confirmed by colony PCR and plasmid sequencing.
To create yeast knockout strains, gene deletion cassettes were amplified using PCR with primers as shown in Additional file 1: Table S1 and pUG6 as the template. The deletion cassettes were then transformed into yeast strains and screened on YPD plates containing 200 μg/mL G418 and verified by PCR.
Growth parameters of engineered strains including growth rate and yield of biomass are shown in Additional file 1: Table S3. Dry cell weight (DCW) was calculated from the optical density at 600 nm (1 OD600 = 0.262 gDCW/L).
Determining FASBEs production
For quantifying FASBEs produced in different engineered yeast strains, single colonies were pre-cultured overnight in 3 mL minimal medium with glucose as carbon source. The cells were then inoculated into 50 mL fresh minimal medium with 1.8 % galactose and 0.2 % glucose in 250 mL glass flasks at an initial OD600 of 0.05. At the stated time points, fatty acid esters were extracted and quantified as follows. Cells (2.5 mL) were centrifuged and the supernatant decanted, and 0.5 mL of autoclaved water was added to re-suspend the cells. Next, the cells were transferred to a 2 mL Fastprep tube (MP Biomedicals, Santa Ana, CA, USA) added with 0.3 g glass beads (425–600 μm). The cells were lyzed using a Fastprep-24 homogenizer, at 6 m/s for 30 s, put on ice for 2 min and repeated for 10 times. Hexane (0.5 mL) spiked with methyl heptadecanoate as internal standard was then added to the lysed cells, followed by vigorous vortex to extract the fatty acid esters. The mixture was centrifuged at maximum speed to separate aqueous and organic layers, where 200 μL of product containing hexane was transferred to GC vials for quantification. GC/MS (7890B GC system, 5977A MSD, Agilent Technologies, Santa Clara, CA, USA) analysis was carried out with a HP-5 ms column (Agilent Technologies) with a 0.25 μm film thickness, 0.25 mm diameter, and 30 m length. The GC program was set as follows: an initial temperature of 45 °C was maintained for 1.5 min, followed by ramping to 180 °C at a rate of 15 °C/min, where it was held for 3 min. The temperature was then ramped to 280 °C at a rate of 10 °C/min, where the temperature was held for 5 min.
Determining alcohol levels and phospholipid content
To quantify alcohols produced by the engineered yeast strains, single colonies were pre-cultured overnight in 1 mL minimal medium with glucose as carbon source. The cells were then inoculated into 5 mL fresh minimal medium with 1.8 % galactose and 0.2 % glucose in 50 mL tubes at an initial OD600 of 0.05. After 48 h growth, alcohols were extracted as follows. Cell culture (0.5 mL) was transferred to a clean 1.5 mL tube, and 0.5 mL hexane spiked with 1-butanol as internal standard was then added to the cells, followed by vigorous vortex to extract the alcohols. The mixture was then centrifuged at maximum speed to separate aqueous and organic layers, where 300 μL of alcohol containing hexane was transferred to GC vials for quantification. GC/MS analysis was carried out with a DB-FFAP column (Agilent Technologies) with a 0.25 μm film thickness, 0.25 mm diameter, and 30 m length. The GC program was set as follows: an initial temperature of 45 °C was maintained for 15 min, followed by ramping to 240 °C at a rate of 30 °C/min, where it was held for 3 min. To analyze the phospholipid concentration, yeast cells were centrifuged and washed with autoclaved deionized water, followed using a phospholipid assay kit (Abnova, Taipei, Taiwan) to determine the concentration of phospholipids according to the manufacturer’s instructions.
High-cell-density fermentation for FASBEs production
To measure the production of FASBEs with fermentation at high cell density, single colonies of the engineered yeast strains were pre-cultured overnight in 3 mL minimal medium with glucose as carbon source. The cells were then rediluted with 50 mL minimal medium with 2 % glucose in 250 mL glass flasks at an initial OD600 of 0.25 and grown for a further 24 h. Next, the cells were centrifuged and inoculated into 50 mL fresh minimal media with 2 % galactose in 250 mL glass flasks (giving an initial OD600 of ~9). After 48 h culture, 2.5 mL of each sample was centrifuged and the FASBEs extracted and quantified as described for FASBEs above.
Abbreviations
- FASBE:
-
fatty acid short- and branched-chain alkyl ester
- FAME:
-
fatty acid methyl ester
- FAEE:
-
fatty acid ethyl ester
- FABE:
-
fatty acid isobutyl ester
- FAIE:
-
fatty acid isoamyl ester
- FAAE:
-
fatty acid active amyl ester
- GC/MS:
-
gas chromatography mass spectrometry
- OD600 :
-
optical density at 600 nm
References
Röttig A, Wenning L, Bröker D, Steinbüchel A. Fatty acid alkyl esters: perspectives for production of alternative biofuels. Appl Microbiol Biotechnol. 2010;85:1713–33.
Lang X, Dalai AK, Bakhshi NN, Reaney MJ, Hertz PB. Preparation and characterization of bio-diesels from various bio-oils. Bioresour Technol. 2001;80:53–62.
Knothe G. Dependence of biodiesel fuel properties on the structure of fatty acid alkyl esters. Fuel Process Technol. 2005;86:1059–70.
Nevoigt E. Progress in metabolic engineering of Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2008;72:379–412.
Shi S, Valle-Rodríguez JO, Khoomrung S, Siewers V, Nielsen J. Functional expression and characterization of five wax ester synthases in Saccharomyces cerevisiae and their utility for biodiesel production. Biotechnol Biofuels. 2012;5:7.
Shi S, Chen Y, Siewers V, Nielsen J. Improving production of malonyl coenzyme A-derived metabolites by abolishing Snf1-dependent regulation of Acc1. MBio. 2014;5:e01130–11144.
Valle-Rodríguez JO, Shi S, Siewers V, Nielsen J. Metabolic engineering of Saccharomyces cerevisiae for production of fatty acid ethyl esters, an advanced biofuel, by eliminating non-essential fatty acid utilization pathways. Appl Energy. 2014;115:226–32.
Shi S, Valle-Rodríguez JO, Siewers V, Nielsen J. Engineering of chromosomal wax ester synthase integrated Saccharomyces cerevisiae mutants for improved biosynthesis of fatty acid ethyl esters. Biotechnol Bioeng. 2014;111:1740–7.
Runguphan W, Keasling JD. Metabolic engineering of Saccharomyces cerevisiae for production of fatty acid-derived biofuels and chemicals. Metab Eng. 2014;21:103–13.
Thompson RA, Trinh CT. Enhancing fatty acid ethyl ester production in Saccharomyces cerevisiae through metabolic engineering and medium optimization. Biotechnol Bioeng. 2014;111:2200–8.
Lian J, Zhao H. Recent advances in biosynthesis of fatty acids derived products in Saccharomyces cerevisiae via enhanced supply of precursor metabolites. J Ind Microbiol Biotechnol. 2015;42:437–51.
Kalscheuer R, Luftmann H, Steinbüchel A. Synthesis of novel lipids in Saccharomyces cerevisiae by heterologous expression of an unspecific bacterial acyltransferase. Appl Environ Microbiol. 2004;70:7119–25.
Kalscheuer R, Stölting T, Steinbüchel A. Microdiesel: Escherichia coli engineered for fuel production. Microbiology. 2006;152(Pt 9):2529–36.
de Kroon AIPM, Rijken PJ, De Smet CH. Checks and balances in membrane phospholipid class and acyl chain homeostasis, the yeast perspective. Progress Lipid Res 2013;52:374–94.
Feng X, Lian J, Zhao H. Metabolic engineering of Saccharomyces cerevisiae to improve 1-hexadecanol production. Metab Eng. 2015;27:10–9.
Avalos JL, Fink GR, Stephanopoulos G. Compartmentalization of metabolic pathways in yeast mitochondria improves the production of branched-chain alcohols. Nat Biotech. 2013;31:335–41.
Yuan J, Ching CB. Combinatorial assembly of large biochemical pathways into yeast chromosomes for improved production of value-added compounds. ACS Synth Biol. 2015;4:23–31.
Barney BM, Wahlen BD, Garner E, Wei J, Seefeldt LC. Differences in substrate specificities of five bacterial wax ester synthases. Appl Environ Microbiol. 2012;78:5734–45.
Peralta-Yahya PP, Zhang F, del Cardayre SB, Keasling JD. Microbial engineering for the production of advanced biofuels. Nature. 2012;488:320–8.
Sandoval CM, Ayson M, Moss N, Lieu B, Jackson P, Gaucher SP, Horning T, Dahl RH, Denery JR, Abbott DA, Meadows AL. Use of pantothenate as a metabolic switch increases the genetic stability of farnesene producing Saccharomyces cerevisiae. Metab Eng. 2014;25:215–26.
Brat D, Weber C, Lorenzen W, Bode HB, Boles E. Cytosolic re-localization and optimization of valine synthesis and catabolism enables inseased isobutanol production with the yeast Saccharomyces cerevisiae. Biotechnol Biofuels. 2012;5:65.
Guo D, Zhu J, Deng Z, Liu T. Metabolic engineering of Escherichia coli for production of fatty acid short-chain esters through combination of the fatty acid and 2-keto acid pathways. Metab Eng. 2014;22:69–75.
Tao H, Guo D, Zhang Y, Deng Z, Liu T. Metabolic engineering of microbes for branched-chain biodiesel production with low-temperature property. Biotechnol Biofuels. 2015;8:92.
Lian J, Si T, Nair NU, Zhao H. Design and construction of acetyl-CoA overproducing Saccharomyces cerevisiae strains. Metab Eng. 2014;24:139–49.
Williams TC, Averesch NJH, Winter G, Plan MR, Vickers CE, Nielsen LK, Krömer JO. Quorum-sensing linked RNA interference for dynamic metabolic pathway control in Saccharomyces cerevisiae. Metab Eng. 2015;29:124–34.
Teo WS, Chang MW. Development and characterization of AND-gate dynamic controllers with a modular synthetic GAL1 core promoter in Saccharomyces cerevisiae. Biotechnol Bioeng. 2014;111:144–51.
Pfleger BF, Gossing M, Nielsen J. Metabolic engineering strategies for microbial synthesis of oleochemicals. Metab Eng. 2015;29:1–11.
Cuellar MC, van der Wielen LA. Recent advances in the microbial production and recovery of apolar molecules. Curr Opin Biotechnol. 2015;33:39–45.
Ling H, Chen B, Kang A, Lee J-M, Chang MW. Transcriptome response to alkane biofuels in Saccharomyces cerevisiae: identification of efflux pumps involved in alkane tolerance. Biotechnol Biofuels. 2013;6:95.
Authors’ contributions
WST and MWC conceived and designed the study. WST, HL and AQY performed plasmid and strain construction, and fermentation experiments. WST and MWC wrote the manuscript. HL and AQY revised the manuscript. All authors read and approved the final manuscript.
Acknowledgements
We thank Jifeng Yuan for the gift of the isobutanol producing strain. This study was funded by the Competitive Research Program of the National Research Foundation of Singapore (NRF-CRP5-2009-03), the Defense Threat Reduction Agency (DTRA, HDTRA1-13-1-0037), Global R&D Project Program, the Ministry of Knowledge Economy, the Republic of Korea (N0000677), and the Synthetic Biology Initiative of the National University of Singapore (DPRT/943/09/14).
Competing interests
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Additional file
13068_2015_361_MOESM1_ESM.pdf
Additional file 1. Supplementary tables and figures. Table S1. List of primers used in this study. Table S2. Codon optimized sequence of wax ester synthase genes used in this study. Table S3. Growth parameters of engineered yeast strains. Figure S1. Relative FAEEs, FABEs or FAIEs production with exogenous alcohol feeding. Figure S2. Production of fatty acid esters in engineered yeast expressing ws2. Figure S3. Relative phospholipid concentration of engineered yeast strains. Figure S4. Production of fatty acid esters in engineered yeast expressing Maqu_0168. Figure S5. Growth curves for engineered cells. Figure S6. OD600 for high cell density fermentation. Figure S7. FASBEs production yield corresponding to Fig. 6.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Teo, W.S., Ling, H., Yu, AQ. et al. Metabolic engineering of Saccharomyces cerevisiae for production of fatty acid short- and branched-chain alkyl esters biodiesel. Biotechnol Biofuels 8, 177 (2015). https://doi.org/10.1186/s13068-015-0361-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13068-015-0361-5